Abstract
Background
Many nuclear receptors are modified by posttranslational modifications.
Objectives
The transcriptional activity of thyroid hormone receptors (TRs) is modified by the influence of its ligand (thyroid hormones T3 and T4), but is also affected by posttranslational modifications. This study focuses on the SUMOylation of TR isoforms and the consequences on transcriptional activity and promoter occupancy.
Methods
SUMOylation of TR wild-type as well as isoform-specific point mutations have been studied in vitro. The promoter occupancy of TR (wild-type and double- or triple-mutated versions) and transcriptional cofactors have been investigated in chromatin immunoprecipitation (ChIP) and Re-ChIP analysis.
Results
TR is modified by SUMO proteins at defined residues: the isoform TRα is mainly modified at lysines 281 and 387, whereas lysines 50 and 443 are major SUMOylation sites of isoform TRβ. Lysine residues K281 (TRα) and K50 (TRβ) are isoform-specific SUMOylation sites influencing differing TR domains, whereas K387 (TRα) and K443 (TRβ) are orthologous residues. TRs are targets of all three SUMO variants (SUMO-1, −2, and −3). The transcriptional activity of SUMOylation-defective mutants of TR alters gene transcription from positively and negatively regulated T3 target genes.
Conclusions
The most pronounced effect is an impaired repressor function of SUMOylation-deficient TR in the absence of T3. The transcriptional properties of SUMOylation-defective TRs can be at least in part ascribed to altered interaction with transcriptional cofactors such as SRC-1 and NCoR. Thus, these data indicate that posttranslational modification of TR by SUMOylation contribute to the fine tuning of its transcriptional response maintaining effects on cellular and physiological homeostasis.
Key Words: Posttranslational modification, SUMO, Gene expression, Ligand-dependent transcription factor, Transrepression
Introduction
Thyroid hormone (TH) has a profound influence on normal development, differentiation, and metabolism. Genomic actions of THs are mainly mediated and regulated by thyroid hormone receptors (TRs); however, TH-transmitted effects are also controlled by nongenomic processes as well as by modifying the ligand itself [1,2,3]. TRs are encoded by two different genes (α and β genes). Due to alternative splicing and alternate translational initiation, at least a dozen functional active TRα and TRβ isoform protein variants are synthesized. The archetypical TR binds to TH response elements which are located in promoter sequences of target genes, but may also be positioned several thousand base pairs up- or downstream of the regulated gene [1,4]. TRs belong to a group of transcription factors whose gene regulation function depends on the presence or absence of their particular ligand (i.e. TH). Liganded TRs commonly recruit a large coactivator complex that possesses or attracts enzymatic activities (e.g. coactivator SRC-1), which alter chromatin of target genes in order to generate an open chromatin structure allowing for proper transcription. In the absence of T3 (triiodothyronine), the unliganded TR undergoes conformational changes and recruits a corepressor complex (e.g. corepressor NCoR). Again, this corepressor complex integrates several enzymatic activities which modify chromatin towards a closed structure, resulting in a transcriptional silent state [5]. Unfortunately, mechanisms of negatively TH-regulated genes in the presence of T3 are less clear [6,7]. However, the balanced recruitment of transcriptional cofactors might also play an essential role for this T3-dependent gene regulation.
TH action has also been described to be modified by posttranslational modifications of TR [3,8]. In this context, TRβ has been described to be phosphorylated at Ser-142 within the DNA-binding domain (DBD) via the MAP kinase pathway [9]. This particular serine residue is not conserved in TRα. Thus, the functional consequences of phosphorylation of different TR isoforms remain controversial [10,11]. Other modifications of TR have also been confirmed to interfere with its transcriptional activity. For example, T3 can trigger increased ubiquitination of TRβ within the ligand-binding domain (LBD), causing proteasomal degradation [12]. Besides phosphorylation and ubiquitination, acetylation at three defined sites within the hinge domain of TRα has also been revealed and probably affects DNA binding and transactivation [13].
Strikingly, TRs have recently also been described to be modified by SUMOylation [14,15]. Three SUMO (small ubiquitin-like modifier) proteins (SUMO-1, −2, and −3) are contained by mammals. These SUMO proteins comprise approximately 100 amino acids and can be covalently attached to the ε-amino group of the amino acid lysine. The SUMO-modified lysine is often found within a minimal consensus motif ψKX(E/D) (ψ: hydrophobic amino acid; K: lysine; X: any amino acid; E/D: glutamate or aspartate). SUMO-1 is characterized as ∼50% identical to SUMO-2/3, whereas SUMO-2 and SUMO-3 are actually almost equal. In contrast to SUMO-1, which is bound as a monomer, SUMO-2 and −3 are able to form polymers. The SUMO modification and also the mode of covalently labeling to the target protein are similar to the posttranslational modification by ubiquitin: SUMO like ubiquitin is activated by an E1 enzyme (activating enzyme) in an ATP-dependent manner, conjugated by an E2 enzyme [e.g. UBC9 (ubiquitin-conjugating enzyme 9)], and linked to a substrate by the action of a E3 ligase family member [16,17]. Posttranslational modification by SUMOylation is not static. In contrast, due to the activity of SUMO-specific proteases, SUMO modification can be detached from the labeled protein without degrading the substrate protein [16,17]. SUMOylation has emerged as a significant regulatory mechanism in cell physiology which is associated with modulating a wide variety of cellular processes, such as transcriptional activity, subcellular localization, or interference with binding partners in a target-specific manner. Modification of a target protein by SUMOylation might also interfere with other posttranslational modifications at the same or a closely adjacent site, and therefore might block or at least attenuate other signal transduction pathways [18,19]. Several transcription factors (e.g. nuclear receptors) and transcriptional cofactors (e.g. coactivators and corepressors) are verified targets of SUMOylation [20,21,22,23]. With this study, I demonstrate that SUMOylation of TRα and TRβ is capable of fine-tuning its corresponding transcriptional activity. Since I could prove that certain SUMOylation sites are isoform-specific, this study suggests that a TR-SUMOylation might offer a sophisticated regulation mechanism for an isoform-specific regulation of TRα and TRβ activity.
