Abstract
In the present work, we demonstrate the potential and versatility of TIMS for the analysis of proteins, DNA-protein complexes and protein-protein complexes in their native and denatured states. In addition, we show that accurate CCS measurement are possible and in good agreement with previously reported CCS values using other IMS analyzers (<5% difference). The main challenges for the analysis of high mass proteins and protein complexes in the mobility and m/z domain are described. That is, the analysis of high molecular weight systems in their native state may require the use of higher electric fields or a compromise in the TIMS mobility resolution by reducing the bath gas velocity in order to effectively trap at lower electric fields. This is the first report of CCS measurements of high molecular weight biomolecules and biomolecular complexes (~ 150 kDa) using TIMS-MS.
Keywords: TIMS, IMS-MS, Ion mobility spectrometry – Mass Spectrometry, proteins, ion—neutral collisional cross sections
Introduction
Experimental determination of biomolecular structures remains a challenging problem despite recent developments in theoretical approaches that are based on available experimental structural observations (e.g., ab initio, molecular dynamics and bioinformatics-based prediction strategies).[1] The elucidation of structural features of interest for biomolecules and biomolecular complexes is challenging because of the highly heterogeneous and dynamic character of biomolecules and their low relative concentrations within physiologically relevant conditions.[2–11] While X-ray crystallography and NMR spectroscopy excel at revealing structures of molecules at the atomic level, these approaches are limited by the fact that they often describes a single state of the biomolecule and/or biomolecular complex structure.[12–17] Moreover, since neither technology involves component separation during analysis, both require highly purified samples.[18, 19]
Recent innovations in speed, accuracy, and sensitivity have established mass spectrometry (MS) based methods as a key technology within the field of structural biology.[20] Specifically native MS techniques, which have been developed over the last two decades, permits the structural interrogation of intact biomolecules and biomolecular complexes at biologically relevant conditions, which are not accessible by other methods.[21–25] Attention has been drawn towards the characterization of intrinsically disordered proteins (IDP)[26–28], antibody therapeutics[29–32], and even massive protein complexes[33–37] that cannot be studied using traditional structural techniques. Most common gas-phase structural probing is based on, or a combination of, tandem MS (ergodic and non-ergodic), gas-phase hydrogen-deuterium exchange, ion spectroscopy, and ion mobility spectrometry.
In particular, ion mobility spectrometry (IMS) is based on the separation of ions as they drift in a bath of inert neutral molecules under the influence of a weak electric field.[38–40] The ion’s mobility gives information on their size and shape via the momentum transfer ion-neutral collision cross section (CCS).[41] This description holds true for most contemporary IMS analyzers (e.g., periodic focusing DC ion guide [42–44], segmented quadrupole drift cell [45], multistage IMS [46–48], traveling wave ion guide (TWIMS) [49, 50], and SLIM devices [51]), and a common pursuit has been to increase IMS resolving power and ion transmission.[52–60] Many different forms of IMS have been used in the analysis of biological molecules for the analysis of isotopomers[61], proteins[62, 63], protein complexes[64–71], folding pathways[72–74], unstructured/intrinsically disordered proteins[75–79], as well as collisionally activated states of peptides and proteins[69, 80–87]. It should be noted that, in the case of structural biology, gas-phase studies take advantage of the desolvation process to effectively reduce sample complexity, permitting molecular characterization in the absence of bulk solvent.
With the recent introduction of a new IMS analyzer - Trapped Ion Mobility Spectrometer (TIMS)- the possibility to decouple the time domain from the IMS separation allows for the study of conformationally trapped molecular ions in the gas-phase as a function of the desolvation time, temperature and bath gas composition. TIMS’ mode of operation and its advantages over traditional IMS are described in ref [88–90]. We have shown the use of TIMS for the study of isomerization kinetics of small molecules[91], peptides[92], and proteins[19, 93–95], the influence of the collision partner on the molecular structure[96], and the factors that affect molecular-adduct complex lifetime and stability during TIMS measurements[97]. In particular, we have shown the combination of simultaneous measurement of first principle derived collision cross sections and back-exchange HDX rates using a TIMS device with theoretical calculations for the assignment of candidate structures of kinetic intermediates as a way to study folding/unfolding pathways.[92]
In this work, we further investigate the potential of trapped ion mobility spectrometry (TIMS) for the separation and characterization of high molecular weight proteins and protein complexes. In particular, the discussion is based on the fundamental factors that affect the molecular ion trapping of high mass molecular systems, the collision cross section calculations and the differences between native and non-native states.
Experimental Methods
Material and reagents
Most proteins utilized in this study were purchased (e.g., ubiquitin, equine holomyoglobin, carbonic anhydrase from bovine erythrocytes, β-lactoglobulin, equine cytochrome C, and bovine serum albumin) from Sigma-Aldrich (St. Louis, MO, USA), or provided by collaborators (e.g., Avastin, HMGA2[98], rat Calmodulin[99] and recombinant mouse DREAM[93]) and used as received. DNA sequences FL875(CCCCCCATATTCGCGATTATTGCCCCCGCAATAATCG CGAATATGGGGGG), FL876 (GGATATTGCCCCCGCAATATCC) were purchased from Eurofin (Ebersberg, Germany). Protein solutions were prepared at a final 1–10 μM concentration in 10–100 mM ammonium acetate, 0–50% methanol, and 0–10% acetic acid. Low concentration Tuning Mix (G2421A; Agilent Technologies, Santa Clara, CA, USA) was used as a mobility calibration standard. All solvents and ammonium acetate salts used in these studies were analytical grade or better and purchased from Fisher Scientific (Pittsburg, PA, USA).
TIMS-MS Analysis
Details regarding the TIMS operation and specifics compared to traditional IMS can be found elsewhere.[88–92] Briefly, mobility separation in TIMS is based on holding the ions stationary using an electric field against a moving gas. The separation in a TIMS device can be described by the center of the mass frame using the same principles as in a conventional IMS drift tube.[41] Since mobility separation is related to the number of ion-neutral collisions (or drift time in traditional drift tube cells), the mobility separation in a TIMS device depends on the bath gas drift velocity, ion confinement and ion elution parameters. The mobility in a TIMS analyzer can be described as:
| (1) |
where υg is the velocity of the bath gas in the mobility cell and Ex(i) is the electric field at which the specific packet of ions elute. These parameters can be related to the elution voltage (Velu(i)) and the exit voltage (Vout) of the region exit. The calibration constant A was determined from previously reported mobility values for Tuning Mix calibration standard (G24221A, Agilent Technologies, Santa Clara, CA) in nitrogen (m/z = 322, K0 = 1.376 cm2 V−1 s−1 and m/z = 622, K0 = 1.013 cm2 V−1 s−1).[90, 100]From the mobility K value, the collisional cross section (CCS) can be determined by the following equation:
| (2) |
Where the charge of the ion is represented by z, kb represents the Boltzman constant, m1 and mb are the masses of molecular ion and the bath gas and N* is the number density.
