Abstract
The MYCN protooncogene is amplified in a number of advanced-stage human tumors such as neuroblastomas. Like other members of Myc family proteins, N-Myc is a transcription factor and its stability and activity are tightly controlled by ubiquitination-dependent proteasome degradation1-4. Although numerous studies demonstrate that N-Myc acts as a driver of neuroblastoma tumorigenesis, therapies that directly suppress N-Myc activity in human tumors are limited. Here, we have identified the herpesvirus-associated ubiquitin-specific protease (HAUSP or USP75-7) as a regulator of N-Myc in neuroblastoma. HAUSP interacts with N-Myc, and HAUSP expression induces deubiquitination and subsequent stabilization of N-Myc. Conversely, RNAi-mediated knockdown of HAUSP in neuroblastoma cancer cell lines, or genetic ablation of Hausp in the mouse brain destabilizes N-Myc, which leads to inhibition of N-Myc function. Notably, HAUSP is more abundant in neuroblastoma patients with poorer prognosis and HAUSP expression significantly correlates with N-Myc transcriptional activity. Furthermore, small molecule inhibitors against HAUSP deubiquitinase activity significantly suppress the growth of MYCN-amplified human neuroblastoma cell lines in xenograft mouse models. Together, our findings demonstrate a crucial role of HAUSP in regulating N-Myc function in vivo and suggest that HAUSP inhibition is a potential therapy for MYCN-amplified tumors.
Originally, the ubiquitin-proteasome pathway was thought to work as “a one-way street” from substrate ubiquitination to degradation by the 26S proteasome. However, the finding that the Herpesvirus-Associated Ubiquitin-Specific Protease (HAUSP, also called USP7) interacts with and deubiquitinates the p53-Mdm2 complex was one of the first examples that deubiquitinases (DUBs) exhibit a specific role in regulating protein stability5-7.
A substantial body of research indicates that HAUSP regulates both p53-dependent and p53-independent signaling networks. Further supporting this notion, HAUSP modulates the stability and activity of several cellular factors8-17. Nevertheless, it remains unclear which cellular factor(s) might contribute to the neonatal lethality of the nestin Cre-recombinase driven Hausp conditional knockout mice18. Here we sought to identify novel substrates of HAUSP, outside the p53 network, that specifically controls embryonic cell growth and development of the mouse brain. N-Myc, which regulates cell cycle progression and neuronal differentiation, predominantly functioning during embryogenesis, emerged as an attractive candidate1,2,19,20. Interestingly, the conventional MYCN mouse knockout21-23 exhibits embryonic lethality, while the nestin-Cre conditional knockout of MYCN24 has a decrease in cerebellum and cerebral cortex mass due to defective cellular proliferation—similar to mice in which floxed Hausp is deleted by nestin-Cre18.
To examine a potential relationship between HAUSP and N-Myc, the mouse cortex was analyzed; HAUSP deletion significantly stabilized p53 (Fig. 1a and Supplementary Fig. 1a) consistent with previous results18. Surprisingly, N-Myc levels decreased compared to HauspFL littermate controls (Fig. 1a and Supplementary Fig. 1a). This decline in the N-Myc protein level was independent of transcriptional regulation as no obvious changes in MYCN messenger RNA (mRNA) was detected (Supplementary Fig. 1b). Moreover, immunohistochemical staining revealed that N-Myc levels are reduced in the Hausp knockout tissues (Fig. 1b); notably, this effect on N-Myc protein levels caused by the Hausp knockout remained the same in the TP53-null background (Fig. 1b). Taken together, our data suggests that HAUSP controls N-Myc protein levels in vivo regardless of p53 status.
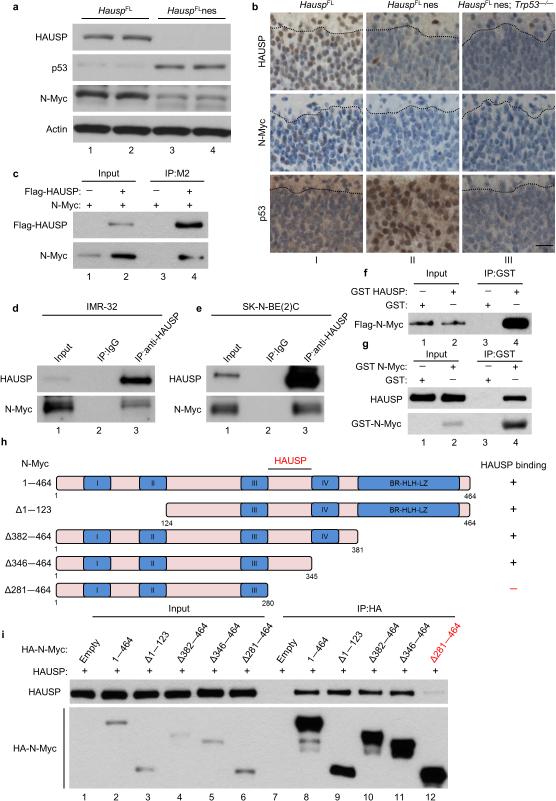
Figure 1. HAUSP affects and directly interacts with N-Myc both in vitro and in vivo.
(a) Western blot of mouse brains collected at day E13.5; n = 2. (b) Representative immunohistochemistry of HAUSP, N-Myc and p53 of E18.5 mouse cortex sections of the marginal zone to the cortical plate separated by a dotted black line; magnification 40 ×; scale bar 25 μm; n = 2 per group. (c) N-Myc expression vector cotransfected with Flag-HAUSP (lane 2) or empty vector (lane 1) in HEK293T cells. Cell lysates were incubated with Flag/M2 beads then subjected to western blot; 10% input; n = 3. (d, e) Endogenous immunoprecipitation of N-Myc with HAUSP from native IMR-32 (d) and SK-N-BE(2)C cells (e); 2% input; n = 3. (f) Direct interaction between purified Flag-N-Myc and GST-HAUSP. Purified N-Myc was incubated with GST protein (lane 1) or GST-HAUSP (lane 2) and immobilized with GST beads then subjected to western blot; 10% input; n = 2. (g) Direct interaction between purified Flag-HAUSP and GST-N-Myc as in Fig. 1f. Proteins were immobilized with GST beads then subjected to western blot; 1% input; n = 3. (h) Schematic representation of N-Myc deletion mutants used for domain mapping. (i) Indicated N-Myc expression vectors cotransfected with HAUSP in HEK293T cells. Lysates were incubated with HA beads then subjected to western blot; 10% input; n = 3.