Material and Methods
Isolation and Characterization of DNA Sequences
Human TSHα promoter from −802 to +22 in pGL3, rat mGPDH promoter B from −316 to +109 in pGL2, the 5xUAS-Luc reporter, and the expression plasmids of chicken TRα1 and human TRβ1 [both N-terminally fused with hemagglutinin (HA) and Flag epitopes] in pSG5 and pcDNA3 have been described previously [24,25]. Expression plasmids of SUMO-1 wild-type in pHH10B, SUMO-1 ΔGG in pHH10B, and UBC9 in pcDNA3 are kind gifts from Frauke Melchior (University of Heidelberg, Germany). The expression plasmid of human TRβ1 is a kind gift from Peter Hofmann (Charité Berlin, Germany). Various Gal4-DBD-TR fusion proteins have been designed by PCR amplification of chicken TRα fragments which were cloned into pcDNA3 (Life Technologies, Darmstadt, Germany). Various GST fusion proteins have been generated by PCR amplification of TRα or TRβ fragments and cloned into pGEX-2T (GE Healthcare, Munich, Germany). Point mutations were introduced using a self-made site-directed mutagenesis system adapted from the QuickChange kit by Stratagene. All clones were confirmed by DNA sequencing. In silico analysis to predict putative SUMOylation sites of TR was performed with SUMOplot (Abgent, San Diego, Calif., USA) and SUMOsp 2.0 [26].
Cell Culture Experiments
Human hepatocarcinoma HepG2 and human embryonic kidney HEK293 cells were cultured under standard conditions. Transient transfection experiments were conducted using a modified calcium phosphate technique demonstrated previously [24,25]. For classical transient transfection experiments on 9.6 cm2 dishes (each containing ∼7 × 105 cells), the first 2 µg of promoter luciferase reporter (pGL3-basic vector) was mixed with 0.2 µg of chicken TRα1 in pSG5 or human TRβ1 in pcDNA3 and then stimulated with 100 nM T3 (Sigma Aldrich, Taufkirchen, Germany). For mammalian one-hybrid experiments, 2 µg of 5xUAS-Luc reporter plasmids were mixed with 0.4 µg of the corresponding Gal4-TRα fusion protein plasmid and stimulated with 100 nM T3. Cells were harvested after a 24-hour incubation, and luciferase activity was determined as depicted previously [24,25] and finally normalized to the total protein concentration of the samples (Bradford, Bio-Rad) or to a normalizing renilla luciferase expression. Luciferase measurements were carried out in duplicate, and each construct was tested in at least three independent transfection experiments with 2-3 culture dishes per experiment.
Immunoprecipitation
For detection of SUMOylated TR in cell culture, 2 µg of Flag-HA-tagged chicken TRα1 in pSG5 or Flag-HA-tagged human TRβ1 in pcDNA3, 2 µg of SUMO-1 (wild-type or mutated version) in pHH10B, and 2 µg of UBC9 in pcDNA3 were transfected into 5 × 106 HepG2 cells. Cells were lysed, incubated with anti-Flag M2 affinity gel (Sigma Aldrich), and immunoprecipitated [27]. After immunoprecipitation, unmodified TRs were detected by Western blot analysis using an anti-HA antibody (No. 6E2; Cell Signaling, Frankfurt am Main, Germany) and SUMO-modified TRs were detected by anti-SUMO-1 antibody (SUMOlink kit, Active Motif). Amounts of immunoprecipitated proteins were quantified using AIDA Image Analyzer v4.15 (Raytest, Straubenhardt, Germany) and expressed as a ratio of modified relative to unmodified protein concentrations.
In vitro SUMOylation Assay
Bacterially expressed GST-TRα and GST-TRβ (wild-type and mutant versions) fusion proteins were produced in Escherichia coli as indicated previously. These fusion proteins contain an HA-tag located C-terminally of the GST and N-terminal of the TR protein portion. The total bacterial lysate derived from 100 ml of E. coli culture was completely loaded on GST GraviTrap columns (GE Healthcare) and purified according to the instructions of the manufacturer. The expression of correctly sized proteins was monitored by SDS-PAGE. 500 ng of the purified proteins were introduced into the SUMOlink kit (Active Motif, Rixensart, Belgium) according to the instructions of the manufacturer. After incubation for 3 h at 30°C, the SUMOylated samples were directly loaded onto an SDS-PAGE and detected by Western blot analysis using an anti-HA antibody (No. 6E2, Cell Signaling).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) analyses have been performed as previously shown [27]. Constructs for cotransfection are as described above. For immunoprecipitation, antibodies against HA-Tag (No. Y-11, Santa Cruz), Flag-tag (M2 affinity gel, Sigma Aldrich), SRC-1 (No. ab84, abcam), NCoR (No. ab24552, abcam), and nonspecific IgGs (No. I5006, Sigma Aldrich) were used. For the detection of mGPDH promoter-bound proteins, the PCR primers mGPDH for 5′-agctggaggttcctgacttcc-3′ and mGPDH rev 5′-ctttatgtttttggcgtcttc-3′ were employed by qPCR (i-cycler, Bio-Rad). For the detection of TSHα promoter-bound proteins, the primers TSHα for 5′-atggtaattacaccaagtaccc-3′ and TSHα rev 5′-ctttatgtttttggcgtcttc-3′ were employed by qPCR (i-cycler, Bio-Rad). After the first immunoprecipitation, Re-ChIP analyses were conducted by incubating the samples with 5 mM DTT for 30 min at 37°C. Samples were then diluted 1:50 with Re-ChIP buffer (150 mM NaCl, 20 mM Tris-HCl, 2 mM EDTA, 1% Triton X-100, pH 8.1), incubated with the second antibody overnight at 4°C, and immunoprecipitated as described. ChIP analyses were normalized in a two-step procedure (1) to the input control of the same sample and (2) to a parallel sample treated with nonspecific IgG. Chromatin occupancy of immunoprecipitated samples was determined relative to the occupancy of TR wild-type without T3 stimulation. The amount of immunoprecipitated samples corresponded to ∼0.06% of those from the input controls. Results of at least three independent ChIP experiments, conducted in duplicates, are shown.
Statistical Analysis
Effects were assessed by one-way analysis of variance. Tests (pair-wise comparisons or comparisons vs. control) were performed by the post hoc Newman-Keuls procedure. Values of p < 0.05 were considered statistically significant. Results are presented as means ± SD. Tests were performed with the SAS statistical package (SAS Institute, Cary, N.C., USA) and Sigma Stat of Jandel Scientific Software (Erkrath, Germany).