TIMS-MS Operation Parameters
TIMS operation can be tuned for either wide range mobility analysis, with a voltage ramp of ΔVramp = 200 V, or for narrow mobility selection, with a narrow voltage ramp of ΔVramp = 10–30 V. In both cases ramp times of up to 500ms are used for analysis. Notice that TIMS mobility resolution depends on both the ramp size as well as the ramp speed, where lower speeds give higher mobility resolutions.[88–90] In addition to the ramp speed, the velocity of the gas also defines the mobility resolution and trapping efficiency of the TIMS analyzer. Increasing the velocity of the gas, by changing the pressure difference between the front (P1 = 1.1 – 4.3 mbar) and the end (P2= 0.6 – 3.0 mbar) of the analyzer region also increases the mobility resolution. As the velocity of the gas increases the ions experience a greater drag force, requiring higher electric fields in order to be trapped. A constant radiofrequency (RF) is applied to the entrance, analyzer and exit region of the TIMS analyzer (frequency of 880kHz with 200– 300 V peak-to-peak). Each funnel electrode is divided into four electrically insulated segments that are used to create a dipole field in the entrance and exit section, to focus the ions downstream, and a quadrupolar field in the separation region to radially confine the ions during the ion trapping and analysis. That is, in the entrance and exit funnel sections, the RF potential applied to the ion funnel is 180° out of phase between adjacent plates. This results in a pseudo-potential, which pushes the ions away from the funnel walls. However, in the analyzer section, the phase of the RF potential does not alternate between adjacent plates but only between adjacent segments. The purpose of the quadrupolar field in the analyzer section is to confine (trap) the ions radially and avoid ion losses due to diffusion. The TIMS analyzer was coupled to a maXis Impact QUHR-TOF (Bruker Daltonics Inc., Billerica, MA). Data acquisition was controlled using in-house software and synchronized with the maXis Impact acquisition program.
Atmospheric Pressure Ionization Sources
Electrospray Ionization Source (ESI)
An orthogonal, commercial ESI source based on the Apollo II design (Bruker Daltonics, Inc., MA) was used. Briefly, sample solutions were introduced into the nebulizer at a rate of 120–180 μL/min using an external syringe pump. Typical operating conditions were 4000–4500 V capillary voltage, 600 V endcap capillary offset voltage, 10 L/min dry gas flow rate, 1.0 bar nebulizer gas pressure, and a dry gas temperature 180 °C. Ions from the ESI source are introduced via a 0.6 mm inner diameter, single-bore glass capillary tube, which is resistively coated across its length, allowing the nebulizer to be maintained at ground potential, while the capillary exit was biased to around 180V.
nanoElectrospray Ionization Source (nanoESI)
A custom-built, pulled capillary orthogonal nanoESI source was utilized for all the experiments. Quartz glass capillaries (O.D.: 1.0 mm and I.D.: 0.70 mm) were pulled utilizing a P-2000 micropipette laser puller (Sutter Instruments, Novato, CA) and loaded with 10 μL aliquot sof the sample solution. A typical nanoESI source voltage of 600–1200 V was applied between the pulled capillary tips and the TIMS-MS instrument inlet. Ions were introduced into the TIMS cell via a stainless steel tube (1/16 x 0.020″, IDEX Health Science, Oak Harbor, WA), which was held at room temperature.
Theoretical Calculations
Theoretical collisional cross sections were calculated from X-ray structures for myoglobin (pdb: 1YMB), cytochrome C (pdb: 1HRC), β-lactoglobulin (pdb:4GNY), and ubiquitin (pdb:1UBQ), and were used as is.[100–103] Ion mobilities were calculated for the X-ray structures using the trajectory method (TM) utilizing IMoS (v.106W64dsoftware.[104–108] This calculation software allows the calculation of collisional cross sections of ions using both traditional methods as well as new methods such as Diffuse Hard Sphere Scattering and Diatomic Trajectory Method. For molecules without defined partial charges the charge is centered at the center of mass of the protein. Gas particles originate from a bounding box from the x, y, and z planes and are diagonalized in order to determine the collisional cross section of the ions. Calculations were performed with 3 TM rotations, with 300,000 gas molecules, and 92% Maxwell distributed remission velocity. Mobility calculations were performed for all experimentally observed charge states.
RESULTS AND DISCUSSION
Ion mobility spectrometry (IMS) combined with molecular dynamic simulations has proven to be a versatile technique for the analysis of intermediate and equilibrium structures of biomolecules enabling the correlation of ion-neutral, collision cross sections (CCS) with candidate structures.[109–115] In particular, it has been shown that by using soft ionization techniques (e.g., ESI) the evaporative cooling of the solvent leads to a freezing of multiple conformations, which has permitted the study of the conformational space dependence on the solvent conditions (e.g., native vs denatured), bath gas collision partner, and temperature.[116, 117] For example, changes in the starting solution conditions (e.g., pH, organic content, etc.) can induce changes in the charge state distribution and the conformational space that is accessible during the IMS-MS measurements.[118–121] In addition, changes in the conformational space can be observed as a function of the desolvation process and upon activation prior to the IMS-MS measurements. Previous work using TIMS-MS has shown the possibility to trap a wide mobility range from small molecules to small proteins[122, 123]; however, it was also demonstrated that the trapping efficiency is a function of the electrode geometry, bath gas profile and electrical confining parameters (e.g., radiofrequency value and amplitude).
Biomolecules can exist in several conformational states inside the cell and their functions are directly related to the folding/unfolding mechanism. Thus, to better understand the structural properties of biomolecules there is a need to accurately measure the CCS, which changes as a function of their conformational state. Biomolecular ions can be introduced in the TIMS analyzer in native and denatured states (see Figure 1). The increase in charge state is generally accompanied by the observation of more denatured conformational states with higher CCS values. That is, the degree of folding/unfolding can be observed as the charge state changes, reflecting biomolecular structural changes and any major transitions. For example, in the case of globular proteins (e.g., cytochrome C 12 kDa and myoglobin 17 kDa), a change from single native state to multiple molten globule and unfolded conformational states are observed. Notice that while the CCS values agree with previously reported CCS using DT-IMS-MS and TWIMS-MS, within 5%, the higher resolution of the TIMS permits, for the first time, the separation of a larger number of conformations. Moreover, in the case of barrel proteins (e.g., Ubiquitin 8.5 kDa and β-Lactoglobulin 18.4kDa) in addition to the unfolding trend, abrupt changes in the CCS can be correlated to significant conformational transitions. When compared to other structural measurements (e.g., solution NMR and X-ray crystallography, solid line in Figure 1), it can be seen that the increase in CCS with the charge state is not merely due to the charge dependence of the CCS, but a consequence of structural changes in the molecular ion (see equation 2). A more detailed list of CCS values as a function of the charge state is shown in Table 1 for protein, DNA-protein complexes, and protein-protein complexes using both ESI and nanoESI sources.
Figure 1.
Ion-neutral collisional cross section dependence on the m/z for globular proteins (myoglobin and cytochrome C) and barrel proteins (β-lactoglobulin and ubiquitin) in their native and denature states. In black, theoretical values obtained from reported tridimensional structures based on X-ray measurements.
Table 1.