To dissect the precise role for HAUSP in regulating N-Myc, we first examined the interaction between HAUSP and N-Myc. To this end, we transfected N-Myc alone or with a Flag-tagged HAUSP expression vector into HEK293 cells. Western blot analysis revealed that N-Myc was detected in HAUSP immunoprecipitates (Fig. 1c) similar to the p53 control (Supplementary Figure 1c). Reciprocally, HAUSP was detected in N-Myc immunoprecipitates (Supplementary Fig. 1d). To elucidate this interaction under physiological settings, cell extracts from neuroblastoma cancer cells IMR-32 (wild-type TP53) and SK-N-BE(2)C (mutant TP53) were immunoprecipitated with the HAUSP-specific antibody. Endogenous N-Myc was readily detected from immunoprecipitates obtained with the anti-HAUSP antibody in both cell lines irrespective of p53 status, but not with the control IgG (Fig. 1d, e). To test whether this interaction is direct, we performed an in vitro glutathione S-transferase (GST) pulldown assay with highly purified GST-tagged HAUSP and Flag-tagged N-Myc recombinant proteins. As shown in Figure 1f and Supplementary Fig. 1e, N-Myc bound to immobilized GST-HAUSP but not to GST alone. To further validate the direct interaction, we performed the reciprocal in vitro binding assay by using bacteria-produced GST-N-Myc and purified Flag-HAUSP. Western blot analysis revealed that HAUSP indeed bound to GST-N-Myc but not to GST (Fig. 1g). To map the minimal domain of N-Myc critical for interacting with HAUSP, we generated N-Myc mutants to narrow down the binding site (Fig. 1h). As shown in Figure 1i and Supplementary Figure 1f, by using the co-immunoprecipitation assays, we found that HAUSP is present in the immune-precipitates containing HA-tagged Full-length N-Myc (1-464), N-Myc Δ1-123, N-Myc Δ382-464, N-Myc Δ346-464 and N-Myc 281-464 immunoprecipitates. In contrast, HAUSP is not detected in the immune-precipitates containing HA-N-Myc Δ281-464 by the same approach. These data demonstrate that the small region of N-Myc (amino acids 281—345) is crucial for the binding with HAUSP (Fig. 1h). Further analyses showed that these differential bindings were not caused by differential subcellular localizations of HAUSP and Myc mutant proteins (Supplementary Fig. 1g and 1h). Sequence analysis also revealed that this region (amino acids 281-345) shares a very low sequence homology with c-Myc—much less than the highly conserved MYC box regions (Supplementary Fig. 1i).
To understand the functional consequence of this interaction, we first examined whether HAUSP expression affects N-Myc protein levels. As shown in Figure 2a and Supplementary Figure 2a, the levels of N-Myc increased upon HAUSP overexpression. Notably, HAUSP was able to stabilize both N-Myc Δ382—464 and N-Myc Δ346—464 but not N-Myc Δ281—464 in the same assay (Fig. 2b), suggesting that direct interaction is required for HAUSP-mediated stabilization of N-Myc. Moreover, the catalytically inactive HAUSP had a markedly reduced impact on N-Myc stabilization (Fig. 2c), independently of affecting MYCN mRNA (Supplementary Fig. 2b) suggesting that HAUSP regulates N-Myc stability through its deubiquitinase activity. Consistent with this conclusion, the half-life of N-Myc was significantly extended in the presence of wild-type HAUSP but not catalytically inactive HAUSP or an empty vector (Fig. 2d and Supplementary Fig. 2c). Next, we sought to determine whether HAUSP is indeed able to catalyze deubiquitination of N-Myc in cells. As shown in Figure 2e, the high levels of ubiquitinated N-Myc were markedly reduced after overexpression of wild-type but not catalytically inactive HAUSP. Collectively, these results demonstrate that N-Myc is a bona fide substrate of HAUSP.
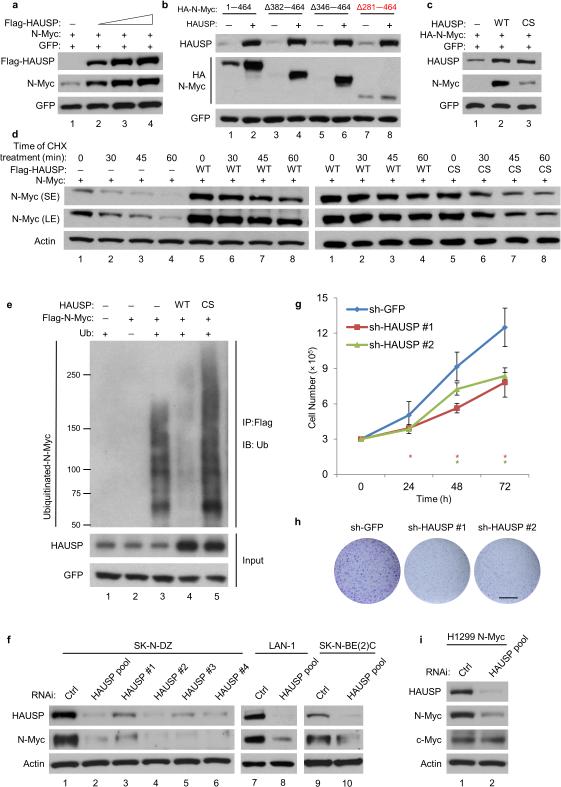
Figure 2. HAUSP regulates N-Myc through deubiquitination.
(a) Western blot of HEK293T cells transfected with N-Myc and empty vector, or increasing Flag-HAUSP; n = 3. (b) Western blot of HEK293T cells transfected with indicated HA-N-Myc constructs and empty vector (—) or HAUSP (+); n = 3. (c) Western blot of HEK293T cells transfected with N-Myc and empty vector, HAUSP wild-type (WT), or HAUSP C223S (CS); n = 3. (d) Western blot of HEK293T cells transfected with N-Myc and empty vector, Flag-HAUSP (WT) (left), or Flag-HAUSPcs (right). Cyclohexamide treatment (CHX, 50 ug/ml) for indicated times; n = 3. (e) Ubiquitin (Ub) and Flag-N-Myc expression vectors transfected with empty vector (lane 3) or in combination with HAUSP WT (lane 4) or HAUSP CS (lane 5) into HEK293T cells. Lanes 1—2 serve as controls. Lysates were incubated with Flag/M2 beads then subjected to western blot; n = 2. (f) Western blot of SK-N-DZ cells transfected with two-rounds of control RNAi, HAUSP pool, or individual HAUSP oligos composing the pool; n = 3. Western blot of LAN-1 (lanes 7—8) or SK-N-BE(2)C (lanes 9—10) cells transfected with three-rounds of control RNAi or HAUSP pool; n = 3. (g) Cell growth assay of SK-N-DZ expressing shGFP, HAUSP-shRNA #1, or HAUSP-shRNA #2. Error bars represent the mean ± SEM of 3 independent experiments. * represents a P value < 0.05 using two-tailed Student's t-test. (h) Colony formation assay using SK-N-DZ cells from Fig. 2g; scale bar 14.5 mm; n = 3. (i) Western blot of H1299 cells stably expressing an N-Myc polypeptide transfected with three-rounds of control RNAi or HAUSP pool; n = 3.