Results
TR Is a SUMOylated Protein in Cell Culture Experiments
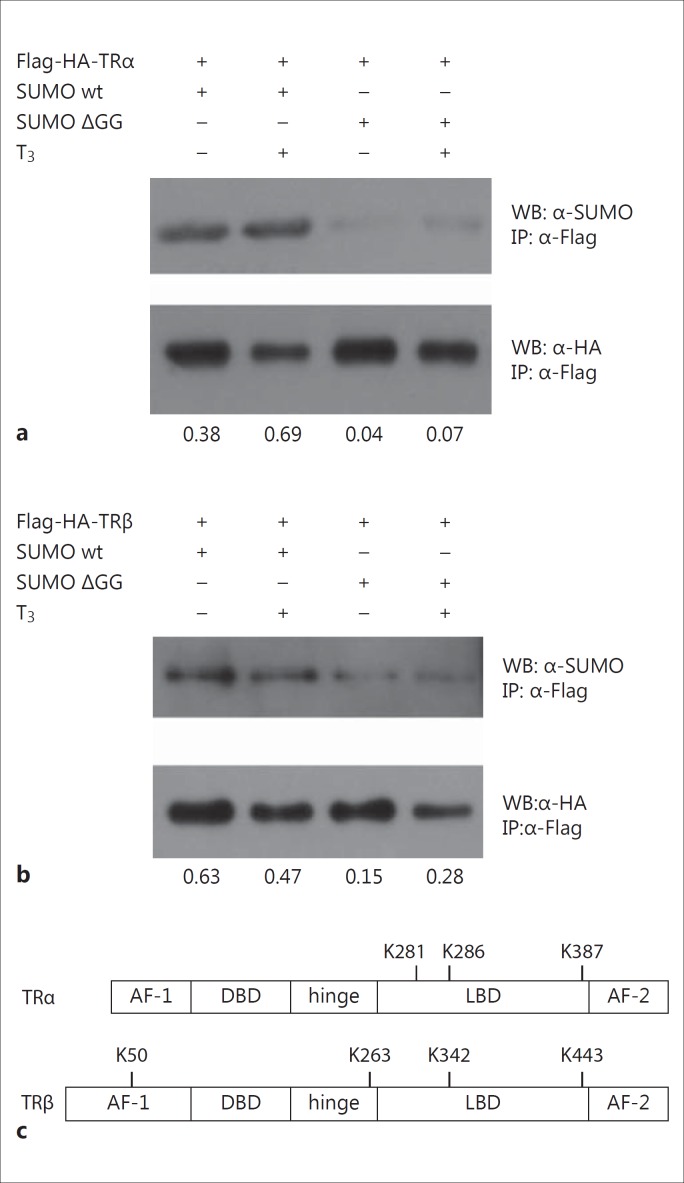
I transfected Flag-HA-tagged TRα1 or Flag-HA-tagged TRβ1 into HepG2 cells to analyze whether TR might be modified by SUMOylation (fig. 1a, b). In addition, a wild-type SUMO-1 protein or a mutated protein version (SUMO ΔGG) was cotransfected. The SUMO ΔGG protein is devoid of two C-terminal glycine residues which prevent conjugating to the lysine residue within the SUMOylation recognition site of target proteins. TR was immunoprecipitated using an antibody directed against a Flag epitope of TR, and Western blot analyses were performed using an anti-HA antibody in order to detect unmodified protein or by using anti-SUMO antibody in order to detect modified versions of TR. Due to the analysis via the Flag- and HA-tags, only preceding cotransfected TR proteins are detectable in this assay, whereas no protein could be detected in Western blot analyses without previous cotransfection of a tagged protein version. As shown in figure 1a and b, both TR isoforms are targets of SUMOylation. The amounts of posttranslationally modified TR appears to by higher in the presence of T3; however, this is rather an effect of reduced concentrations of unmodified TR in the presence of T3. This data indicates that TR is a target of SUMOylation in cell culture experiments.
Fig. 1.
TR is a SUMOylated protein. Expression plasmids of Flag-HA-tagged TRα wild-type (Flag-HA-TRα) (a) or Flag-HA-tagged TRβ wild-type (Flag-HA-TRβ) (b), SUMO wild-type (SUMO wt), or SUMO ΔGG were transfected into HepG2 cells and stimulated with or without T3. Cells were immunoprecipitated with an anti-Flag antibody (α-Flag) and Western blot analyses were performed using an anti-HA antibody (α-HA) in order to detect unmodified TR or anti-SUMO antibody (α-SUMO) in order to detect SUMOylated protein versions. Numbers indicate the quantification of the SUMOylated relative to the unmodified protein version. c In silico analysis identified 3 putative SUMOylation sites in chicken TRα (K281, K286 and K387) and 4 putative SUMOylation sites in human TRβ (K50, K263, K342 and K443). Abbreviations: AF-1 (-2) = activation function-1 (-2); IP = immunoprecipitation; WB = Western blot.
Further, sequence inspections identified three putative SUMOylation sites in chicken TRα1 at K281, K286, and K387, all located in the LBD of the protein (fig. 1c). In the human TRβ1 protein, four putative SUMOylation sites were detected. These lysine residues are located within the N-terminal activation domain AF-1 (K50), the hinge domain (K263), and the LBD (K342 and K443) (fig. 1c). Two of the SUMOylation sites positioned in the LBD are conserved between the two TR isoforms (i.e. K286 and K387 of TRα correspond to K342 and K443 of TRβ). In contrast, putative SUMOylation sites K281 of TRα and K50/K263 of TRβ are unique residues within their particular isoforms (fig. 1c).
SUMOylation of TRα and TRβ at Defined Residues
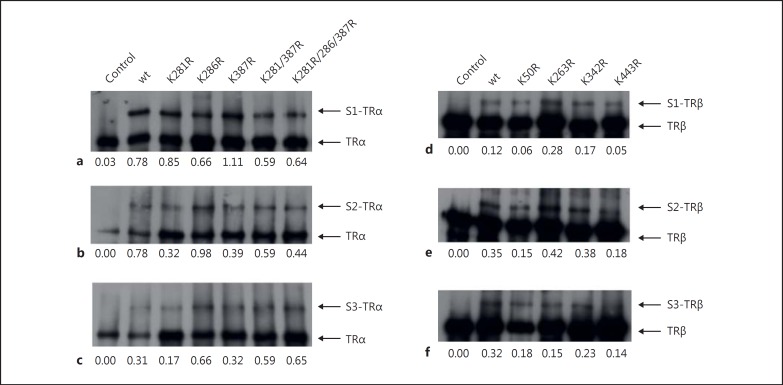
To evaluate the SUMOylation pattern of TRα and TRβ, I investigated several single, double, and triple mutations in an in vitro SUMOylation assay. In this context, the central lysine residue within the SUMOylation motif was mutated into the amino acid arginine. This mutation is the most conservative mutation, changing one basic amino acid into the second one. However, it has been shown previously that arginine (in contrast to lysine) is not able to be modified by SUMOylation. Furthermore, three single point mutations in the TRα protein were integrated, changing the lysine residues 281, 286, and 387 into arginine (mutants K281R, K286R, and K387R, respectively). Moreover, a double mutant (K281,387R) and a triple mutant (K281,286,387R) of TRα were generated. The wild-type and mutated proteins were introduced to an in vitro SUMOylation assay. Quantification of SUMOylated proteins was performed by comparing the upper band (SUMOylated protein) relative to the lower band (unmodified protein). Again, the in vitro SUMOylation assay not only demonstrated that TRα is a target of SUMOylation (fig. 2) but also that TRα is SUMOylated by all three SUMOylation isoforms: SUMO-1, SUMO-2, and SUMO-3 (fig. 2a-c). Additionally, the major SUMOylation sites appear to be K281 and K387, which were most apparent when SUMO-2/3 modification of TR occurred (fig. 2b, c). However, noncanonic SUMOylation sites might exist since even the triple mutant is a target of SUMOylation in the in vitro assay.
