Ceritinib resulted in a significant, durable response of a metastatic inflammatory myofibroblastic tumor (IMT) after failure of crizotinib. A chromoplectic TPM3–ALK rearrangement involving many known oncogenes was found in the residual IMT. Ceritinib may be useful for patients with IMT after failure of crizotinib, and chromoplexy may have a role in the oncogenesis or treatment resistance of IMTs.
Keywords: ALK, IMT, chromoplexy, ceritinib, resistance
Abstract
Background
Inflammatory myofibroblastic tumors (IMTs) are rare sarcomas that can occur at any age. Surgical resection is the primary treatment for patients with localized disease; however, these tumors frequently recur. Less commonly, patients with IMTs develop or present with metastatic disease. There is no standard of care for these patients and traditional cytotoxic therapy is largely ineffective. Most IMTs are associated with oncogenic ALK, ROS1 or PDGFRβ fusions and may benefit from targeted therapy.
Patient and methods
We sought to understand the genomic abnormalities of a patient who presented for management of metastatic IMT after progression of disease on crizotinib and a significant and durable partial response to the more potent ALK inhibitor ceritinib.
Results
The residual IMT was resected based on the recommendations of a multidisciplinary tumor sarcoma tumor board and analyzed by whole-genome mate pair sequencing. Analysis of the residual, resected tumor identified a chromoplectic TPM3–ALK rearrangement that involved many other known oncogenes and was confirmed by rtPCR.
Conclusions
In our analysis of the treatment-resistant, residual IMT, we identified a complex pattern of genetic rearrangements consistent with chromoplexy. Although it is difficult to know for certain if these chromoplectic rearrangements preceded treatment, their presence suggests that chromoplexy has a role in the oncogenesis of IMTs. Furthermore, this patient's remarkable response suggests that ceritinib should be considered as an option after progression on crizotinib for patients with metastatic or unresectable IMT and ALK mutations.
introduction
Inflammatory myofibroblastic tumors (IMTs) are distinctive neoplasms composed of myofibroblasts and fibroblasts with inflammatory infiltrates including many plasma cells. IMTs predominantly affect children and young adults but can occur at any age. IMTs occur predominantly in the lung, soft tissue and viscera. Surgical resection is the primary means of management for patients with localized disease; however, these tumors may recur. Rarely, patients with IMTs develop or present with metastatic disease. There is no standard of care for these patients and traditional cytotoxic therapy is largely ineffective.
Chromosomal rearrangements are frequently identified in IMTs. ALK rearrangements are identified in approximately half of patients, and ROS1 or PDGFRβ fusions are found in about one-third of patients [1]. Responses to crizotinib have been observed in patients with metastatic IMT and ALK or ROS1 rearrangements [2]. Currently, we do not have an understanding of the mechanisms of progression in patients with IMTs on targeted kinase inhibitors, or how to treat them afterwards.
Herein, we describe the case of a patient who presented for management of a metastatic IMT after progression of disease on crizotinib then celecoxib. A significant partial response was observed with ceritinib (previously known as LDK-378) obtained on a compassionate use basis. The residual disease was resected and interrogated for genetic rearrangements. We identified complex chromosomal rearrangements with many known oncogenes, including ALK, in a chromoplectic pattern.
methods
compassionate use ceritinib
Arrangements were made with Novartis, the Mayo Clinic Institutional Review Board and the Food and Drug Administration to provide ceritinib on a compassionate use basis (FDA IND 120983). The patient provided informed consent for compassionate use. The National Cancer Institute's Common Terminology Criteria for Adverse Events v4.0 were used to describe adverse events and Response Evaluation Criteria in Solid Tumors version 1.1 were used to assess his response.
next-generation sequencing
Mate Pair sequencing (MPSeq) tiles the whole genome with large spanning (∼3 kb) fragments to increase the probability of spanning a genomic breakpoint. The MP library was assembled WGA DNA, according to a previously published protocol [3] using the Illumina Nextera Mate Pair kit (Illumina, CA; FC-132-1001). The MPSeq library was sequenced in half a lane of an Illumina flow cell and sequenced to 101×2 paired-end reads on an Illumina HiSeq2000. Base calling was carried out using Illumina Pipeline v1.5.