The removal of ubiquitin and subsequent stabilization of N-Myc suggests HAUSP prevents N-Myc degradation. Therefore, we reasoned that ablation of HAUSP may drive N-Myc proteolysis. A MYCN-amplified neuroblastoma cell line, SK-N-DZ, was used to ablate endogenous HAUSP levels. As shown in Figure 2f, reducing HAUSP (RNAi/HAUSP pool) decreased N-Myc protein levels; moreover, the levels of N-Myc were consistently decreased by using each of the four individual oligos composing the HAUSP RNAi pool, excluding off-target effects (Fig. 2f). Similar results were obtained using other MYCN-amplified neuroblastoma cell lines: LAN-1 and SK-N-BE(2)C (Fig. 2f) whereas MYCN mRNA levels remained unaffected upon HAUSP knockdown (Supplementary Fig. 2d). Many studies indicate that neuroblastoma cells have a crucial dependency on N-Myc expression for cell proliferation4,25. Indeed, HAUSP knockdown suppressed BrdU incorporation in a manner analogous to N-Myc knockdown in both SK-N-DZ and IMR-32 cells (Supplementary Fig. 2e,f). Moreover, SK-N-DZ expressing HAUSP shRNA (Supplementary Fig. 2g) displayed a significant growth defect when compared with sh-GFP control cells without any detectable signs of apoptosis (Fig. 2e and Supplementary Fig. 2h,i). Finally, we tested whether HAUSP could affect the oncogenic properties of neuroblastoma cells. As shown in Figure 2h and Supplementary Figure 2j, HAUSP knockdown in SK-N-DZ cells suppressed colony formation. Collectively, these data implicate that HAUSP is critical for both the stability and activity of endogenous N-Myc in neuroblastoma cells.
Although their spatial and temporal expression differs, N-Myc is closely related to c-Myc in structure and function19,20; therefore, we investigated whether HAUSP ablation also affects c-Myc stability. To examine the effects of HAUSP on c-Myc and N-Myc in the same cellular context, we generated a TP53-null, lung carcinoma H1299 cell line stably expressing an exogenous N-Myc polypeptide (Supplementary Fig. 3a). HAUSP knockdown reduced levels of N-Myc but had no obvious impact on c-Myc levels (Fig. 2i); similar results were obtained in native H1299 and human colon cancer HCT116 cell lines (Supplementary Fig. 3b). Moreover, in mouse embryonic fibroblasts generated from the HauspFL embryos, deletion of HAUSP did not alter the expression levels of endogenous c-Myc (Supplementary Fig. 3c). Consistent with this, HAUSP knockdown has no obvious cell growth defects in native H1299 cells (Supplementary Figs. 3d,e). Lastly, unlike with N-Myc, HAUSP was unable to remove the ubiquitin moieties from c-Myc (Supplementary Fig. 3f). These data suggest that HAUSP-mediated effects are specific for N-Myc but not c-Myc.
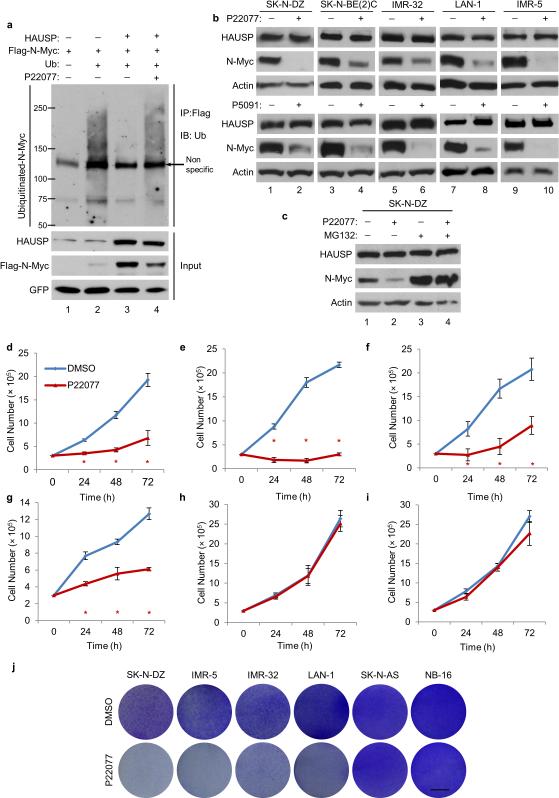
Recent studies described small molecules that specifically inhibit HAUSP deubiquitination activity in vivo and in vitro8,26-29. To further extend our findings, we tested whether HAUSP-mediated stabilization of N-Myc is also suppressed by HAUSP-specific inhibitors. We first determined whether the deubiquitinase activity of HAUSP on N-Myc is indeed suppressed by the inhibitor. As shown in Figure 3a, the ability of HAUSP to remove ubiquitin moieties from N-Myc was completely blocked by the commercially available HAUSP inhibitor, P2207716. Similar results were also obtained with another specific HAUSP inhibitor P509118 (Supplementary Fig. 4a). Using a panel of neuroblastoma cell lines, we demonstrate that both P22077 (Fig. 3b, top) and P5091 (Fig. 3b, bottom) destabilize N-Myc protein levels without significantly altering MYCN mRNA levels (Supplementary Figs. 4b,c). Similar to HAUSP knocking down, P22077 did not affect c-Myc ubiquitination or protein levels (Supplementary Figs. 4d,e). Consistently, the reduction of N-Myc protein levels was reverted by addition of the proteasome inhibitor, MG132 (Fig. 3c), without affecting MYCN mRNA (Supplementary Fig. 4f), suggesting that the HAUSP inhibitor, like HAUSP knockdown, promotes N-Myc proteolysis. Moreover, the functional consequence HAUSP exerts on neuroblastomas through regulating N-Myc was examined by analyzing BrdU incorporation and cell growth assays. Indeed, treatment with P22077 reduced incorporated BrdU in SK-N-DZ cells (Supplementary Fig. 4g) and also inhibited cell growth in all MYCN-amplified neuroblastoma cells tested (Fig. 3d-g and Supplementary Fig. 4h). In contrast to SK-N-DZ (mutant TP53) cells, IMR-5 (TP53-proficient) cells induced PARP1 cleavage after treatment with P22077 (Supplementary Fig 4i), suggestion that HAUSP inhibition can destabilize N-Myc as well as active p53-mediated responses in TP53-profiecient cells. As expected, p53 is indeed stabilized in LAN-1 neuroblastoma cells (TP53 wild-type) after treatment with P22077 (Supplementary Fig. 4j). While N-Myc levels are significantly reduced upon the same treatment, the MDM2 inhibitor Nutlin-3a only affected the stability of p53 but not N-Myc (Supplementary Fig. 4j). To further validate the specificity of HAUSP-mediated regulation on N-Myc, we examined the effects of the HAUSP inhibitor on neuroblastoma cell lines expressing mutant TP53 and lacking N-Myc: SK-N-AS and NB-16 (Supplementary Fig. 4k). Treatment with P22077 failed to induce any obvious effects on cell growth in SK-N-AS and NB-16 cells (Fig. 3h,i and Supplementary Fig. 4h). Finally, as shown in Figure 3j, the treatment with the HAUSP inhibitor suppressed colony formation in all MYCN-amplified cells tested whereas no obvious effects were observed in the MYCN non-amplified context. These data collectively demonstrate that HAUSP inhibition is very effective in suppressing cell growth of neuroblastomas by destabilizing N-Myc protein.