Fig. 2.
In vitro SUMOylation of TRα and TRβ. a-c SUMOylation of TRα by SUMO-1 (S1) (a), SUMO-2 (S2) (b), and SUMO-3 (S3) (c) was identified. TRα wild-type (wt), the indicated single point mutants, a double mutant K281/387R, and a triple mutant K281/286/387R were applied to an in vitro SUMOylation assay. SUMOylated and unmodified TR were detected by Western blot analysis and are indicated by arrows. d-f SUMOylation of TRβ by SUMO-1 (S1) (d), SUMO-2 (S2) (e), and SUMO-3 (S3) (f) was also recorded. TRβ wt and the indicated single point mutants were applied to an in vitro SUMOylation assay and detected by Western blot analysis and quantified. Numbers indicate the quantification of the upper (SUMOylated) relative to the lower (unmodified) band. SUMOylated and unmodified TR are denoted by arrows.
In a similar approach, I established four single point mutations into TRβ, yielding the mutants K50R, K263R, K342R and K443R, respectively. The in vitro SUMOylation assay confirmed the SUMOylation of TRβ by all three SUMOylation isoforms (fig. 2). Further on, the major SUMOylation sites seem to be K50 and K443, and - again - this modification was mostly pronounced by SUMO-2/3 conjugation of TR (fig. 2e, f).
Effect of SUMOylation-Defective Mutants in Cell Culture Experiments
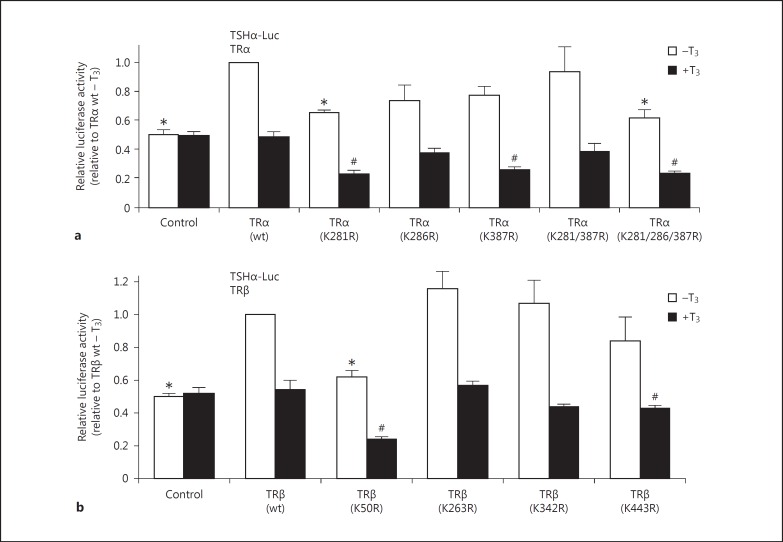
To test whether SUMOylation-defective mutants might have altered transactivation capacity, I performed classical transient transfection assays on natural promoter constructs upon stimulation by TH. In this assay a reporter construct of a T3 target gene was transfected together with wild-type or mutated variants of TR into HepG2 cells and subsequently stimulated with T3. Without TR cotransfection, no T3-mediated gene expression was observed (fig. 3). As shown in figure 3, TRα SUMOylation-defective mutants K281 and K387 and TRβ SUMOylation-defective mutants K50 and K443 show alterations on a negatively T3-regulated TSHα promoter. This observation is in agreement with a previous report on GH and TSHβ promoters [14].
Fig. 3.
Regulation of a TSHα promoter by TR isoforms and mutated protein versions in response to T3. A TSHα promoter luciferase reporter was transfected together with TRα wild-type (wt) or the indicated mutants thereof (a), or together with TRβ wild-type and mutated versions (b) into HepG2 cells and subsequently stimulated with T3. Expression rates are illustrated relative to those of the corresponding wild-type TR isoform without T3 stimulation ± SD. * Indicates p < 0.05 relative to wild-type and unstimulated and # indicates p < 0.05 relative to wild-type and stimulated levels.
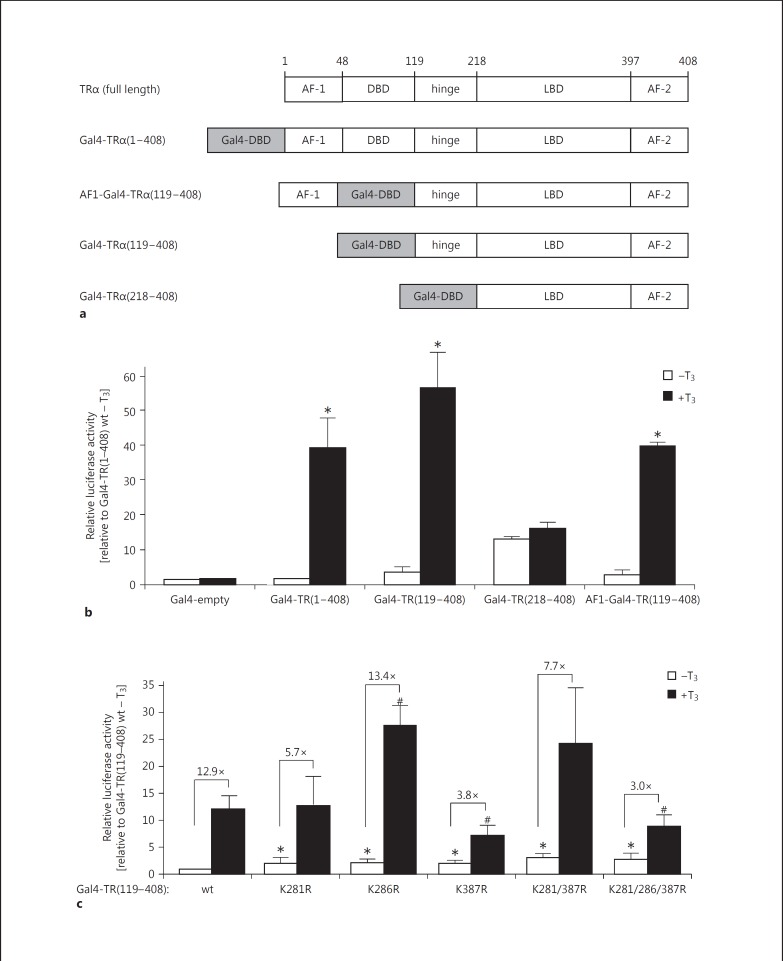
To further address this issue and to reduce the complexity of the cell system, I conducted mammalian one-hybrid assays by investigating the T3-dependent effect of Gal4-DBD-TR-fusion proteins on a UAS-Luc reporter. In a first approach I designed several Gal4-TRα fusion proteins (fig. 4a). Firstly, the full-length TRα1 protein (from amino acid 1 to 408) was fused C-terminally to the DBD of Gal4 [construct Gal4-TR(1-408)]. Secondly, for the construct AF1-Gal4-TR(119-408), the endogenous DBD of TRα was replaced by the Gal4-DBD. Thirdly, the N-terminal activation domain AF-1 and the endogenous DBD of TR was compensated by Gal4-DBD [Gal4-TR(119-408)]. Finally, for the construct Gal4-TR(218-408), the AF-1, TR-DBD, and the hinge domain were substituted by Gal4-DBD. These four constructs are schematically summarized in figure 4a. As shown in figure 4b, the constructs Gal4-TR(1-408), AF1-Gal4-TR(119-408), and Gal4-TR(119-408) responded properly to T3 stimulation, whereas Gal4-TR(218-408) apparently failed. Hence, I decided to integrate SUMOylation-defective point mutants into the context of the construct Gal4-TR(119-408).