data analysis
Bioinformatics protocols to rapidly and efficiently process NGS MPSeq data using a 32-bit binary indexing of the Hg19 reference genome have been previously published from our laboratory [4, 5]. The algorithm maps both MPSeq reads successively to the whole genome, selecting reads <15 kb apart allowing up to 10 mismatches, with the lowest cumulative mismatch count sent to the output. Discordant MPs mapping >30 kb apart or in different chromosomes were selected for further analysis. Algorithmic filters to determine lineage relationships were set to minimize the effects of both false positives (FP) and false negatives (FN). Namely, the lowest limit of MP associates to call an event was set at 7, where the FP rate was practically zero, and a mask of breakpoints was used to eliminate common variants and discordant events from experimental or algorithmic errors. The combined nucleotide distance to cluster associates of a single event was set to 3000 bp's, thereby eliminating closely related but not identical breakpoints from being called as shared. Breakpoints near gaps of reference genome sequence were also eliminated. The FN rate was estimated to be <15%, dictated by the incompleteness of the reference genome and by regions that are difficult to map. The landscape of genomic rearrangements is presented as a genome plot. The genome plot details all chromosomes (stacked horizontally with chr.1–12 and X on the left and chr.13–22 and Y, on the right. Copy number levels are presented across each chromosome with normal two-copy levels presented in gray, gains in blue and losses in red. Solid lines linking distal chromosomal regions represent intra- and interchromosomal breakpoint fusion junctions. Conversely, the circos plot represents all chromosomes on the periphery of a continuous circle; with copy numbers represented in an inner circle and breakpoint fusion junction links across the center. The circos plot was created as described elsewhere [6].
validation of rearrangements and fusion transcript
Primers spanning the detected fusion junctions were used in PCR validations (25 µl reaction volumes, 50 ng Template, 35 cycles) using EasyA high fidelity polymerase (Stratagene, La Jolla, CA; #600404). PCR validations were carried out on tumor DNA and a human genomic DNA control (C) (Promega, Madison, WI; G304A), as well as germline blood DNA (B) where available. GAPDH control PCRs were carried out using standard primers.
immunohistochemistry
Four micrometer sections cut from formalin-fixed, paraffin-embedded blocks were placed on charged slides, then dried and melted at 62°C for 20 min. Slides were placed on a Ventana BenchMark XT (Ventana Medical Systems Inc., Tucson, AZ) for staining. The staining protocol includes online deparaffinization, heat-induced epitope retrieval with Ventana Cell Conditioning 1 for 32 min, primary ALK antibody incubation for 32 min at 37°C (clone D5F3, a rabbit monoclonal antibody, 1:100 dilution, Cell Signaling Technology, Danvers, MA). Antigen–antibody reactions were visualized using Ventana Optiview Universal DAB Detection Kit. Counterstaining was carried out on the Ventana BenchMark XT using Ventana Hematoxylin II for 8 min, followed by bluing reagent for 4 min. The positive control was a known ALK-rearranged lung adenocarcinoma and the negative control was a mouse IgG1 serum substitution for the primary ALK antibody.
results
patient presentation
A 32-year-old male was diagnosed with metastatic IMT involving his left lung and chest wall, medial right thigh, right gluteal muscle and omentum ∼1 year after he presented with dyspnea. He was initially treated with prednisone with improvement of his dyspnea for possible eosinophilic pneumonia because of a prominent eosinophil component identified in a bronchioloalveolar lavage specimen. He was then lost to follow-up. Approximately 1 year later, his dyspnea worsened and was accompanied by a palpable mass in his medial right thigh. A biopsy of this lesion was pursued. Multiple pathologists reviewed the specimen and agreed on the diagnosis of IMT. Immunohistochemistry was carried out and identified intense, extensive expression of ALK. Treatment was initiated with crizotinib and resulted in a partial response. Eight months later, he experienced worsening chest pain and dyspnea and imaging studies confirmed disease progression. He was placed on celecoxib, and over the course of 1 month, he experienced significant worsening of his symptoms and performance status decline. He then presented to our center for further management. This patient did not qualify for available clinical trials (NCT01742286, NCT01283516) at the time of his presentation based on his performance status.
Due to this patient's previous response to crizotinib and ALK expression, arrangements were made with Novartis, the Mayo Clinic Institutional Review Board and the United States Food and Drug Administration to administer ceritinib 750 mg daily on a compassionate use basis. His symptoms of dyspnea and pain significantly improved within 2 weeks of therapy. He developed mild nausea (grade 1) that was controlled with ondansetron and a dose reduction in ceritinib to 600 mg daily. Imaging studies after 8 weeks of treatment demonstrated a significant partial response to treatment that was confirmed 8 weeks later (Figure 1). Given the minimal residual disease burden after 6 months of therapy with ceritinib, surgical resection of his pulmonary lesion and ablation of his gluteal lesion were recommended by a multi-institutional Sarcoma Tumor Board to consolidate his significant response. This patient continued with ceritinib in an adjuvant fashion for 18 additional months after its initiation until a left-sided, 18F-fluorodeoxyglucose-avid pleural lesion was identified and confirmed to be recurrent IMT by biopsy.