Figure 3. Pharmacologically blocking HAUSP deubiquitination in neuroblastoma cells.
(a) Ubiquitin and Flag-N-Myc vectors transfected with empty vector (lane 2) or in combination with HAUSP (lane 3—4) into HEK293T cells. Flag-N-Myc was transfected with empty vector (lane 1). Cells were treated with 20 μm of P22077 for the last 8 hours (lane 4). Lanes 1—3 were treated with DMSO. Cell lysates were incubated with Flag/M2 beads then subjected to western blot; n = 3. (b) Western blot analysis of indicated neuroblastoma cells treated with 20 μm of DMSO (—) or P22077 (+) (upper) and 12.5 μm of P5091 (+) (bottom) for 8 hours; n = 4. (c) Western blot analysis of SK-N-DZ cells treated for 6 hours with DMSO (—) or 10 μm of P22077 (+) or additionally treated with MG132 (lanes 3—4); n = 3. (d—i) Cell growth assay of SK-N-DZ (d), IMR-5 (e), IMR-32 (f), LAN-1 (g), SK-N-AS (h) NB-16 (i) cells treated either with DMSO or 10 μm of P22077. Error bars represent the mean ± SEM of 3 independent experiments. * represents a P value < 0.05 using two-tailed Student's t-test. (j) Colony assay using indicated neuroblastoma cells singly treated with DMSO or 10 μm of P22077; scale bar 8 mm; n = 3.
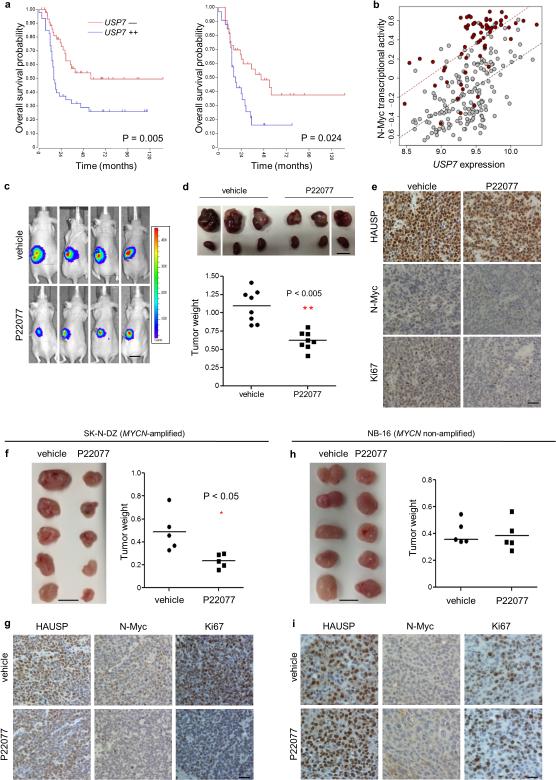
To assess the function of HAUSP in the pathogenesis of human neuroblastoma, we analyzed gene expression profiles from a cohort of 250 neuroblastoma primary tumors30. The levels of USP7 were significantly increased from patients with High risk (MYCN-amplified and MYCN non-amplified) neuroblastoma compared to patients categorized as Low risk (Stage 1) (p = 1.38e-07) (Supplementary Fig. 5a). Using three independent databases, we demonstrate that USP7 expression was prognostic for disease outcome in neuroblastoma; patients with elevated USP7 levels showed significantly shorter overall survival compared with patients bearing tumors that express lower levels of USP7 (Fig. 4a and Supplementary Fig. 5b). To determine if USP7 correlates with N-Myc function in patient tumor samples, we quantified N-Myc transcriptional activity by studying the expression of an N-Myc specific signature in each sample31. The correlation between USP7 expression and the N-Myc signature (see methods) is significantly higher than the correlation between USP7 expression and MYCN mRNA expression (p = 0.004) reminiscent of the correlation of MYCN expression to N-Myc activity (Fig. 4b and Supplementary Fig. 5c), further validating the conclusion that HAUSP regulates N-Myc stability and thereby N-Myc activity in human neuroblastoma. The same trend was observed using three different MYC signatures (Supplementary Table 1). As a further control, six N-Myc-specific transcriptional targets from the core signature were confirmed to be downregulated upon P22077 treatment in IMR-5 cells (Supplementary Fig. 5d).
Figure 4. The role of HAUSP on the pathogenesis of neuroblastomas.
(a) Kaplan-Meier analyses for SEQC RNA-seq database (left) and Kocak database (right) of overall survival for only MYCN-amplified samples. (b) Correlation of USP7 expression versus N-Myc transcriptional activity in neuroblastoma patients; MYCN-amplification (red) MYCN non-amplification (grey). (c—e) Pre-clinical neuroblastoma mouse model (see methods) (c) Representative in vivo bioluminescence imaging of tumor-bearing mice from day 12. Luciferase activity: low = blue, high = red; scale bar 13 mm. (d; top) Representative images of tumor-bearing kidneys versus normal kidney at day 15; scale bar 10 mm. (d; bottom) Tumor weight from day 15; n = 8 mice per group; ** represents a P value < 0.005 using two-tailed Student's t-test. (e) Representative immunohistochemistry of SK-N-DZ tumors for HAUSP, N-Myc, and Ki-67 from day 15. Magnification 40 ×; scale bar 25 μm; n = 3. (f—i) Subcutaneous xenograft models (see methods); (f) SK-N-DZ tumor image and weight; scale bar 10 mm; n = 5 mice per group; * represents a P value < 0.05 using two-tailed Student's t-test. (g) Representative immunohistochemistry of tumors harvested at day 12 stained for HAUSP, N-Myc, and Ki-67; Magnification 40 ×; scale bar 12.5 μm; n = 3. (h) NB-16 tumor image and weight; scale bar 10 mm; n = 5 mice per group. (i) Representative immunohistochemistry of tumors harvested at day 12 stained for HAUSP, N-Myc, and Ki-67; Magnification 40 ×; scale bar 12.5 μm; n = 3. **Note: Because NB-16 derived tumors grow faster in the subcutaneous model compared with SK-N-DZ, treatment started two weeks post implantation while with SK-N-DZ derived tumors, treatment began three weeks after implantation to more closely account for tumor size. Both SK-N-DZ and NB-16 subcutaneous models were treated with the same dose and time period (See Supplementary Fig. 6g).