Ion-neutral collisional cross sections measured by nanoESI-TIMS-MS in nitrogen as a bath gas for proteins, protein – DNA complexes and protein-protein complexes.
| Protein | m/z (charge), CCS; |
| Avastin | 5960 (+25), 6709; 5731 (+26), 6997; 5519 (+27), 7031; |
| BSA Dimer | 5870 (+23), 6208; 5625 (+24), 6454; 5400 (+25), 6699; |
| BSA | 4467 (+15), 4200, 4188 (+16), 4291, 3941 (+17), 4351; 3722 (+18), 4476; |
| β-Lactoglobulin Dimer | 2831 (+13), 3430; |
| DREAM* | 2455 (+12), 3025, 3181; 2266 (+13), 3561, 3756, 4078; 2104 (+14), 4029, 4195, 4315, 4526; 1964 (+15), 4243, 4459, 4809; 1841 (+16), 4526, 4829, 5024; 1733 (+17), 4663, 4829, 5102; 1636 (+18), 4751, 5317; 1473 (+20), 5151, 5639; 1403 (+21), 5756, 5912, 6253, 6467; 1339 (+22), 5951, 6184, 6340,6526; 1281 (+23), 6223, 6585, 6818; 1227 (+24), 6760, 6917, 7062 |
| Carbonic Anhydrase | 2887 (+10), 2530; 2625 (+11), 2606; 1444 (+20), 5422; 1375(+21), 5510; 1312 (+22), 5863; 1255 (+23), 6625; 1203 (+24), 6809; 1155 (+25), 6962; 1110 (+26), 7144, 1069 (+27), 7317; 1031 (+28), 7475; 996 (+29), 7596; 962 (+30), 7710; 931 (+31), 7831; 902 (+32), 7943; 875 (+33), 8139; 849 (+34), 8276; 825 (+35), 8380; 802 (+36), 8559; 780 (+37), 8692; 760 (+38), 8755; 740 (+39), 8906 |
| Myoglobin | 2147 (+8), 2187; 1908 (+9), 2399,2465; 1717 (+10), 2749, 2805, 2882, 2937; 1561 (+11), 2992, 3052, 3125, 3228, 3300; 1431 (+12), 3123, 3192, 3209, 3300, 3337, 3385, 3446, 3374, 3446, 3531, 3591; 1227 (+13), 3809; 1145 (+14), 3945; 1073 (+15), 4084; 1010 (+16), 4245; 954 (+16), 4222; 904 (+17), 4266 |
| β-Lactoglobulin | 3680 (+5), 1422; 3067 (+6), 1636; 2629 (+7), 1773; 2300 (+8), 1995; 2044 (+9), 2155, 2381; 1840 (+10), 2867, 1673 (+11), 3343; 1533 (+12), 3449; 1415 (+13), 3518; 1314 (+14), 3655; 1227 (+15), 3795; 1150 (+16), 3937; 1082 (+17), 4076; 1022 (+18), 4207; 968 (+19), 4867; 876 (+21), 5152 |
| Ubiquitin | 2125 (+4), 1149; 1700 (+5), 1229; 1417 (+6), 1699; 1214 (+7), 1794; 1063 (+8), 1975; 944 (+9), 2069; 850 (+10), 2213; 773 (+11), 2292; 708 (+12), 2468; 654 (+13), 2624 |
| Cytochrome C | 2064 (+6), 1289, 1438; 1769 (+7), 1307, 1574, 1922, 2083; 1548 (+8), 1686, 1925, 2249, 2468, 2496; 1376 (+9), 1637, 2024, 2258, 2358, 2388, 2631; 1238 (+10), 2181, 2365, 2508, 2652, 2717; 1125 (+11) 2403, 2553, 276, 2813, 2861; 1032 (+12), 2678, 2805, 2877, 3189; 952 (+13), 2794, 2969, 2998, 3044; 825.6 (+14), 3279, 3318, 3358; 774 (+16), 3036, 3311, 3382, 3465, 3557; 728 (+17), 3516, 3611, 3663, 3724; 688 (+18), 3598, 3689, 3743, 3778; 651 (+19), 3727, 3798; 619 (+20), 3885, 589 (+21), 3907,3954 |
| Calmodulin | 2405 (+7), 1860; 2104 (+8), 1915, 2103, 2378; 1870 (+9), 2256, 2396, 2750, 2933; 1684 (+10), 2741, 2981; 1530, (+11), 2957, 3097, 3256, 3323; 1403 (+12), 3170, 3323, 3402; 1295 (+13), 3512, 3591, 3707; 1202 (+14), 3695, 3749, 3847; 1122 (+15), 3841, 3944; 1052 (+16), 1091; 990 (+17) 4212, 935 (+18), 4341 |
| HMGA2 | 1477 (+8), 2114; 1313 (+9), 2353; 1181 (+10), 2511; 1074 (+11), 2704; 984 (+12), 2869, 2890; 909 (+13), 2998; 844 (+14), 2967, 3059, 3130; 787 (+15), 3236, 3334; 739 (+16), 3339 |
| FL876+HMGA2 (GGATATTGCCCCCGCAATATCC) | 2639 (+7), 1624; 2309 (+8), 1658; 2053 (+9), 1741, 2003; 1847 (+10), 1769, 1621, 2082, 2169, 2286; 1680 (+11), 2148, 2423; 1540 (+12), 2582, 2748; 1421 (+13), 2706, 2886; 1320 (+14), 2810, 3031, 3258; 1232 (+15), 2921, 3204; 1155 (+16), 3300, 3548; 1087 (+17) 3651, 4003; |
| FL875+HMGA2 (FL875: CCCCCCATATTCGCGATTATTGCCCCCGCAATAATCGCGAATATGGGGGG) | 3401 (+8), 1991; 3023 (+9), 2069; 2721 (+10), 2098, 2140, 2246, 2459 |
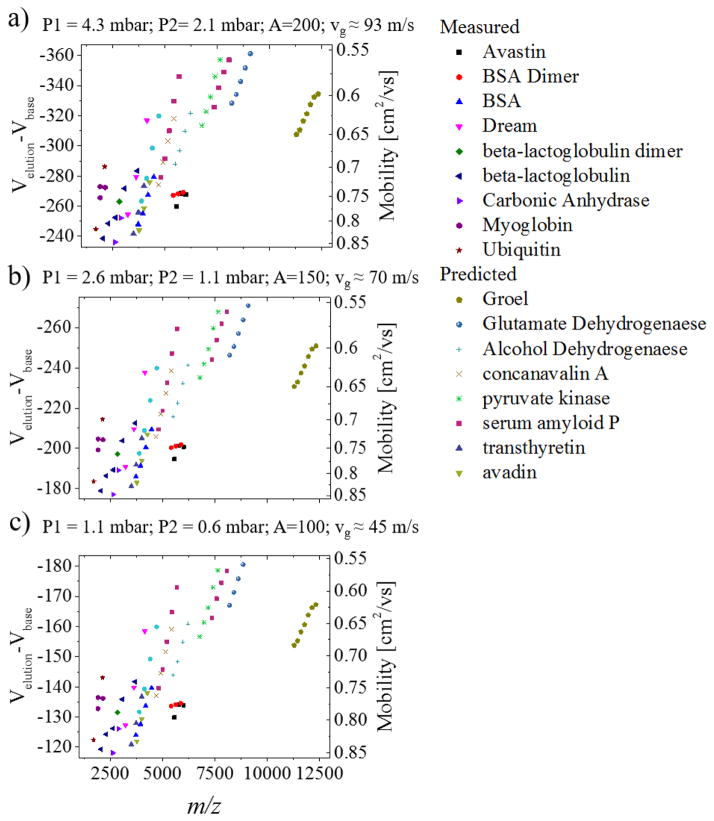
The charge state dependence with the CCS shows that, as the protein mass increases, there is a need to measure higher m/z in order to sample native conformations. Previous work has shown that with extended mass range MS analyzers, larger proteins and protein complexes can be studied by IMS-MS.[124] In the case of TIMS-MS, a similar approach can be taken but different from other IMS variants, the experimental conditions for molecular ion trapping and efficiency in the TIMS analyzer plays a major role. For example, nanoESI-TIMS-MS can separate larger molecular ions based on both mobility and m/z (see example in Figure 2 for the BSA dimer 132 kDa and the Avastin monomer 149 kDa) under native conditions. However, it should be noted that the TIMS-MS experiment can be performed under multiple trapping conditions. That is, in TIMS ion trapping is based on compensating the drift force (defined by the pressured difference in the tunnel region or bath gas velocity) with an electric field (see equation 1). However, there are practical limitations on the value of the applicable voltage difference across the tunnel region and therefore the mobility range that can be studied (see examples in Figure 3 for TIMS measured and for predicted K values based on other IMS techniques [50, 70]). As we mentioned before, the higher the velocity of the gas the higher the electric field required for the analysis. For example, when TIMS-MS is operated at an A ~200 (vg ≈ 93 m/s), the required field strength to trap the high mass protein ions in their native state is Velution − Vbase < 360 V. Although this electric field is technically achievably, the trapping efficiency is typically low for high mass species. Nevertheless, under these conditions, ion mobility resolutions (R= CCS/ΔCCS) of up to 400 have been achieved for lower mass systems (see example in the separation of hydroxylated polybrominated-diphenyl ethers[125]). In the case of larger mass systems, a factor of <2x in the resolving power can be compromised in order to achieved higher trapping efficiencies. For example, when TIMS-MS is operated at an A ~150 (vg ≈ 70 m/s) and A ~100 (vg ≈ 45 m/s), the required field strength to trap the high mass protein ions in their native state is Velution − Vbase < 275 and <185 V, respectively. Under these conditions large mass proteins like glutamate dehydrogenase, and massive protein complexes, such as Groel can be analyzed. Notice that although certain proteins and protein complexes typically have a very high mass (~800 kDa for Groel), a m/z range of up to 12,500 will suffice to perform TIMS-MS experiments of proteins and protein complexes in their native state.