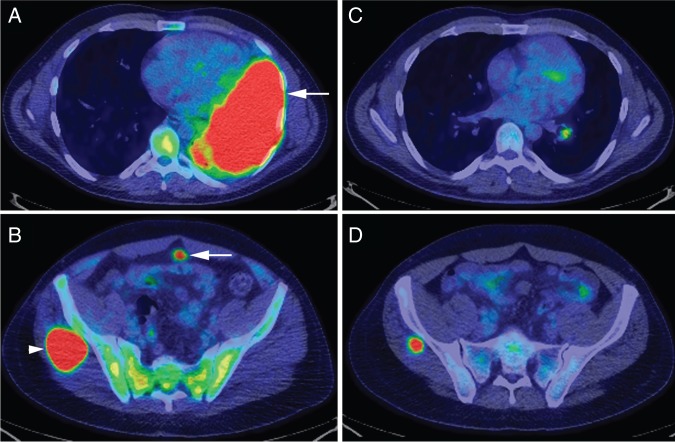
Figure 1.
Radiologic response to ceritinib. 18F-fluorodeoxyglucose positron emission tomography/computed tomography (PET/CT) was obtained after progression on crizotinib and celecoxib, before initiation of treatment with ceritinib (A and B) and after 8 weeks of therapy (C and D). The tumor involved the majority of this patient's left lung (A, arrow), omentum (B, arrow) and right gluteal muscle (B, arrowhead). Imaging obtained 8 weeks after therapy started with ceritinib demonstrated a partial response with significant reduction in the pulmonary (C) and gluteal lesions (D). No additional changes were identified by PET/CT obtained 8 weeks later (after 16 weeks of treatment), confirming the partial response to treatment.
characterization of resected IMT specimen
whole-genome mate pair sequencing
The resected pulmonary lesion was used for genomic characterization. In order to characterize the potential ALK rearrangement, the Illumina mate pair sequencing (MPSeq) protocol was used to elucidate genome-wide structural variations within the tumor. MPSeq spans the genome with larger spanning genomic fragments than conventional genomic sequencing, increasing the potential of detecting discordantly mapping breakpoints from rearrangements. The MPSeq data yielded over 121 million paired reads, of which 97.4% successfully mapped to the human reference genome (hg38) (supplementary Table S1, available at Annals of Oncology online). The biallelic bridged coverage of spanning fragments [7], important for breakpoint calling, was 91X, while absolute sequenced base coverage was just 3X. With an average fragment size of 3169 bp, 85.5% mapped concordantly to the reference genome with the correct distal span separating the paired reads. Conversely, 3.3% of paired reads mapped discordantly. Following algorithmic filtering and masking of known germline events, 209 unique discordant genomic breakpoints were detected. Of these events, 141 (67%) were intra-chromosomal events, while the remaining 68 (33%) were translocations between different chromosomes (supplementary Figure S1, available at Annals of Oncology online).
genomic rearrangements at the ALK locus
A genome plot of all structural variations in the tumor, together with the conventional circos plot, is presented in Figure 2A and B. The genome plot reveals a complex chromoplectic event of intricately weaved genomic rearrangements occurring in concert between chromosomes 1, 2, 9 and 20. While the chromoplectic regions are associated with focal copy gains of chromosomal sequence, the majority of the tumor genome is structurally intact, with minimal discordance from the reference genome. The chromoplectic event involves two focal regions on the q-arms of chromosomes 1 and 9, and one region of chromosome 20, together with multiple regions of chromosome 2, with one containing the ALK locus (Figure 2C).