Fig. 4.
Influence of TR SUMOylation-defective mutants in a mammalian one-hybrid assay. a Schematic overview of different Gal4-DBD-TR fusion proteins. Numbers indicate the position of the diverse functional domains within the protein sequence. * p < 0.05 relative to unstimulated, corresponding wild-type levels; # p < 0.05 relative to stimulated wild-type (wt) levels. b, c Expression vectors of the indicated Gal4-TR fusion proteins (wild-type or mutated versions) together with an UAS-Luc reporter vector have been transfected into HepG2 cells and stimulated with T3. c The induction rates ± T3 are indicated. Expression rates are displayed in relation to those of the corresponding wild-type Gal4-TR version without T3 stimulation ± SD.
As illustrated in figure 4c, Gal4-TR(119-408) wild-type responded appropriately to T3 stimulation. T3-mediated activation of this construct led to a 12.9-fold induction of reporter gene activity compared to unstimulated expression rates. For the majority of SUMOylation-defective mutants, the activation rates were reduced in comparison to the wild-type Gal4-TR(119-408) fusion protein. Intriguingly, the most pronounced effects were detected for the single mutants K281R (5.7-fold) and K387R (3.8-fold), as well as for the triple mutant K281,286,387R (3.0-fold), whereas the single mutant K286R remained unaffected (fig. 4c). Remarkably, a second distinct effect on the transactivation capacity of the SUMOylation-defective mutants became apparent in this assay: in the absence of ligand T3 the expression rates of SUMOylation-defective mutants are markedly increased compared to wild-type (p < 0.05). This T3-absence-triggered impact could be demonstrated for all investigated single mutants. However, this enhanced transcription activity was most evident in the double as well as triple mutants (fig. 4c). Thus, it has to be hypothesized that SUMOylation-deficient TRs are restrained in their function to suppress T3-dependent gene expression in the absence of T3. This suggests that TR-SUMOylation is significant for repressor activity in the unliganded state.
Occupancy of TR Wild-Type and SUMOylation-Defective Mutants on Natural Promoters
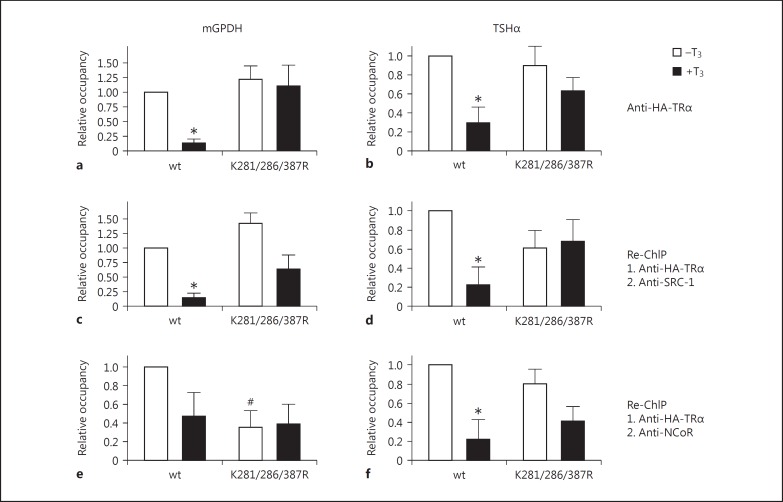
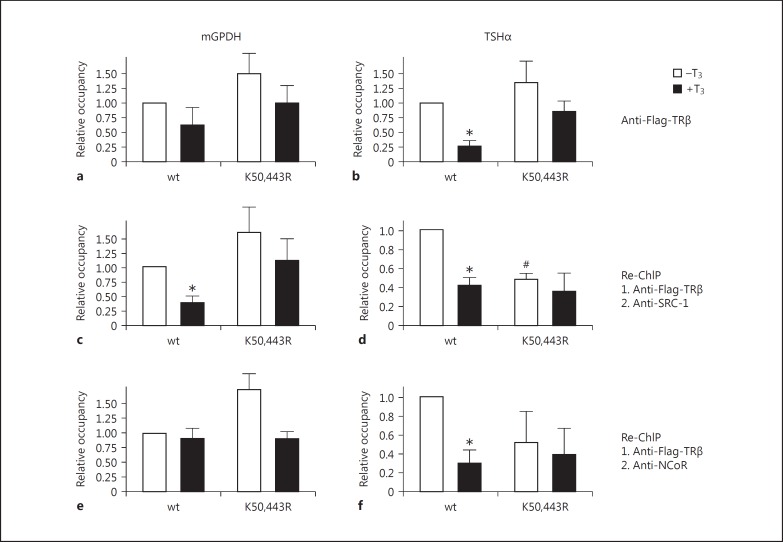
Since the triple mutant TRα(K281,286,387R) delivered the most striking effects in cell-based transactivation assays (fig. 3a, 4c), I analyzed the occupancy of this mutated protein version relative to the wild-type protein on natural promoters of T3 targets. Promoter occupancy was normalized in a two-step procedure. In a first normalization step the expression levels were normalized to the expression levels of the input controls within the same samples. In a second normalization step the expression levels were normalized to the expression levels detected by using an unspecific IgG from the same starting sample. As shown in figure 5, a TR wild-type version bound to promoter fragments of T3-regulated target genes in ChIP assays. This holds true not only for a positively regulated T3 target gene promoter (mGPDH, fig. 5a, c, e), but also for a negatively regulated one (TSHα, fig. 5b, d, f). Surprisingly, the occupancy of TRα wild-type was reduced in the presence of T3, both on mGPDH and TSHα genes (fig. 5a, b). On the contrary, this diverse promoter occupancy is completely abrogated by the triple mutant of TRα (fig. 5a, b). In a similar experimental setting, wild-type and the double mutant K50/443R of TRβ were analyzed (fig. 6).