These results prompted us to test the therapeutic effects of HAUSP inhibitors utilizing a kidney implantation xenograft mouse model of neuroblastoma32. Luciferase-expressing SK-N-DZ cells were injected into the left kidney and monitored for tumor growth; 14 days after injection, the cohort was randomized into two treatment groups: vehicle or the HAUSP inhibitor P22077. Mice tolerated the daily intraperitoneal injection (20 mg/kg) treatment regimen for 15 days as no weight loss or other health problems were observed during treatment or upon full necropsies (Supplementary Fig. 6a). Similarly, wild-type mice treated with the same daily-dose for a one month period did not show an obvious signs of weight loss, lethargy, or microscopic tissue damage (Supplementary Fig. 6b,c). Tumor-bearing mice receiving the HAUSP inhibitor had a significantly smaller tumor weight (day 15) compared with vehicle-treated mice (p < 0.005) (Fig. 4c,d). Further, immunohistological analysis showed that although HAUSP levels were unaffected upon P22077 treatment, N-Myc levels markedly decreased in treated tumors with an observable reduction for the proliferation marker Ki-67 (Fig. 4e and Supplementary Fig. 6f). Moreover, consistent results were obtained by adopting the commonly used subcutaneous xenograft system: P22077-treated SK-N-DZ tumors were significantly smaller than vehicle-treated tumors (p < 0.05) and expressed less N-Myc and incorporated less Ki67 (Fig. 4f,g and Supplementary Fig. 6f). Notably, as shown in Figure 4h,i and Supplementary Figure 6f, the MYCN non-amplified NB-16 tumors did not show any significant differences in weight or in cellular proliferation upon treatment with P22077. These data corroborate our cell culture findings demonstrating that HAUSP inhibition regulates neuroblastoma by affecting N-Myc stability and activity. Collectively, our data demonstrate that HAUSP expression can act as a biomarker for poor prognosis of human neuroblastomas and that small molecule HAUSP inhibitors may represent a therapeutic option for MYCN amplified neuroblastoma.
N-Myc is a short-lived protein and its degradation is mediated by several E3 ligases through the ubiquitin-proteasome system4,33-36. Recent work identified small molecules that increase N-Myc proteolysis through modulating the efficacy of FBXW7, a major E3 ligase for N-Myc37,38. Modulation of deubiquitination catalysis process may provide a quicker and more efficient way for reducing the stability of cellular proteins since inhibition of deubiquitinase activities is more direct than activating the E3 ligases involving ubiquitination of those proteins.17. A growing number of mammalian deubiquitination enzymes involved in tumorigenesis has been recently revealed including HAUSP8. Previous studies demonstrate that HAUSP inhibition leads to p53 activation through destabilizing Mdm26,18,39,40. Furthermore, several studies indicate that HAUSP is a potential target for therapeutic purpose by activating p53 activities26-29. In this study, we have identified N-Myc as a critical target for HAUSP. Like Mdm2, N-Myc is overexpressed in several different types of human cancers including neuroblastomas. Thus, HAUSP inhibitors are likely very effective for the treatment of human neuroblastomas by suppressing N-Myc activities regardless of p53 status.
Methods
General methodology
Unless otherwise noted, each sample was assayed in triplicate. No samples, mice or data points were excluded from the reported analyses except non-engrafted xenograft mice, as indicated below. Samples were not randomized to experimental groups except for the treatment allocation in the xenograft models, where indicated below. Analyses were not performed in a blinded fashion; for xenograft models, investigators were not blinded to group allocation.
Mice
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at Columbia University Health Sciences Center under the supervision of the Institute of Comparative Medicine. The conditional knockout of Hausp was designed to flox exon 6, which upon nestin-driven Cre recombinase, resulted in the removal of the conserved catalytic site (cysteine 206). This caused a truncation in the HAUSP protein due to a frame-shift which ensured loss of all HAUSP function. All animals (both female and male) were included in the analysis and no randomization was used. The Hausp-floxed (HauspFL) mice were maintained on a mixed background of 129Sv and C57BL/6J as described previously18. No statistical methods were used to predetermine sample size for the analysis of HauspFL mice. For the pre-clinical neuroblastoma mouse model, four-week-old female homozygous nude mice (nu/nu mice; Charles River) were housed for one week before being implanted with 1.7 × 106 SK-N-DZ cells (in 0.1 ml of PBS) expressing a luciferase construct into the left kidney. The flank muscles were then closed with absorbable suture and the skin closed with staples (as in ref. 32). After two weeks, engrafted tumors were visualized and tumor-bearing mice were randomized into two groups: vehicle (DMSO) and P22077. Mice with a non-detectable engraftment were excluded. Mice were treated at 20 mg/kg with daily intraperitoneal injections. At day 15 post-treatment, mice were killed; tumor bearing kidneys and the right kidney were resected, weighed, and photographed. For tumor weight, tumor-free kidney weight was subtracted from tumor-bearing kidney weight. Sample size (n = 8 per treatment group) was chosen to verify satisfactory interanimal reproducibility. For the toxicity assay, ten 1.5 month old wild-type C57BL/6J mice were separated into two groups: vehicle (DMSO) and P22077. Mice were treated at 20 mg/kg with daily intraperitoneal injects for 30 days. For sub-cutaneous xenografts, either SK-N-DZ cells or NB-16 cells (1.5 × 106) were mixed at a 1:1 ratio (volume) with Matrigel (BD Biosciences) and injected subcutaneous into 5 week old nude mice (Charles River). Two weeks after injection of NB-16 and three weeks after injection of SK-N-DZ cells, mice were treated for 12 days of either vehicle of P22077 at 20 mg/kg with daily intraperitoneal injections; investigators were not blinded to group allocation.
Embryo Collection and Histology
Embryos were collected from timed pregnancy for histological analysis as described18. Embryos were fixed in PBS-buffered 4% paraformaldehyde overnight, dehydrated through ethanol, and embedded in paraffin for histological analysis. Sagittal sections (6 μm) were incubated with indicated antibodies. Horseradish peroxidase-conjugated secondary antibodies were applied per the manufacturer's recommendations. A 3,3’-diaminodbenzidine (DAB)-chromogen substrate mixture was applied. Slides were subjected to hematoxylin-eosin (H&E) staining and examined by light microscopy.
Western Blot
For Western blot analysis, cells were lysed in cold FLAG lysis buffer [50 mM Tris-HCl (pH 7.3), 137 mM NaCl, 10 mM NaF, 0.5 mM EDTA, 1% Triton X-100, 0.2% Sarkosyl, 10% glycerol, and freshly supplemented protease inhibitor cocktail (Sigma)]. Protein concentration was determined by the Bradford method using Bio-Rad protein assay before proteins were equally loaded and separated in polyacrylamide gels. Nitrocellulose membranes were blocked with 5% (w/v) nonfat dry milk in Tris-buffered saline with Tween-20 [20 mM Tris-HCl (pH 7.6), 137 mM NaCl, 0.1% Tween-20], incubated with indicated primary antibodies, then secondary HRP-conjugated antibodies (GE Healthcare) (1:3000; diluted in 2% (w/v) nonfat dry milk in Tris-buffered saline with 0.1% Tween-20), and detected on autoradiographic films after incubating with ECL (GE Healthcare) or SuperSignal West Dura reagents (Thermo scientific).