Figure 2.
a) 2D-IMS-MS contour plot for a mixture of Avastin antibody and BSA dimer. Mass spectra projections for Region 1 (Avastin) and Region 2 (BSA dimer).
Figure 3.
Dependence of the trapping potential for a m/z up to 12,500 for several TIMS measured and reported biomolecular systems as a function of the bath gas velocity. Notice that as the velocity of the gas decreases, the lower the trapping potential required in the TIMS analyzer.
CONCLUSIONS
In the present work we demonstrate the potential and versatility of TIMS for the analysis of proteins, DNA-proteins, and protein-protein complexes in their native and denatured states. In addition, it was shown that accurate CCS measurements are possible and are in good agreement with previously reported CCS values using other IMS analyzers (<5% difference). The main challenges for the analysis of high mass proteins and protein complexes in the mobility and m/z domain were described. That is, the analysis of high mass systems in their native state will require the use of higher electric fields or a compromise in the TIMS mobility resolution by reducing the bath gas velocity. This is the first report of CCS measurements of high mass biomolecules and biomolecular complexes (~ 150 kDa) using TIMS-MS.
Acknowledgments
The authors would like to acknowledge Dr. Fenfei Leng (FIU), Dr. Jaroslava Miksovska (FIU), and Dr. Walter Gonzalez (FIU-Caltech) for their support during the protein expression, separation and purification processes, as well as Dr. Peter Sander (Bruker) for the development of DataAnalysis v. 5.0. IMoS is freely available from Dr. Carlos Larriba-Anadaluz (UIPUI). This work was supported by National Institute of Health support (Grant No. R00GM106414 to FF-L).
References
- 1.Zhang Y. Progress and challenges in protein structure prediction. Current opinion in structural biology. 2008;18(3):342–8. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts B. The Cell as a Collection of Protein Machines: Preparing the Next Generation of Molecular Biologists. Cell. 1998;92(3):291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 3.Dinant C, Luijsterburg MS, Höfer T, von Bornstaedt G, Vermeulen W, Houtsmuller AB, van Driel R. Assembly of multiprotein complexes that control genome function. The Journal of cell biology. 2009;185(1):21–26. doi: 10.1083/jcb.200811080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung E, Heller M, Sanchez JC, Hochstrasser DF. Proteomics meets cell biology: The establishment of subcellular proteomes. ELECTROPHORESIS. 2000;21(16):3369–3377. doi: 10.1002/1522-2683(20001001)21:16<3369::AID-ELPS3369>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 5.Hegyi H, Gerstein M. The relationship between protein structure and function: a comprehensive survey with application to the yeast genome 1. Journal of Molecular Biology. 1999;288(1):147–164. doi: 10.1006/jmbi.1999.2661. [DOI] [PubMed] [Google Scholar]
- 6.Todd AE, Orengo CA, Thornton JM. Evolution of function in protein superfamilies, from a structural perspective1. Journal of Molecular Biology. 2001;307(4):1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- 7.Whisstock JC, Lesk AM. Prediction of protein function from protein sequence and structure. Quarterly Reviews of Biophysics. 2003;36(03):307–340. doi: 10.1017/s0033583503003901. [DOI] [PubMed] [Google Scholar]
- 8.Loo TW, Bartlett MC, Clarke DM. Drug binding in human P-glycoprotein causes conformational changes in both nucleotide-binding domains. Journal of Biological Chemistry. 2003;278(3):1575–1578. doi: 10.1074/jbc.M211307200. [DOI] [PubMed] [Google Scholar]
- 9.Dauber-Osguthorpe P, Roberts VA, Osguthorpe DJ, Wolff J, Genest M, Hagler AT. Structure and energetics of ligand binding to proteins: Escherichia coli dihydrofolate reductase-trimethoprim, a drug-receptor system. Proteins: Structure, Function, and Bioinformatics. 1988;4(1):31–47. doi: 10.1002/prot.340040106. [DOI] [PubMed] [Google Scholar]
- 10.Teague SJ. Implications of protein flexibility for drug discovery. Nat Rev Drug Discov. 2003;2(7):527–541. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- 11.Jefferis R. Recombinant antibody therapeutics: the impact of glycosylation on mechanisms of action. Trends in Pharmacological Sciences. 2009;30(7):356–362. doi: 10.1016/j.tips.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450(7172):973–82. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 13.Sali A, Glaeser R, Earnest T, Baumeister W. From words to literature in structural proteomics. Nature. 2003;422(6928):216–25. doi: 10.1038/nature01513. [DOI] [PubMed] [Google Scholar]
- 14.Ketchem RR, Lee K-C, Huo S, Cross TA. Macromolecular structural elucidation with solid-state NMR-derived orientational constraints. Journal of Biomolecular NMR. 8(1):1–14. doi: 10.1007/BF00198135. [DOI] [PubMed] [Google Scholar]
- 15.Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature. 2002;420(6911):98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 16.Opella SJ, Marassi FM. Structure Determination of Membrane Proteins by NMR Spectroscopy. Chemical Reviews. 2004;104(8):3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y. A Glimpse of Structural Biology through X-Ray Crystallography. Cell. 2014;159(5):995–1014. doi: 10.1016/j.cell.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 18.Schenk ER, Nau F, Fernandez-Lima F. Theoretical predictor for candidate structure assignment from IMS data of biomolecule-related conformational space. International Journal for Ion Mobility Spectrometry. 2015;18(1–2):23–29. doi: 10.1007/s12127-015-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schenk ER, Almeida R, Miksovska J, Ridgeway ME, Park MA, Fernandez-Lima F. Kinetic intermediates of holo- and apo-myoglobin studied using HDX-TIMS-MS and molecular dynamic simulations. J Am Soc Mass Spectrom. 2015;26(4):555–63. doi: 10.1007/s13361-014-1067-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scarff CA, Thalassinos K, Hilton GR, Scrivens JH. Travelling wave ion mobility mass spectrometry studies of protein structure: biological significance and comparison with X-ray crystallography and nuclear magnetic resonance spectroscopy measurements. Rapid communications in mass spectrometry : RCM. 2008;22(20):3297–304. doi: 10.1002/rcm.3737. [DOI] [PubMed] [Google Scholar]
- 21.Feng X, Liu X, Luo Q, Liu BF. Mass spectrometry in systems biology: an overview. Mass Spectrom Rev. 2008;27(6):635–60. doi: 10.1002/mas.20182. [DOI] [PubMed] [Google Scholar]
- 22.Simoneit BR. A review of current applications of mass spectrometry for biomarker/molecular tracer elucidation. Mass Spectrom Rev. 2005;24(5):719–65. doi: 10.1002/mas.20036. [DOI] [PubMed] [Google Scholar]