Fig. 5.
ChIP and Re-ChIP analysis of TRα on mGPDH and TSHα gene promoters. mGPDH or TSHα promoter constructs were transfected together with HA-TRα wild-type (wt) or triple mutant HA-TRα(K281/286/387R) into HepG2 cells. a, b Chromatin was immunoprecipitated with an anti-HA antibody and TR-bound DNA was quantified by qPCR. After the first immunoprecipitation the antigen-antibody complex was released and a second immunoprecipitation was performed using an anti-SRC-1 antibody (c, d) or an anti-NCoR antibody (e, f). DNA occupancy was calculated relative to the occupancy of the TR wild-type version without T3 stimulation ± SD. * p < 0.05 relative to unstimulated wild-type levels; # p < 0.05 relative to unstimulated wild-type levels.
Fig. 6.
ChIP and Re-ChIP analysis of TRβ on mGPDH and TSHα gene promoters. mGPDH or TSHα promoter constructs were transfected together with Flag-TRβ wild-type (wt) or double mutant Flag-TRβ(K50,443R) into HepG2 cells. a, b Chromatin was immunoprecipitated with an anti-Flag antibody and TR-bound DNA was quantified by qPCR. After the first immunoprecipitation the antigen-antibody complex was released and a second immunoprecipitation was performed using an anti-SRC-1 antibody (c, d) or an anti-NCoR antibody (e, f). DNA occupancy was calculated relative to the occupancy of the TR wild-type version without T3 stimulation ± SD. * p < 0.05 relative to unstimulated wild-type levels; # p < 0.05 relative to unstimulated wild-type levels.
For the next series of experiments, I conducted Re-ChIP analysis in order to examine the influence of SUMOylation on TR-coactivator-binding (e.g. to SRC-1) and on TR-corepressor-binding (e.g. to NCoR). To this end, after immunoprecipitation of TR (and release of the first antibody), a second immunoprecipitation was performed by precipitation with an anti-SRC-1 antibody (fig. 5c, d) or anti-NCoR antibody (fig. 5e, f), respectively. Again, these data unveiled that the promoter occupancy could be altered for the triple mutant relative to the wild-type version of TR. This effect was mostly noticeable for Re-ChIP analysis using an anti-SRC-1 antibody on the negatively regulated TSHα gene (fig. 5d) and an anti-NCoR antibody on the positively regulated mGPDH gene (fig. 5e). Very similar data have been observed by comparing TRβ wild-type to the double mutant TRβ(K50,443R), indicating identical molecular mechanisms of TRα and TRβ isoforms (fig. 6).
Discussion
THR belong to a family of transcription factors whose transcriptional activity is modulated by the absence or presence of the corresponding ligand, but additionally via various posttranslational modifications. For example, TRβ has been demonstrated to be phosphorylated at serine-142 within the DBD [9]. This particular serine residue is not conserved in TRα, thus a specific transcriptional regulation by TRβ due to phosphorylation appears possible. In line with this argumentation, different DNA-binding and/or transactivation properties of phosphorylated TRα and TRβ have been described [9,10,11]. Furthermore, acetylation of TR has been disclosed only for TRα at lysines 128, 132, and 134 within the hinge domain [13]. However, since these lysine residues are conserved in the β-isoform, a similar acetylation of TRβ appears likely. Although the lysine residue mentioned above does not reside in classical SUMOylation recognition elements, cross-talk between lysine acetylation and lysine SUMOylation cannot be excluded.
In the present study I described the SUMOylation of TRα and TRβ at defined lysine residues. These findings suggest the major SUMOylation sites of chicken TRα1 to be K281 and K387 (corresponds to K283 and K389 of rat TRα1), both being located in the LBD (fig. 1c). The main SUMOylation sites of TRβ1 appear to be K50 (positioned in the N-terminal activation domain AF-1) and K443 (situated in the LBD) (fig. 2). A preferential SUMOylation of TRα by SUMO-1 and SUMO-2 and preferential SUMOylation of TRβ by SUMO-3 as described recently [14] could not be observed in the present study (fig. 2). Furthermore, a preferential SUMOylation of TRβ in the presence of T3 could also not be observed (fig. 1b). Of note, the SUMOylation sites K387 (TRα) and K443 (TRβ) are known to represent orthologous sequences, thus being conserved in both isoforms (fig. 1c). Hence, targeting this particular site might simultaneously affect both isoforms. Methionine-386 of chicken TRα1 (which corresponds to methionine-388 of rat TRα1) directly contacts the ligand within the ligand-binding pocket of TRα [28]. Thus, it is tempting to speculate that modification of the immediately adjacent lysine-387 with a bulky SUMO protein will impede the ligand binding of TR to T3. In contrast to the isoform-conserved lysines, lysine-281 is only detectable in all available TRα variants (e.g. human, mouse, rat, pig, chicken, frog, salmon, Japanese medaka), but in none of the TRβ proteins. The other way round, lysine-50 is part of the extended AF-1 domain of TRβ and therefore not included in TRα. Therefore, specifically targeting K281 in TRα or selectively aiming K50 in TRβ might offer a way to differentially regulate the two TR isoforms. Interestingly, a yeast two-hybrid screen has identified the ubiquitin E3 ligase cullin 1 as a physical interaction partner of TRα, but not as a binding partner of TRβ [29]. Thus, specifically targeting isoform-specific SUMOylation sites might be a way to differentially regulate one particular TR isoform without effecting the SUMOylation (and therefore activity) of the second isoform.