Antibodies
Antibodies used in this study include N-Myc (OP-13; 1:1000) from EMD Biosciences; p53 (DO-1; 1:1000), and c-Myc (9E10 and C33; 1:500) from Santa Cruz Biotechnology; Ki-67 (SP6; 1:500) from Lab Vision; β-actin (AC-15; 1:10,000), Vinculin (V284; 1:1000), α-Tubulin (B-5-1-2; 1:4000) and Flag (M2; 1:1000) from Sigma; p53 (CM5; 1:1000) from Leica Microsystems; HA (3F10; 1:1000) and BrdU (1:500) from Roche Applied Science; GFP (JL-8; 1:1000) from Clontech; N-Myc (9405; 1:500), Lamin B1 (12586; 1:1000), and Parp1 (9532; 1:1000) from Cell Signaling; For IHC, HAUSP (1:500) Bethyl; For westerns, HAUSP antibody as described before5 (1:1000). All primary antibodies were diluted in 5% (w/v) nonfat dry milk in Tris-buffered saline with 0.1% Tween-20, except N-Myc (9405) which was diluted in 5% (w/v) BSA in Tris-buffered saline with 0.1% Tween-20.
Immunoprecipitation
For immunoprecipitation of ectopically expressed FLAG-tagged proteins, cells were lysed in BC100 buffer (20 mM Tris-HCl (pH 7.3), 100 mM NaCl, 0.2 mM EDTA, 0.2% Triton X-100, 10% glycerol, and freshly supplemented protease inhibitor). Cell extracts were incubated with the monoclonal M2/Flag-agarose beads (Sigma) at 4°C overnight. After five washes with the lysis buffer, the bound proteins were eluted with Flag-peptide (Sigma) in BC100 for 2 hours at 4°C. pcDNA3.1-HA-N-Myc plasmid or mutant constructs were cotransfected with HAUSP for 24 hours and lysed in BC100 buffer. Cell extracts were incubated with HA-agarose beads (Sigma) at 4°C overnight. After five washes with the lysis buffer, the bound proteins were eluted with HA-peptide (Sigma).
For endogenous immunoprecipitation assays, cells were lysed in cold BC100 buffer with sonication. 5% of cell extracts were saved for input, and the rest was incubated with either HAUSP antibody or control IgG at 4°C overnight. A/G PLUS agarose beads (Santa Cruz Biotechnology) were then added for 2 hours at 4°C. After five washes with the lysis buffer, the bound proteins were eluted by boiling with SDS sample buffer.
For cell based deubiquitination assay, Flag-N-Myc or Flag-c-Myc and HA-HIS-ubiquitin were cotransfected with empty vector or HAUSP (WT or CS) constructs for 36 hours. Cells were lysed in BC100 buffer and an M2/Flag immunoprecipitation was performed as describe above. For the deubiquitination assay with HAUSP inhibitor (either P5091 or P22077), 8 hours before harvesting the samples, the HAUSP inhibitor, or DMSO (for controls) were added at indicated doses.
GST Pulldown
pcDNA3.1-Flag-N-Myc plasmid or pcDNA3.1-Flag-HAUSP constructs were purified under highly stringent conditions (BC300). Purified N-Myc was incubated with either purified GST alone or purified GST-HAUSP. Purified HAUSP was incubated with either purified GST alone or purified GST-N-Myc. 20 μl GST slurry beads were added in BC100 buffer at 4°C overnight with gentle rotation. The GST beads were washed 4 times with BC100 buffer and eluted with 40 μl binding buffer plus 20 mM reduced glutathione for 2 hours with gentle rotation. Half of the elute was resolved on a 4-20% SDS-PAGE gel and detected by western blot.
Plasmids, Cell Culture, and Transfection
N-Myc expression vectors were constructed by polymerase chain reaction (PCR) amplification from pGEM7Z(+)-pTH-N-Myc vector (Addgene plasmid 35416) and subcloned either into a pCIN4-Flag-HA expression vector or pcDNA3.1 vector (Invitrogen). All HA-N-Myc deletion mutants were generated in pcDNA3.1 vectors. To construct the GST-N-Myc vector, cDNA sequences corresponding to the full-length protein were amplified by PCR from other expression vectors and cloned into a pGEX (GST) vector for expression in bacteria. The HAUSP and HAUSP-CS (C223S) plasmids were described before5. Full-length human p53 cDNA was subcloned into the pCIN4-Flag expression vector. To confirm equal transfection, a pCMV-GFP plasmid was used.
All cell lines were maintained in DMEM supplemented with 10% fetal bovine serum and 100 units per ml penicillin and 100 μg per ml streptomycin. H1299, HEK293T, HCT116, IMR-32, and SK-N-AS were all obtained from the American Type Culture Collection (ATCC); SK-N-DZ, LAN-1, IMR-5, NB-16, and SK-N-BE(2)C were provided by Dr. John Maris41; Shep21N were provided by Darrell Yamashiro; and MEF cells were described previously18. Cells were verified negative for mycoplasma contamination before experiments. All transfections were performed with Lipofectamine2000 (Invitrogen) according to the manufacture's protocol. To generate the Flag-HA-N-Myc expressing H1299 cell line, H1299 cells were transfected with pCIN4-Flag-HA-N-Myc vector and selected with 1 mg/ml G418 (GIBCO). To generate control and stable HAUSP knockdown cells, SK-N-DZ or H1299 were infected with pGIPZ empty vector or pGIPZ encoding HAUSP short hair pin RNA purchased from Thermo Open Biosystems (shRNA #1: 5’-GCATTAAAGCAGCGTATC-3’ and shRNA #2: 5’-GCATTAAAGCAGCGTATC-3’). Lentiviral vectors were packaged with pCMV-dR8.91 and pMDG-VSV-G vectors and transfected into HEK293T cells. 48 hours later, the viral supernatant was incubated with Polybrene and added to SK-N-DZ or H1299 cells. Puromycin (5ug/mL) was used to select positive clones.
siRNA Knock Down
Considering HAUSP is a stable protein, for the required efficient reduction of HAUSP, we transfected target cells with two to three-consecutive rounds of siRNA transfection. Cells were seeded around 30-40% confluency, and 24 hours later transfected with the first round. 24 hours later cells were split back to 30-40% confluency. 24 hours later, the second round of siRNA was transfected. Ablation of HAUSP was performed by transfecting indicated adhesive cell lines with Silencer select pre-designed siRNA duplex oligos (Oligo 1: 5’-AAGCGUCCCUUUAGCAUUA-3’; Oligo 2: 5’-GCAUAGUGAUAAACCUGUA-3’; Oligo 3: 5’-UAAGGACCCUGCAAAUUAU-3’; Oligo 4: 5’-GUAAAGAAGUAGACUAUCG-3’) synthesized by Dharmacon. siRNA duplexes for N-Myc (L-003913-01-0005) and control (D-001810-10-0050) were also purchased from Dharmacon. RNAi transfections were performed using Lipofectamine2000 following the manufacturer's protocol.