- 23.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 1997;16(1):1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 24.Winston RL, Fitzgerald MC. Mass spectrometry as a readout of protein structure and function. Mass Spectrometry Reviews. 1997;16(4):165–179. doi: 10.1002/(SICI)1098-2787(1997)16:4<165::AID-MAS1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 25.Miranker A, Robinson CV, Radford SE, Aplin RT, Dobson CM. Detection of transient protein folding populations by mass spectrometry. Science. 1993;262(5135):896–900. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- 26.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41(21):6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 27.Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC genomics. 2008;9(Suppl 2):S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nature reviews Molecular cell biology. 2005;6(3):197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 29.Ferguson CN, Gucinski-Ruth AC. Evaluation of Ion Mobility-Mass Spectrometry for Comparative Analysis of Monoclonal Antibodies. Journal of The American Society for Mass Spectrometry. 2016;27(5):822–833. doi: 10.1007/s13361-016-1369-1. [DOI] [PubMed] [Google Scholar]
- 30.Beck A, Sanglier-Cianférani S, Van Dorsselaer A. Biosimilar, Biobetter, and Next Generation Antibody Characterization by Mass Spectrometry. Analytical Chemistry. 2012;84(11):4637–4646. doi: 10.1021/ac3002885. [DOI] [PubMed] [Google Scholar]
- 31.Burkitt W, Domann P, O’Connor G. Conformational changes in oxidatively stressed monoclonal antibodies studied by hydrogen exchange mass spectrometry. Protein Science. 2010;19(4):826–835. doi: 10.1002/pro.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marcoux J, Champion T, Colas O, Wagner-Rousset E, Corvaïa N, Van Dorsselaer A, Beck A, Cianférani S. Native mass spectrometry and ion mobility characterization of trastuzumab emtansine, a lysine-linked antibody drug conjugate. Protein Science. 2015;24(8):1210–1223. doi: 10.1002/pro.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Percipalle P, Fomproix N, Cavellán E, Voit R, Reimer G, Krüger T, Thyberg J, Scheer U, Grummt I, Östlund Farrants AK. The chromatin remodelling complex WSTF–SNF2h interacts with nuclear myosin 1 and has a role in RNA polymerase I transcription. EMBO reports. 2006;7(5):525–530. doi: 10.1038/sj.embor.7400657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasten MM, Dorland S, Stillman DJ. A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Molecular and Cellular Biology. 1997;17(8):4852–4858. doi: 10.1128/mcb.17.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Enzymatic Activity Associated with Class II HDACs Is Dependent on a Multiprotein Complex Containing HDAC3 and SMRT/N-CoR. Molecular Cell. 2002;9(1):45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 36.Trip EN, Scheffers DJ. A 1 MDa protein complex containing critical components of the Escherichia coli divisome. Scientific Reports. 2015;5:18190. doi: 10.1038/srep18190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaplan M, Cukkemane A, van Zundert GCP, Narasimhan S, Daniels M, Mance D, Waksman G, Bonvin AMJJ, Fronzes R, Folkers GE, Baldus M. Probing a cell-embedded megadalton protein complex by DNP-supported solid-state NMR. Nat Meth. 2015;12(7):649–652. doi: 10.1038/nmeth.3406. [DOI] [PubMed] [Google Scholar]
- 38.Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH., Jr Ion mobility-mass spectrometry. Journal of mass spectrometry : JMS. 2008;43(1):1–22. doi: 10.1002/jms.1383. [DOI] [PubMed] [Google Scholar]
- 39.Maurer MM, Donohoe GC, Valentine SJ. Advances in ion mobility-mass spectrometry instrumentation and techniques for characterizing structural heterogeneity. Analyst. 2015;140(20):6782–98. doi: 10.1039/c5an00922g. [DOI] [PubMed] [Google Scholar]
- 40.Wyttenbach T, Pierson NA, Clemmer DE, Bowers MT. Ion mobility analysis of molecular dynamics. Annu Rev Phys Chem. 2014;65:175–96. doi: 10.1146/annurev-physchem-040513-103644. [DOI] [PubMed] [Google Scholar]
- 41.McDaniel EW, Mason EA. Mobility and diffusion of ions in gases. John Wiley and Sons, Inc; New York: 1973. p. 381. [Google Scholar]
- 42.Gillig KJ, Russell DH. A periodic field focusing ion mobility spectrometer. The Texas A & M University System. 2001:36. [Google Scholar]
- 43.Gillig KJ, Ruotolo BT, Stone EG, Russell DH. An electrostatic focusing ion guide for ion mobility-mass spectrometry. Int J Mass Spectrom. 2004;239(1):43–49. [Google Scholar]
- 44.Silveira JA, Gamage CM, Blase RC, Russell DH. Gas-phase ion dynamics in a periodic-focusing DC ion guide. International Journal of Mass Spectrometry. 2010;296(1–3):36–42. [Google Scholar]
- 45.Guo Y, Wang J, Javahery G, Thomson BA, Siu KW. Ion mobility spectrometer with radial collisional focusing. Anal Chem. 2005;77(1):266–75. doi: 10.1021/ac048974n. [DOI] [PubMed] [Google Scholar]
- 46.Koeniger SL, Merenbloom SI, Valentine SJ, Jarrold MF, Udseth HR, Smith RD, Clemmer DE. An IMS-IMS analogue of MS-MS. Anal Chem. 2006;78(12):4161–74. doi: 10.1021/ac051060w. [DOI] [PubMed] [Google Scholar]
- 47.Kurulugama RT, Nachtigall FM, Lee S, Valentine SJ, Clemmer DE. Overtone mobility spectrometry: part 1. Experimental observations. J Am Soc Mass Spectrom. 2009;20(5):729–37. doi: 10.1016/j.jasms.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glaskin RS, Valentine SJ, Clemmer DE. A scanning frequency mode for ion cyclotron mobility spectrometry. Anal Chem. 2010;82(19):8266–71. doi: 10.1021/ac1017474. [DOI] [PubMed] [Google Scholar]
- 49.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. An investigation of the mobility separation of some peptide and protein ions using a new hybrid quadrupole/travelling wave IMS/oa-ToF instrument. Int J Mass Spectrom. 2007;261(1):1–12. [Google Scholar]
- 50.Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision Cross Sections of Proteins and Their Complexes: A Calibration Framework and Database for Gas-Phase Structural Biology. Analytical Chemistry. 2010;82(22):9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 51.Webb IK, Garimella SV, Tolmachev AV, Chen TC, Zhang X, Norheim RV, Prost SA, LaMarche B, Anderson GA, Ibrahim YM, Smith RD. Experimental evaluation and optimization of structures for lossless ion manipulations for ion mobility spectrometry with time-of-flight mass spectrometry. Anal Chem. 2014;86(18):9169–76. doi: 10.1021/ac502055e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dugourd P, Hudgins RR, Clemmer DE, Jarrold MF. High-resolution ion mobility measurements. Review of Scientific Instruments. 1997;68(2):1122–1129. [Google Scholar]