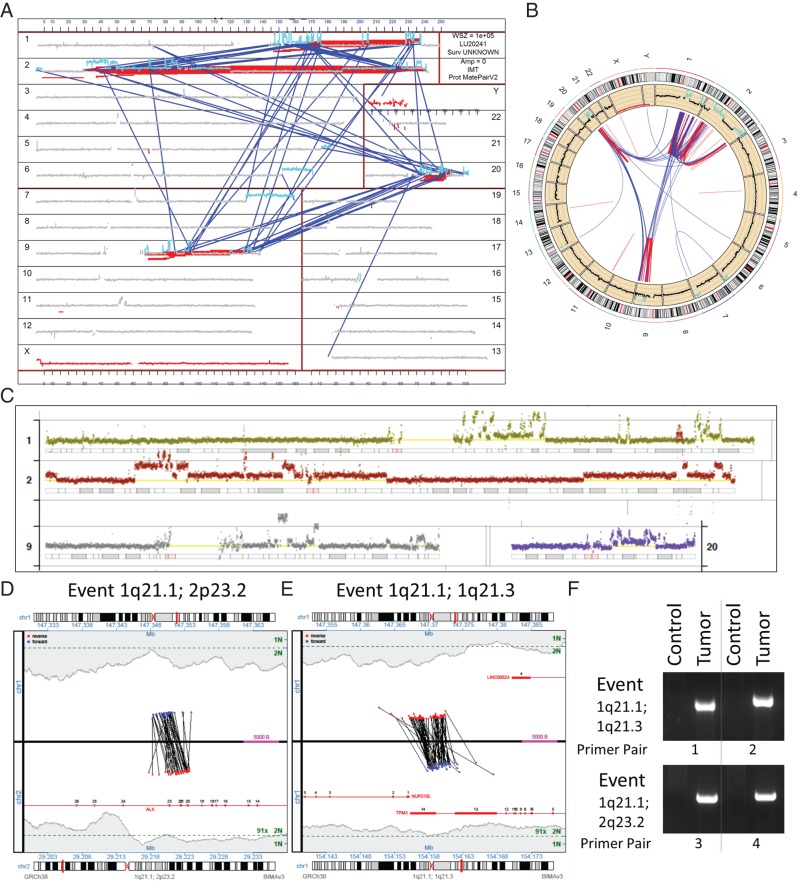
Figure 2.
Molecular characterization of treatment-resistant tumor sample. The multiple rearrangements are demonstrated in a genome plot of all structural variations in the tumor (A) and a conventional circos plot (B). The genome plot presents coverage (y-axis) across each chromosome (x-axis) with a window size (WSZ) of 1 × 105. Gray indicates normal diploid coverage, with gains indicated in cyan. Inter- and intrachromosomal rearrangements are presented as blue and red lines linking chromosome positions, analogous to the circos plot. The q-arms of chromosomes 1, 9 and 20, together with multiple regions of chromosome 2 including one containing the ALK locus were involved in a chromoplectic rearrangement, with more detailed coverage across each chromosome presented in (C). The normal diploid two-copy level is presented by the yellow lines for each chromosome. Junction plots present the mate pair reads supporting the translocation between intron 19 of ALK at 2p23.2 (lower panel of plot) and chromosome 1 (1q21.1) at a non-genic region between CHD1L and LINC00624 (upper panel of plot) (D). A second junction plot describes an intra-chromosomal rearrangement adjacent to this chromosome one position at 1q21.1, linking to the final intron of TPM3 at 1p21.1 (E). Red and blue dots represent mate pair reads mapping to the positive or negative DNA strands, respectively, with coverage levels across each position presented in gray-shaded regions, with single (1N) and two-copy levels (2N) indicated (green). Gene regions are presented in red with exons numerated. Both the ALK (1q21.1; 2p23.2) and TPM3 (1p21.1; 1p21.3) events were verified by PCR on tumor DNA using different primer combinations spanning the breakpoint junctions (F).
While a number of breakpoints hit directly on the ALK gene, no individual event predicted a fusion that could be driving the observed ALK overexpression in the patient's tumor. However, the extensive chromosomal shuffling at the ALK locus made interpretation of the rearrangements more complex, with three events hitting the ALK gene directly on 2p23.2. Two breakpoints were positioned on introns 12 and 19 of ALK, while the third was located just downstream of the 3′UTR of ALK (supplementary Table S2, available at Annals of Oncology online). The event hitting intron 19 involved a translocation to chromosome 1 (1q21.2) at a non-genic region between CHD1L and LINC00624 (Figure 2D). An additional breakpoint was observed adjacent to this site, linking to the final intron of Topomyosin 3 (TPM3) (Figure 2E). TPM3 encodes for an actin-binding protein, which provides stability to actin filaments and regulates access of other actin-binding proteins. As pathogenic translocations have previously been reported in anaplastic large cell lymphomas [8] and IMTs [9] linking TPM3-driven transcription of a TPM3–ALK fusion protein, we hypothesized these two events could result in a similar outcome. Both the ALK (1:2)(q21.1;p23.2) and TPM3 (1:1)(p21.1;p21.3) events were verified by PCR on tumor DNA (Figure 2F).
validation of the TPM3–ALK fusion transcript in RNA
The complex rearrangement linking a potential fusion between TPM3 and ALK is depicted in Figure 3A. Forward and reverse primers were designed in exonic sequences of ALK and TPM3 to investigate the presence of a fusion transcript in cDNA generated from RNA extracted from the patient's tumor. Subsequent rtPCR generated a clear DNA band (Figure 3B) indicating the predicted fusion of exon 8 of TPM3 to exon 20 of ALK (Figure 3C). Sanger sequencing of the PCR product confirmed this as a functional, in-frame fusion transcript (Figure 3D). No mutations were detected between exon 20 of ALK and the stop codon (Figure 3D and supplementary Figure S2, available at Annals of Oncology online). Intense, extensive ALK expression was observed with immunohistochemistry (Figure 3E and F).