BrdU Incorporation
Cells were grown in medium containing 5μg/mL BrdU (Calbiochem) for 3 hours and fixed in 4% paraformaldehyde. DNA was denatured and cells were premeabilized in 2 N HCL with 0.5% Triton X-100 (Sigma) and then blocked with 5% BSA in PBS. Anti-BrdU was added following the manufacture's protocol (Amersham). After washing with 5% BSA in PBS, the cells were incubated with AlexaFluor 594 conjugated anti-mouse IgG (Molecular Probes). Finally, cells were counterstained with DAPI to visualize the nuclei.
Cell Counting and Colony Formation Assay
For all cell counting experiments, cells were initially seeded into 6-well plates at 3.0 × 105 cells overnight. Where applicable, the next morning either DMSO or 10 μm of P22077 (stock is dissolved at 20 mM in DMSO) was added. 24 hours after initial seeding (as in Fig. 2g and Supplementary Fig. 3e), or drug addition (as in Figs. 3d—i), cells were trypsinized and counted. For colony formation assay, 5000 cells were seeded in 6 cm2 plates and kept under selection for 14 days (as in Fig. 2h). Cells were stained for crystal violet solution. With addition of P22077, 3.0 × 105 cells were plated in 6-well plates. The next morning, either DMSO or P22077 was added at 10 μm. 96 hours later the cells were stained for crystal violet solution (as in Fig. 3j). Each experiment described above was repeated at least three independent times.
RNA Isolation and Quantitative real-time PCR
Total RNA was isolated from cells using TRIzol (Invitrogen) according to the manufacturer's protocol. 2 μg total RNA was reverse-transcribed using the ProtoScrpt First Stand cDNA Synthesis Kit (NEB). Quantitative PCR was done using a 7500 Fast Real-Time PCR System (Applied Biosystems) with a standard protocol. All mouse mRNA expression levels were normalized to β-actin while all human mRNA expression levels were normalized to GAPDH or HPRT.
RT-PCR primers
Human:
MYCN forward: 5′-ACCACAAGGCCCTCAGTACCTC-3’
MYCN reverse: 5′-GATGACACTCTTGAGCGGACGTGG-3’
MYCN forward: 5’-CCACAAGGCCCTCAGTACC-3’
MYCN reverse: 5’-TCTTCCTCTTCATCATCTTCATCA-3’
SKP2 forward: 5’-CCCACGGAAACGGCTGAAGA-3’
SKP2 reverse: 5’-CGCTAGGCGATACCACCTCTTACAA-3’
TWIST1 forward: 5’-AGCTACGCCTTCTCGGTCT-3’
TWIST1 reverse: 5’-CCTTCTCTGGAAACAATGACATC-3’
CCNE1 forward: 5’-GAACTGTGTCAAGTGGATGGT-3’
CCNE1 reverse: 5’-CCGCTGCTCTGCTTCTTAC-3’
PHOX2b forward: 5’-CTACCCCGTCGGCATTTT-3’
PHOX2b reverse: 5’-CAAAGCAGTGCCAAAAAGTTC-3’
MCM3 forward: 5’-GGGTGGAACGAGACCTAGAA-3’
MCM3 reverse: 5’-AGACTTGGCAACGGATGG-3’
MAD2L forward: 5’-CGCGTGCTTTTGTTTGTGT-3’
MAD2L reverse: 5’-GCTGTTGATGCCGAATGAG-3’
HPRT forward: 5´-TTGCTCGAGATGTGATGAAGGA-3´
HPRT reverse: 5´-TTCCAGTTAAAGTTGAGAGATCA-3’
GAPDH forward: 5’-ATCAATGGAAATCCCATCACCA-3’
GAPDH reverse: 5’-GACTCCACGACGTACTCAGCG-3’
Mouse:
MYCN forward: 5´-GTGTCTGTTCCAGCTACTGC-3´
MYCN reverse: 5´-CATCTTCCTCCTCGTCATC-3´
β-actin forward: 5´-GATGACGATATCGCTGCGCTG-3´
β-actin reverse, 5´-GTACGACCAGAGGCATACAGG-3’
Statistical Analysis
Results are presented as the means ± SEM. Differences were determined using a two-tailed, unpaired Student's t-test with CI of 95%. A P value ≤ 0.05 was denoted as statistically significant.
Bioinformatic Analysis
For the analysis of primary neuroblastoma tumors we studied a cohort of 250 patients organized and compiled by the TARGET (Therapeutically Applicable Research To Generate Effective Treatments, http://target.nci.nih.gov) consortium. Gene expression profiles where obtained from affimetrix HumanExon 1.0ST platform. We applied Robust Multichip Analisys (RMA) normalization and summarization with the core probeset annotation provided Netaffix (http://www.affymetrix.com/analysis/index.affx) with affimetrix power tools (apt). Clinical data was available including censored survival data, stage and MYCN amplification status.
To categorize patients into different stages, we used the International Neuroblastoma Staging System (INSS) definitions for Stage 1 and 4 which represent the most distant stages in terms of their histopathology and clinical outcome. Additionally both groups where filtered to fit into the risk stratification groups as defined by the Children's Oncology Group (COG). Therefore in this study, Stage 1 is equivalent to low risk while Stage 4 is equivalent to high risk tumors. We evaluated whether HAUSP expression is different between these groups (Supplementary Fig. 5a) and assessed the significance of this difference using a Kruskal-wallis test.
In order to understand the association of HAUSP with patient survival we made use of clinical information provided by TARGET which included overall survival (OS). We performed two statistical tests for the association between gene expression and censored survival data. We used a cox proportional hazards models to estimate the significance of the association between a continuous variable (USP7 expression) and censored (survival) data. We used the implementation provided by the R survival package for both tests (http://cran.r-project.org/web/packages/survival/index.html). Analysis for survival curves from SEQC and Kocak were obtained from R2 Genomics Analysis and Visualization Platform (http://r2.amc.nl). We extracted from the tumor cohort the 1/3 higher and lower quantiles for USP7 expression and performed a Log-rank test, Fig. 4a.