- 53.Merenbloom SI, Glaskin RS, Henson ZB, Clemmer DE. High-resolution ion cyclotron mobility spectrometry. Anal Chem. 2009;81(4):1482–7. doi: 10.1021/ac801880a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kemper PR, Dupuis NF, Bowers MT. A new, higher resolution, ion mobility mass spectrometer. International Journal of Mass Spectrometry. 2009;287(1–3):46–57. [Google Scholar]
- 55.Blase RC, Silveira JA, Gillig KJ, Gamage CM, Russell DH. Increased ion transmission in IMS: A high resolution, periodic-focusing DC ion guide ion mobility spectrometer. International Journal of Mass Spectrometry. 2011;301(1–3):166–173. [Google Scholar]
- 56.May JC, Russell DH. A mass-selective variable-temperature drift tube ion mobility-mass spectrometer for temperature dependent ion mobility studies. J Am Soc Mass Spectrom. 2011;22(7):1134–45. doi: 10.1007/s13361-011-0148-2. [DOI] [PubMed] [Google Scholar]
- 57.Kemper PR, Bowers MT. A hybrid double-focusing mass spectrometer—High-pressure drift reaction cell to study thermal energy reactions of mass-selected ions. Journal of the American Society for Mass Spectrometry. 1990;1(3):197–207. [Google Scholar]
- 58.Wu C, Siems WF, Asbury GR, Hill HH. Electrospray ionization high-resolution ion mobility spectrometry-mass spectrometry. Anal Chem. 1998;70(23):4929–38. doi: 10.1021/ac980414z. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, Clemmer DE. Characterizing oligosaccharides using injected-ion mobility/mass spectrometry. Anal Chem. 1997;69(13):2504–9. doi: 10.1021/ac9701344. [DOI] [PubMed] [Google Scholar]
- 60.Jarrold MF, Constant VA. Silicon cluster ions: Evidence for a structural transition. Physical review letters. 1991;67(21):2994–2997. doi: 10.1103/PhysRevLett.67.2994. [DOI] [PubMed] [Google Scholar]
- 61.Kaszycki JL, Bowman AP, Shvartsburg AA. Ion Mobility Separation of Peptide Isotopomers. Journal of The American Society for Mass Spectrometry. 2016;27(5):795–799. doi: 10.1007/s13361-016-1367-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hogan CJ, Mora JF. Ion Mobility Measurements of Nondenatured 12–150 kDa Proteins and Protein Multimers by Tandem Differential Mobility Analysis–Mass Spectrometry (DMA-MS) Journal of The American Society for Mass Spectrometry. 2011;22(1):158–172. doi: 10.1007/s13361-010-0014-7. [DOI] [PubMed] [Google Scholar]
- 63.de la Mora JF, Ude S, Thomson BA. The potential of differential mobility analysis coupled to MS for the study of very large singly and multiply charged proteins and protein complexes in the gas phase. Biotechnology Journal. 2006;1(9):988–997. doi: 10.1002/biot.200600070. [DOI] [PubMed] [Google Scholar]
- 64.Duijn Ev, Barendregt A, Synowsky S, Versluis C, Heck AJR. Chaperonin Complexes Monitored by Ion Mobility Mass Spectrometry. Journal of the American Chemical Society. 2009;131(4):1452–1459. doi: 10.1021/ja8055134. [DOI] [PubMed] [Google Scholar]
- 65.Uetrecht C, Barbu IM, Shoemaker GK, van Duijn E, HeckAlbert JR. Interrogating viral capsid assembly with ion mobility–mass spectrometry. Nat Chem. 2011;3(2):126–132. doi: 10.1038/nchem.947. [DOI] [PubMed] [Google Scholar]
- 66.Wang SC, Politis A, Di Bartolo N, Bavro VN, Tucker SJ, Booth PJ, Barrera NP, Robinson CV. Ion Mobility Mass Spectrometry of Two Tetrameric Membrane Protein Complexes Reveals Compact Structures and Differences in Stability and Packing. Journal of the American Chemical Society. 2010;132(44):15468–15470. doi: 10.1021/ja104312e. [DOI] [PubMed] [Google Scholar]
- 67.Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJR. Ion mobility mass spectrometry of proteins and protein assemblies. Chemical Society Reviews. 2010;39(5):1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- 68.Politis A, Park AY, Hyung SJ, Barsky D, Ruotolo BT, Robinson CV. Integrating Ion Mobility Mass Spectrometry with Molecular Modelling to Determine the Architecture of Multiprotein Complexes. PLoS ONE. 2010;5(8):e12080. doi: 10.1371/journal.pone.0012080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hyung SJ, Robinson CV, Ruotolo BT. Gas-Phase Unfolding and Disassembly Reveals Stability Differences in Ligand-Bound Multiprotein Complexes. Chemistry & Biology. 2009;16(4):382–390. doi: 10.1016/j.chembiol.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 70.Salbo R, Bush MF, Naver H, Campuzano I, Robinson CV, Pettersson I, Jørgensen TJD, Haselmann KF. Traveling-wave ion mobility mass spectrometry of protein complexes: accurate calibrated collision cross-sections of human insulin oligomers. Rapid Communications in Mass Spectrometry. 2012;26(10):1181–1193. doi: 10.1002/rcm.6211. [DOI] [PubMed] [Google Scholar]
- 71.Loo JA, Berhane B, Kaddis CS, Wooding KM, Xie Y, Kaufman SL, Chernushevich IV. Electrospray Ionization Mass Spectrometry and Ion Mobility Analysis of the 20S Proteasome Complex. Journal of the American Society for Mass Spectrometry. 2005;16(7):998–1008. doi: 10.1016/j.jasms.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 72.Ruotolo BT, Hyung SJ, Robinson PM, Giles K, Bateman RH, Robinson CV. Ion Mobility–Mass Spectrometry Reveals Long-Lived, Unfolded Intermediates in the Dissociation of Protein Complexes. Angewandte Chemie International Edition. 2007;46(42):8001–8004. doi: 10.1002/anie.200702161. [DOI] [PubMed] [Google Scholar]
- 73.Scott D, Layfield R, Oldham NJ. Ion mobility–mass spectrometry reveals conformational flexibility in the deubiquitinating enzyme USP5. PROTEOMICS. 2015;15(16):2835–2841. doi: 10.1002/pmic.201400457. [DOI] [PubMed] [Google Scholar]
- 74.Smith DP, Giles K, Bateman RH, Radford SE, Ashcroft AE. Monitoring Copopulated Conformational States During Protein Folding Events Using Electrospray Ionization-Ion Mobility Spectrometry-Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2007;18(12):2180–2190. doi: 10.1016/j.jasms.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Knapman WT, Valette MN, Warriner LS, Ashcroft EA. Ion Mobility Spectrometry-Mass Spectrometry of Intrinsically Unfolded Proteins: Trying to Put Order into Disorder. Current Analytical Chemistry. 2013;9(2):181–191. doi: 10.2174/1573411011309020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jurneczko E, Cruickshank F, Porrini M, Nikolova P, Campuzano Iain DG, Morris M, Barran Perdita E. Intrinsic disorder in proteins: a challenge for (un)structural biology met by ion mobility–mass spectrometry. Biochemical Society Transactions. 2012;40(5):1021–1026. doi: 10.1042/BST20120125. [DOI] [PubMed] [Google Scholar]