Generally, SUMOylation of proteins is a significant regulator of cell physiology. This posttranslational modification is capable of interfering with multiple cellular processes such as transcriptional activity, subcellular localization, or interference with binding partners. As shown in figures 3a and b, SUMOylation-defective mutants of TRα (K281 and K387) and TRβ (K50 and K443) revealed alterations when binding to natural promoters in cell culture experiments. This observation is partially consistent and partially contradictory to recently published data. For example, a TSHβ reporter construct has been shown to be activated by the mutant TRβ (K50Q) in the absence of T3[14]. In contrast, a TSHα reporter construct has been shown to be repressed by the mutant TRβ (K50R) in the absence of T3 in our experiments (fig. 3b). Since both experiments have been performed using HepG2 cells, differences in the TSH subunit reporter constructs or the investigated mutants (lysine → glutamine exchange in the study by Liu et al. [14] versus lysine → arginine substitution in this study) might account for some differences. The transactivation capacity of TR did not completely depend on an appropriate posttranslational modification by SUMOylation. Detailed cell-based analyses indicate that SUMOylation-defective TR mutants elicit an altered repressor function in the absence of the ligand (fig. 4c). Specifically, transactivation of the mutant versions is already increased in the absence of the ligand (fig. 4c). Admittedly, the transcriptional activity of these mutants was increased after stimulation with the ligand T3. However, the x-fold T3-mediated activation rates of the SUMOylation-defective mutants K281R and K387R were markedly reduced compared to the expression rates of the wild-type protein version (fig. 4c). These diminished activation rates are mainly attributed to the observed increased transcription activity in the absence of T3. This implies a decreased repressor function of un-SUMOylated TR for T3 dependently regulated target genes. This contradicts a previous report which showed a differential SUMOylation in response to ligand binding to the receptor [14]. Additionally, I did not detect an altered subcellular localization of SUMOylation-defective mutants (not shown), as this has been described for some SUMOylated nuclear receptors such as AR, PR, or RAR [30,31,32]. Thus, I have to assume that the primarily effect of SUMOylation incompetence of TR might be an inappropriate repression of gene expression in the absence of T3.
Further on, other results presented above indicate that a SUMOylation of TR within the LBD might interfere with the ligand binding. As the SUMOylation sites K281/K387 (TRα) and K443 (TRβ) are all located within the LBD of TR (fig. 1c), these residues could be attractive candidates for such a regulation mechanism. Additionally, SUMOylation at these sites might interfere with interacting binding partners in a ligand-dependent manner. ChIP analysis underscored this assumption (fig. 5, 6). Concerning promoter occupancy of wild-type and SUMOylation-defective mutant TR, it could be disclosed that the wild-type TR binding on natural promoters, regardless whether on positively or negatively regulated target genes, is reduced following T3 delivery (fig. 5a, b, 6a, b). This interesting observation has been noticed in ChIP analyses of TR by several groups [33,34,35,36] and might be connected to an altered half-life time depending on the T3 concentration. The differential DNA occupancy of wild-type TR in dependency of T3 is abrogated in the triple mutant TRα(K281,286,387R) and in the double mutant TRβ(K50,443R), underlining the potential significance of posttranslational TR-SUMOylation (fig. 5a, b; 6a, b). Re-ChIP analyses demonstrated that the TR binding to the coactivator SRC-1 and to the corepressor NCoR is altered in the SUMOylation-defective mutant relative to the wild-type version (fig. 5c-f, 6c-f). Consequently, SUMOylation of TR appears to be important for its proper interaction with transcriptional cofactors.
Taken together, the data presented in this paper indicate that TRα and TRβ are SUMOylated not only at orthologous but also at isoform-specific lysine residues. Furthermore, it could be revealed that SUMOylation of TR in general might interfere with the ligand binding as well as could exert a substantial impact on transcriptional cofactor attachment. Thus, posttranslational modification by SUMOylation contributes to the fine-tuning of transcriptional activity of TR isoforms both on positively as well as on negatively regulated T3 target genes.
Disclosure Statement
There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported. This research did not receive any specific grant from any funding agency in the public, commercial or not-for-profit sector. J.M.W. designed the study, performed the research, and wrote the manuscript.
Acknowledgements
The technical assistance of Ursula Antkewitz and Swanhild Rodewald is gratefully acknowledged. I am also indebted to Frauke Melchior and Peter Hofmann for their kind gifts of plasmid DNA, and to Alexander Sobczak for his helpful comments on the manuscript.
References
- 1.Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31:139–170. doi: 10.1210/er.2009-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cioffi F, Senese R, Lanni A, Goglia F. Thyroid hormones and mitochondria: with a brief look at derivatives and analogues. Mol Cell Endocrinol. 2013;379:51–61. doi: 10.1016/j.mce.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Davis PJ, Lin HY, Tang HY, Davis FB, Mousa SA. Adjunctive input to the nuclear thyroid hormone receptor from the cell surface receptor for the hormone. Thyroid. 2013;23:1503–1509. doi: 10.1089/thy.2013.0280. [DOI] [PubMed] [Google Scholar]
- 4.Weitzel JM, Iwen KA. Coordination of mitochondrial biogenesis by thyroid hormone. Mol Cell Endocrinol. 2011;342:1–7. doi: 10.1016/j.mce.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Astapova I, Hollenberg AN. The in vivo role of nuclear receptor corepressors in thyroid hormone action. Biochim Biophys Acta. 2013;1830:3876–3881. doi: 10.1016/j.bbagen.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weitzel JM. To bind or not to bind - how to down-regulate target genes by liganded thyroid hormone receptor? Thyroid Res. 2008;1:4. doi: 10.1186/1756-6614-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos GM, Fairall L, Schwabe JW. Negative regulation by nuclear receptors: a plethora of mechanisms. Trends Endocrinol Metab. 2011;22:87–93. doi: 10.1016/j.tem.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pascual A, Aranda A. Thyroid hormone receptors, cell growth and differentiation. Biochim Biophys Acta. 2013;1830:3908–3916. doi: 10.1016/j.bbagen.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 9.Lin HY, Zhang S, West BL, Tang HY, Passaretti T, Davis FB, Davis PJ. Identification of the putative MAP kinase docking site in the thyroid hormone receptor-beta1 DNA-binding domain: functional consequences of mutations at the docking site. Biochemistry. 2003;42:7571–7579. doi: 10.1021/bi0273967. [DOI] [PubMed] [Google Scholar]