Since protein data is not available for human samples, we designed an analysis which allows for the measurement of the transcriptional activity of a transcription factor (N-Myc) by studying expression variation of its down-stream targets. Therefore, to measure the impact of HAUSP on N-Myc protein stabilization in human primary tumors, we assume that changes in N-Myc protein stability will directly impact its transcriptional activity without changing N-Myc mRNA levels. The changes in N-Myc activity that cannot be explained by N-Myc mRNA changes must be controlled by N-Myc regulation at the protein level. To measure N-Myc transcription factor activity we followed the method described in the VIPER Bioconductor package (http://bioconductor.org/packages/release/bioc/html/viper.html)42 which uses genome wide transcriptional networks to infer transcription factor activity by implementing a modification of the GSEA algorithm on a single sample level. However, instead of a newly inferred N-Myc target signature we used a validated list of targets from literature (Pubmed ID in Supplementary Table 1). As shown in Fig. 4b and Supplementary Fig. 5c, the correlation between MYCN expression and N-Myc activity in the cohort of MYCN-amplified neuroblastoma samples is 0.62, while the correlation between MYCN and USP7 expression is 0.36. Further, the correlation between USP7 expression and N-Myc activity is 0.61 (highlighted in Supplementary Table 1). This result suggests that changes in USP7 expression explains N-Myc activity changes that cannot be explained by N-Myc expression. In order to calculate the significance of the difference between two correlation coefficients, we applied the Fisher's transformation of r to Z-score. This allows us to parameterize our test as the difference between two normally distributed values Z1 and Z2, which difference returns another Z normalized score from which an analytical p-value can be calculated.
Fisher's transform of r to z equation:
Supplementary Material
Acknowledgements
This work was supported by the National Cancer Institute of the National Institutes of Health under Award 5R01CA193890, 5RO1CA190477, 5RO1CA085533 and 2P01CA080058 (W.G.) The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work is also partially supported by NIH cancer biology training grant T32-CA09503 (O.T.).
Footnotes
Author Contributions:
The whole project was conceived and designed by O.T. and W.G. Experiments were performed mainly by O.T., D.L. and C.D. The Bioinformatic Analysis was performed by G.L. and A.C. Some of the experiments were performed with the help from D.B., N.K., C.C, H.S., and D.J.Y. The paper was written by O.T. and W.G.
Competing financial interests:
The authors declare no competing financial interests.
References
- 1.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harbor perspectives in medicine. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Popov N, et al. The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;9:765–774. doi: 10.1038/ncb1601. [DOI] [PubMed] [Google Scholar]
- 4.Otto T, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Li M, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 6.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Molecular cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 7.Hu M, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 8.Nicholson B, Suresh Kumar KG. The multifaceted roles of USP7: new therapeutic opportunities. Cell biochemistry and biophysics. 2011;60:61–68. doi: 10.1007/s12013-011-9185-5. [DOI] [PubMed] [Google Scholar]
- 9.van der Horst A, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 10.Song MS, et al. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du Z, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Science signaling. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faesen AC, et al. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol Cell. 2011;44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 13.Lee HR, et al. Bilateral inhibition of HAUSP deubiquitinase by a viral interferon regulatory factor protein. Nat Struct Mol Biol. 2011;18:1336–1344. doi: 10.1038/nsmb.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma H, et al. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Natl Acad Sci U S A. 2012;109:4828–4833. doi: 10.1073/pnas.1116349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colleran A, et al. Deubiquitination of NF-kappaB by Ubiquitin-Specific Protease-7 promotes transcription. Proc Natl Acad Sci U S A. 2013;110:618–623. doi: 10.1073/pnas.1208446110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng C, Niu C, Yang Y, Wang Y, Lu M. Expression of HAUSP in gliomas correlates with disease progression and survival of patients. Oncology reports. 2013;29:1730–1736. doi: 10.3892/or.2013.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfoh R, Lacdao IK, Saridakis V. Deubiquitinases and the new therapeutic opportunities offered to cancer. Endocrine-related cancer. 2015;22:T35–54. doi: 10.1530/ERC-14-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kon N, et al. Roles of HAUSP-mediated p53 regulation in central nervous system development. Cell death and differentiation. 2011;18:1366–1375. doi: 10.1038/cdd.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harbor perspectives in medicine. 2013;3 doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charron J, et al. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1992;6:2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- 22.Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- 23.Sawai S, et al. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development. 1993;117:1445–1455. doi: 10.1242/dev.117.4.1445. [DOI] [PubMed] [Google Scholar]
- 24.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Seminars in cancer biology. 2006;16:313–317. doi: 10.1016/j.semcancer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Colland F, et al. Small-molecule inhibitor of USP7/HAUSP ubiquitin protease stabilizes and activates p53 in cells. Molecular cancer therapeutics. 2009;8:2286–2295. doi: 10.1158/1535-7163.MCT-09-0097. [DOI] [PubMed] [Google Scholar]
- 27.Altun M, et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chemistry & biology. 2011;18:1401–1412. doi: 10.1016/j.chembiol.2011.08.018. [DOI] [PubMed] [Google Scholar]
- 28.Reverdy C, et al. Discovery of specific inhibitors of human USP7/HAUSP deubiquitinating enzyme. Chemistry & biology. 2012;19:467–477. doi: 10.1016/j.chembiol.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Chauhan D, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345–358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pugh TJ, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valentijn LJ, et al. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc Natl Acad Sci U S A. 2012;109:19190–19195. doi: 10.1073/pnas.1208215109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim ES, et al. Potent VEGF blockade causes regression of coopted vessels in a model of neuroblastoma. Proc Natl Acad Sci U S A. 2002;99:11399–11404. doi: 10.1073/pnas.172398399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi SH, Wright JB, Gerber SA, Cole MD. Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 2010;24:1236–1241. doi: 10.1101/gad.1920310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sjostrom SK, Finn G, Hahn WC, Rowitch DH, Kenney AM. The Cdk1 complex plays a prime role in regulating N-myc phosphorylation and turnover in neural precursors. Dev Cell. 2005;9:327–338. doi: 10.1016/j.devcel.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Welcker M, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao X, et al. The HECT-domain ubiquitin ligase Huwe1 controls neural differentiation and proliferation by destabilizing the N-Myc oncoprotein. Nat Cell Biol. 2008;10:643–653. doi: 10.1038/ncb1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brockmann M, et al. Small Molecule Inhibitors of Aurora-A Induce Proteasomal Degradation of N-Myc in Childhood Neuroblastoma. Cancer Cell. 2013 doi: 10.1016/j.ccr.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gustafson WC, et al. Drugging MYCN through an Allosteric Transition in Aurora Kinase A. Cancer Cell. 2014 doi: 10.1016/j.ccr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cummins JM, Vogelstein B. HAUSP is required for p53 destabilization. Cell Cycle. 2004;3:689–692. [PubMed] [Google Scholar]
- 40.Kon N, et al. Inactivation of HAUSP in vivo modulates p53 function. Oncogene. 2010;29:1270–1279. doi: 10.1038/onc.2009.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods only references
- 41.Rader J, et al. Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin Cancer Res. 2013;19:6173–6182. doi: 10.1158/1078-0432.CCR-13-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alvarez MJ, et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet. 2016 doi: 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.