- 77.Pagel K, Natan E, Hall Z, Fersht AR, Robinson CV. Intrinsically Disordered p53 and Its Complexes Populate Compact Conformations in the Gas Phase. Angewandte Chemie International Edition. 2013;52(1):361–365. doi: 10.1002/anie.201203047. [DOI] [PubMed] [Google Scholar]
- 78.Zhou M, Politis A, Davies RB, Liko I, Wu KJ, Stewart AG, Stock D, Robinson CV. Ion mobility–mass spectrometry of a rotary ATPase reveals ATP-induced reduction in conformational flexibility. Nat Chem. 2014;6(3):208–215. doi: 10.1038/nchem.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shepherd Dale A, Holmes K, Rowlands David J, Stonehouse Nicola J, Ashcroft Alison E. Using Ion Mobility Spectrometry–Mass Spectrometry to Decipher the Conformational and Assembly Characteristics of the Hepatitis B Capsid Protein. Biophysical Journal. 2013;105(5):1258–1267. doi: 10.1016/j.bpj.2013.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pierson NA, Chen L, Valentine SJ, Russell DH, Clemmer DE. Number of Solution States of Bradykinin from Ion Mobility and Mass Spectrometry Measurements. Journal of the American Chemical Society. 2011;133(35):13810–13813. doi: 10.1021/ja203895j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shi L, Holliday AE, Bohrer BC, Kim D, Servage KA, Russell DH, Clemmer DE. “Wet” Versus “Dry” Folding of Polyproline. Journal of The American Society for Mass Spectrometry. 2016:1–11. doi: 10.1007/s13361-016-1372-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi L, Holliday AE, Glover MS, Ewing MA, Russell DH, Clemmer DE. Ion Mobility-Mass Spectrometry Reveals the Energetics of Intermediates that Guide Polyproline Folding. Journal of The American Society for Mass Spectrometry. 2015;27(1):22–30. doi: 10.1007/s13361-015-1255-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shi L, Holliday AE, Shi H, Zhu F, Ewing MA, Russell DH, Clemmer DE. Characterizing Intermediates Along the Transition from Polyproline I to Polyproline II Using Ion Mobility Spectrometry-Mass Spectrometry. Journal of the American Chemical Society. 2014;136(36):12702–12711. doi: 10.1021/ja505899g. [DOI] [PubMed] [Google Scholar]
- 84.Valentine SJ, Anderson JG, Ellington AD, Clemmer DE. Disulfide-Intact and -Reduced Lysozyme in the Gas Phase: Conformations and Pathways of Folding and Unfolding. The Journal of Physical Chemistry B. 1997;101(19):3891–3900. [Google Scholar]
- 85.Niu S, Ruotolo BT. Collisional unfolding of multiprotein complexes reveals cooperative stabilization upon ligand binding. Protein Science. 2015;24(8):1272–1281. doi: 10.1002/pro.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hopper JTS, Oldham NJ. Collision induced unfolding of protein ions in the gas phase studied by ion mobility-mass spectrometry: The effect of ligand binding on conformational stability. Journal of the American Society for Mass Spectrometry. 2009;20(10):1851–1858. doi: 10.1016/j.jasms.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 87.Zhong Y, Han L, Ruotolo BT. Collisional and Coulombic Unfolding of Gas-Phase Proteins: High Correlation to Their Domain Structures in Solution. Angewandte Chemie International Edition. 2014;53(35):9209–9212. doi: 10.1002/anie.201403784. [DOI] [PubMed] [Google Scholar]
- 88.Fernandez-Lima FA, Kaplan DA, Park MA. Note: Integration of trapped ion mobility spectrometry with mass spectrometry. Rev Sci Instr. 2011;82(12):126106. doi: 10.1063/1.3665933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fernandez-Lima F, Kaplan DA, Suetering J, Park MA. Gas-phase separation using a trapped ion mobility spectrometer. International journal for ion mobility spectrometry : official publication of the International Society for Ion Mobility Spectrometry. 2011;14(2–3):93–98. doi: 10.1007/s12127-011-0067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hernandez DR, Debord JD, Ridgeway ME, Kaplan DA, Park MA, Fernandez-Lima F. Ion dynamics in a trapped ion mobility spectrometer. Analyst. 2014;139(8):1913–21. doi: 10.1039/c3an02174b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schenk ER, Mendez V, Landrum JT, Ridgeway ME, Park MA, Fernandez-Lima F. Direct observation of differences of carotenoid polyene chain cis/trans isomers resulting from structural topology. Anal Chem. 2014;86(4):2019–24. doi: 10.1021/ac403153m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schenk ER, Ridgeway ME, Park MA, Leng F, Fernandez-Lima F. Isomerization kinetics of AT hook decapeptide solution structures. Anal Chem. 2014;86(2):1210–4. doi: 10.1021/ac403386q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gonzalez WG, Ramos V, Diaz M, Garabedian A, Molano-Arevalo JC, Fernandez-Lima F, Miksovska J. Characterization of the Photophysical, Thermodynamic, and Structural Properties of the Terbium (III)–DREAM Complex. Biochemistry. 2016;55(12):1873–1886. doi: 10.1021/acs.biochem.6b00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu FC, Kirk SR, Bleiholder C. On the structural denaturation of biological analytes in trapped ion mobility spectrometry - mass spectrometry. Analyst. 2016 doi: 10.1039/c5an02399h. [DOI] [PubMed] [Google Scholar]
- 95.Ridgeway ME, Silveira JA, Meier JE, Park MA. Microheterogeneity within conformational states of ubiquitin revealed by high resolution trapped ion mobility spectrometry. Analyst. 2015;140(20):6964–6972. doi: 10.1039/c5an00841g. [DOI] [PubMed] [Google Scholar]
- 96.Molano-Arevalo JC, Hernandez DR, Gonzalez WG, Miksovska J, Ridgeway ME, Park MA, Fernandez-Lima F. Flavin adenine dinucleotide structural motifs: from solution to gas phase. Anal Chem. 2014;86(20):10223–30. doi: 10.1021/ac5023666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McKenzie-Coe A, DeBord JD, Ridgeway M, Park M, Eiceman G, Fernandez-Lima F. Lifetimes and stabilities of familiar explosive molecular adduct complexes during ion mobility measurements. Analyst. 2015;140(16):5692–9. doi: 10.1039/c5an00527b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cui T, Wei S, Brew K, Leng F. Energetics of binding the mammalian high mobility group protein HMGA2 to poly (dA-dT) 2 and poly (dA)-poly (dT) Journal of molecular biology. 2005;352(3):629–645. doi: 10.1016/j.jmb.2005.07.048. [DOI] [PubMed] [Google Scholar]
- 99.George SE, Su Z, Fan D, Means AR. Calmodulin-cardiac troponin C chimeras. Effects of domain exchange on calcium binding and enzyme activation. Journal of Biological Chemistry. 1993;268(33):25213–25220. [PubMed] [Google Scholar]
- 100.Evans SV, Brayer GD. High-resolution study of the three-dimensional structure of horse heart metmyoglobin. Journal of Molecular Biology. 1990;213(4):885–897. doi: 10.1016/S0022-2836(05)80270-0. [DOI] [PubMed] [Google Scholar]