- 10.Chen SL, Chang YJ, Wu YH, Lin KH. Mitogen-activated protein kinases potentiate thyroid hormone receptor transcriptional activity by stabilizing its protein. Endocrinology. 2003;144:1407–1419. doi: 10.1210/en.2002-220911. [DOI] [PubMed] [Google Scholar]
- 11.Mochizuki K, Sakaguchi N, Takabe S, Goda T. De-phosphorylation of TRalpha-1 by p44/42 MAPK inhibition enhances T(3)-mediated GLUT5 gene expression in the intestinal cell line Caco-2 cells. Biochem Biophys Res Commun. 2007;359:979–984. doi: 10.1016/j.bbrc.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Dace A, Zhao L, Park KS, Furuno T, Takamura N, Nakanishi M, West BL, Hanover JA, Cheng S. Hormone binding induces rapid proteasome-mediated degradation of thyroid hormone receptors. Proc Natl Acad Sci USA. 2000;97:8985–8990. doi: 10.1073/pnas.160257997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Pacheco A, Martinez-Iglesias O, Mendez-Pertuz M, Aranda A. Residues K128, 132, and 134 in the thyroid hormone receptor-alpha are essential for receptor acetylation and activity. Endocrinology. 2009;150:5143–5152. doi: 10.1210/en.2009-0117. [DOI] [PubMed] [Google Scholar]
- 14.Liu YY, Kogai T, Schultz JJ, Mody K, Brent GA. Thyroid hormone receptor isoform-specific modification by small ubiquitin-like modifier (SUMO) modulates thyroid hormone-dependent gene regulation. J Biol Chem. 2012;287:36499–36508. doi: 10.1074/jbc.M112.344317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu YY, Ayers S, Milanesi A, Teng X, Rabi S, Akiba Y, Brent GA. Thyroid hormone receptor sumoylation is required for preadipocyte differentiation and proliferation. J Biol Chem. 2015;290:7402–7415. doi: 10.1074/jbc.M114.600312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meulmeester E, Melchior F. Cell biology: SUMO. Nature. 2008;452:709–711. doi: 10.1038/452709a. [DOI] [PubMed] [Google Scholar]
- 17.Muller S, Hoege C, Pyrowolakis G, Jentsch S. SUMO, ubiquitin's mysterious cousin. Nat Rev Mol Cell Biol. 2001;2:202–210. doi: 10.1038/35056591. [DOI] [PubMed] [Google Scholar]
- 18.Treuter E, Venteclef N. Transcriptional control of metabolic and inflammatory pathways by nuclear receptor SUMOylation. Biochim Biophys Acta. 2011;1812:909–918. doi: 10.1016/j.bbadis.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Wadosky KM, Willis MS. The story so far: post-translational regulation of peroxisome proliferator-activated receptors by ubiquitination and SUMOylation. Am J Physiol Heart Circ Physiol. 2012;302:H515–H526. doi: 10.1152/ajpheart.00703.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung SS, Ahn BY, Kim M, Kho JH, Jung HS, Park KS. SUMO modification selectively regulates transcriptional activity of peroxisome-proliferator-activated receptor gamma in C2C12 myotubes. Biochem J. 2011;433:155–161. doi: 10.1042/BJ20100749. [DOI] [PubMed] [Google Scholar]
- 21.Rytinki MM, Palvimo JJ. SUMOylation attenuates the function of PGC-1alpha. J Biol Chem. 2009;284:26184–26193. doi: 10.1074/jbc.M109.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hentschke M, Süsens U, Borgmeyer U. Transcriptional ERRgamma2-mediated activation is regulated by sentrin-specific proteases. Biochem J. 2009;419:167–176. doi: 10.1042/BJ20081556. [DOI] [PubMed] [Google Scholar]
- 23.Wilson BJ, Tremblay AM, Deblois G, Sylvain-Drolet G, Giguere V. An acetylation switch modulates the transcriptional activity of estrogen-related receptor alpha. Mol Endocrinol. 2010;24:1349–1358. doi: 10.1210/me.2009-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wulf A, Harneit A, Kroger M, Kebenko M, Wetzel MG, Weitzel JM. T3-mediated expression of PGC-1alpha via a far upstream located thyroid hormone response element. Mol Cell Endocrinol. 2008;287:90–95. doi: 10.1016/j.mce.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 25.Wulf A, Wetzel MG, Kebenko M, Kroger M, Harneit A, Merz J, Weitzel JM. The role of thyroid hormone receptor DNA binding in negative thyroid hormone-mediated gene transcription. J Mol Endocrinol. 2008;41:25–34. doi: 10.1677/JME-08-0023. [DOI] [PubMed] [Google Scholar]
- 26.Ren J, Gao X, Jin C, Zhu M, Wang X, Shaw A, Wen L, Yao X, Xue Y. Systematic study of protein sumoylation: development of a site-specific predictor of SUMOsp 2.0. Proteomics. 2009;9:3409–3412. doi: 10.1002/pmic.200800646. [DOI] [PubMed] [Google Scholar]
- 27.Wulf A, Harneit A, Weitzel JM. T3-mediated gene expression is independent of PGC-1alpha. Mol Cell Endocrinol. 2007;270:57–63. doi: 10.1016/j.mce.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Wagner RL, Apriletti JW, McGrath ME, West BL, Baxter JD, Fletterick RJ. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 29.Albers M, Kranz H, Kober I, Kaiser C, Klink M, Suckow J, Kern R, Koegl M. Automated yeast two-hybrid screening for nuclear receptor-interacting proteins. Mol Cell Proteomics. 2005;4:205–213. doi: 10.1074/mcp.M400169-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Rytinki M, Kaikkonen S, Sutinen P, Paakinaho V, Rahkama V, Palvimo JJ. Dynamic SUMOylation is linked to the activity cycles of androgen receptor in the cell nucleus. Mol Cell Biol. 2012;32:4195–4205. doi: 10.1128/MCB.00753-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu L, Santos NC, Kim KH. Small ubiquitin-like modifier-2 modification of retinoic acid receptor-alpha regulates its subcellular localization and transcriptional activity. Endocrinology. 2009;150:5586–5595. doi: 10.1210/en.2009-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Man JH, Li HY, Zhang PJ, Zhou T, He K, Pan X, Liang B, Li AL, Zhao J, Gong WL, Jin BF, Xia Q, Yu M, Shen BF, Zhang XM. PIAS3 induction of PRB sumoylation represses PRB transactivation by destabilizing its retention in the nucleus. Nucleic Acids Res. 2006;34:5552–5566. doi: 10.1093/nar/gkl691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharma P, Thakran S, Deng X, Elam MB, Park EA. Nuclear corepressors mediate the repression of phospholipase A2 group IIa gene transcription by thyroid hormone. J Biol Chem. 2013;288:16321–16333. doi: 10.1074/jbc.M112.445569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Umezawa R, Yamada M, Horiguchi K, Ishii S, Hashimoto K, Okada S, Satoh T, Mori M. Aberrant histone modifications at the thyrotropin-releasing hormone gene in resistance to thyroid hormone: analysis of F455S mutant thyroid hormone receptor. Endocrinology. 2009;150:3425–3432. doi: 10.1210/en.2008-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SW, Ho SC, Hong SJ, Kim KM, So EC, Christoffolete M, Harney JW. A novel mechanism of thyroid hormone-dependent negative regulation by thyroid hormone receptor, nuclear receptor corepressor (NCoR), and GAGA-binding factor on the rat cD44 promoter. J Biol Chem. 2005;280:14545–14555. doi: 10.1074/jbc.M411517200. [DOI] [PubMed] [Google Scholar]
- 36.Mendez-Pertuz M, Sanchez-Pacheco A, Aranda A. The thyroid hormone receptor antagonizes CREB-mediated transcription. EMBO J. 2003;22:3102–3112. doi: 10.1093/emboj/cdg295. [DOI] [PMC free article] [PubMed] [Google Scholar]