- 101.Bushnell GW, Louie GV, Brayer GD. High-resolution three-dimensional structure of horse heart cytochrome c. Journal of Molecular Biology. 1990;214(2):585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 102.Gutiérrez-Magdaleno G, Bello M, Portillo-Téllez MC, Rodríguez-Romero A, García-Hernández E. Ligand binding and self-association cooperativity of β-lactoglobulin. Journal of Molecular Recognition. 2013;26(2):67–75. doi: 10.1002/jmr.2249. [DOI] [PubMed] [Google Scholar]
- 103.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8Å resolution. Journal of Molecular Biology. 1987;194(3):531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 104.Czerwinska I, Far J, Kune C, Larriba-Andaluz C, Delaude L, De Pauw E. Structural analysis of ruthenium-arene complexes using ion mobility mass spectrometry, collision-induced dissociation, and DFT. Dalton Transactions. 2016;45(15):6361–6370. doi: 10.1039/c6dt00080k. [DOI] [PubMed] [Google Scholar]
- 105.Larriba C, Hogan CJ. Ion Mobilities in Diatomic Gases: Measurement versus Prediction with Non-Specular Scattering Models. The Journal of Physical Chemistry A. 2013;117(19):3887–3901. doi: 10.1021/jp312432z. [DOI] [PubMed] [Google Scholar]
- 106.Larriba C, Hogan CJ. Free molecular collision cross section calculation methods for nanoparticles and complex ions with energy accommodation. Journal of Computational Physics. 2013;251:344–363. [Google Scholar]
- 107.Larriba-Andaluz C, Fernández-García J, Ewing MA, Hogan CJ, Clemmer DE. Gas molecule scattering & ion mobility measurements for organic macro-ions in He versus N 2 environments. Physical Chemistry Chemical Physics. 2015;17(22):15019–15029. doi: 10.1039/c5cp01017a. [DOI] [PubMed] [Google Scholar]
- 108.Ouyang H, Larriba-Andaluz C, Oberreit DR, Hogan CJ., Jr The collision cross sections of iodide salt cluster ions in air via differential mobility analysis-mass spectrometry. Journal of the American Society for Mass Spectrometry. 2013;24(12):1833–1847. doi: 10.1007/s13361-013-0724-8. [DOI] [PubMed] [Google Scholar]
- 109.Mao Y, Ratner MA, Jarrold MF. Molecular Dynamics Simulations of the Charge-Induced Unfolding and Refolding of Unsolvated Cytochrome c. The Journal of Physical Chemistry B. 1999;103(45):10017–10021. [Google Scholar]
- 110.Onufriev A, Case DA, Bashford D. Structural details, pathways, and energetics of unfolding apomyoglobin. Journal of molecular biology. 2003;325(3):555–567. doi: 10.1016/s0022-2836(02)01207-x. [DOI] [PubMed] [Google Scholar]
- 111.Arteca GA, Tapia O. On the nature of the unfolded state: competing mechanisms in the unfolding of anhydrous protein ions. Chemical physics letters. 2004;383(5):462–468. [Google Scholar]
- 112.Chen L, Shao Q, Gao YQ, Russell DH. Molecular dynamics and ion mobility spectrometry study of model β-hairpin peptide, trpzip1. The Journal of Physical Chemistry A. 2011;115(17):4427–4435. doi: 10.1021/jp110014j. [DOI] [PubMed] [Google Scholar]
- 113.Zakharova NL, Crawford CL, Hauck BC, Quinton JK, Seims WF, Hill HH, Jr, Clark AE. An assessment of computational methods for obtaining structural information of moderately flexible biomolecules from ion mobility spectrometry. Journal of The American Society for Mass Spectrometry. 2012;23(5):792–805. doi: 10.1007/s13361-012-0339-5. [DOI] [PubMed] [Google Scholar]
- 114.D’Atri V, Porrini M, Rosu F, Gabelica V. Linking molecular models with ion mobility experiments. Illustration with a rigid nucleic acid structure. Journal of Mass Spectrometry. 2015;50(5):711–726. doi: 10.1002/jms.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hall Z, Politis A, Bush MF, Smith LJ, Robinson CV. Charge-state dependent compaction and dissociation of protein complexes: insights from ion mobility and molecular dynamics. Journal of the American Chemical Society. 2012;134(7):3429–3438. doi: 10.1021/ja2096859. [DOI] [PubMed] [Google Scholar]
- 116.Breuker K, McLafferty FW. Stepwise evolution of protein native structure with electrospray into the gas phase, 10–12 to 102 s. Proceedings of the National Academy of Sciences. 2008;105(47):18145–18152. doi: 10.1073/pnas.0807005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kebarle P, Tang L. From ions in solution to ions in the gas phase-the mechanism of electrospray mass spectrometry. Analytical chemistry. 1993;65(22):972A–986A. [Google Scholar]
- 118.Shi H, Atlasevich N, Merenbloom SI, Clemmer DE. Solution dependence of the collisional activation of ubiquitin [M+ 7H] 7+ ions. Journal of The American Society for Mass Spectrometry. 2014;25(12):2000–2008. doi: 10.1007/s13361-014-0834-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Vahidi S, Stocks BB, Konermann L. Partially disordered proteins studied by ion mobility-mass spectrometry: implications for the preservation of solution phase structure in the gas phase. Analytical chemistry. 2013;85(21):10471–10478. doi: 10.1021/ac402490r. [DOI] [PubMed] [Google Scholar]
- 120.Going CC, Williams ER. Supercharging with m-Nitrobenzyl Alcohol and Propylene Carbonate: Forming Highly Charged Ions with Extended, Near-Linear Conformations. Analytical Chemistry. 2015;87(7):3973–3980. doi: 10.1021/acs.analchem.5b00071. [DOI] [PubMed] [Google Scholar]
- 121.Shi H, Pierson NA, Valentine SJ, Clemmer DE. Conformation types of ubiquitin [M+ 8H] 8+ ions from water: methanol solutions: evidence for the N and A states in aqueous solution. The Journal of Physical Chemistry B. 2012;116(10):3344–3352. doi: 10.1021/jp210797x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Castellanos A, Benigni P, Hernandez DR, DeBord JD, Ridgeway ME, Park MA, Fernandez-Lima F. Fast Screening of Polycyclic Aromatic Hydrocarbons using Trapped Ion Mobility Spectrometry - Mass Spectrometry. Analytical methods : advancing methods and applications. 2014;6(23):9328–9332. doi: 10.1039/C4AY01655F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Benigni P, Thompson CJ, Ridgeway ME, Park MA, Fernandez-Lima F. Targeted high-resolution ion mobility separation coupled to ultrahigh-resolution mass spectrometry of endocrine disruptors in complex mixtures. Anal Chem. 2015;87(8):4321–5. doi: 10.1021/ac504866v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sobott F, Hernández H, McCammon MG, Tito MA, Robinson CV. A Tandem Mass Spectrometer for Improved Transmission and Analysis of Large Macromolecular Assemblies. Analytical Chemistry. 2002;74(6):1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 125.Adams KJ, Montero D, Aga D, Fernandez-Lima F. Isomer separation of polybrominated diphenyl ether metabolites using nanoESI-TIMS-MS. International Journal for Ion Mobility Spectrometry. 2016:1–8. doi: 10.1007/s12127-016-0198-z. [DOI] [PMC free article] [PubMed] [Google Scholar]