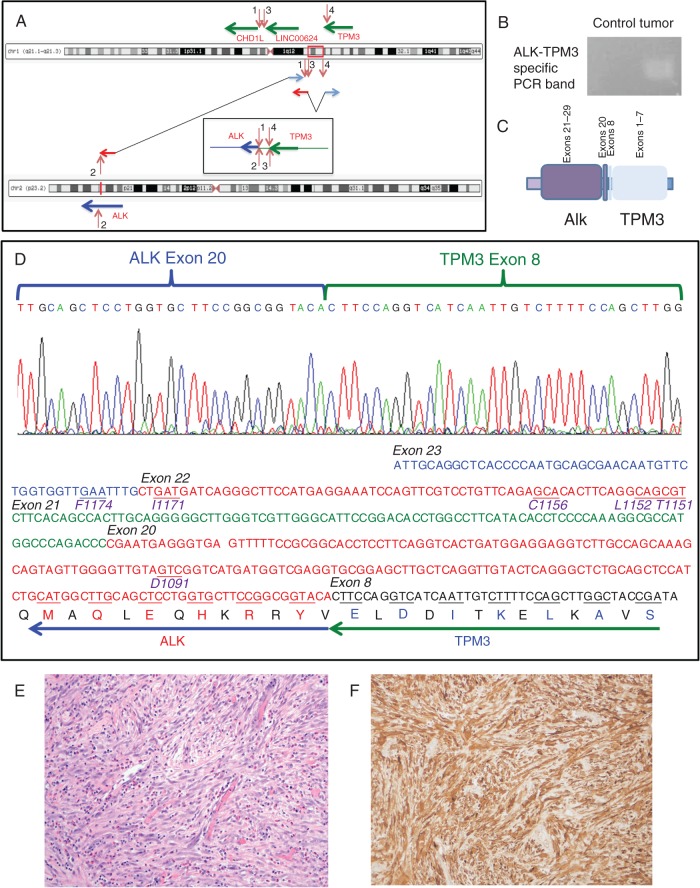
Figure 3.
Transcriptomic and immunohistochemical analysis of TPM3–ALK rearrangement. The chromoplectic rearrangement linking a potential fusion between TPM3 and ALK is depicted (A). This rearrangement was confirmed by rtPCR in cDNA generated from tumor RNA using forward and reverse primers of exonic sequences of ALK and TPM3 (B), indicating the predicted fusion of exon 8 of TPM3 to exon 20 of ALK (C). Sanger sequencing of the PCR product confirmed this as a functional, in-frame fusion transcript. Amino acid positions of previously reported resistance mutations in the ALK kinase domain covered in the cDNA PCR amplicon sequenced are indicated in purple below the underlined codons in the DNA sequence and were demonstrated to be wild-type in this patient. (D) Histopathologic examination of the tumor revealed myofibroproliferation with increased inflammatory infiltrates including many plasma cells and eosinophils, diagnostic of inflammatory myofibroblastic tumor (hematoxylin and eosin stain, 200× original magnification) (E). Immunohistochemical staining with ALK antibody (D5F3 clone, Cell Signaling Technology) demonstrated diffuse positivity in the myofibroblastic tumor cells (200× original magnification) (F).
additional potentially oncogenic chromoplectic genomic events
The complex chromoplectic event linking chromosomes 1, 2, 9 and 20 involved 201 (96%) of the 209 breakpoints detected in the patient's tumor. Of these events, 64 (32%) of them were interchromosomal translocations between the four chromosomes (supplementary Figure S3, available at Annals of Oncology online). Chromosomes 5, 6 and 7 were also linked by just three rearrangements to the chromoplectic event. Specifically, a breakpoint within the chromoplectic region of chromosome 1 translocated to chromosome 7 (7q32.2), with additional translocations linking 7q36.3 to 6q25.1 and 5q35.2 to 20q13.33. Of the 209 breakpoint junctions detected, 185 (89%) hit at least one gene directly, with 74 (34%) hitting two genes and 20 (10%) of these in the right orientation for potential fusion gene products (supplementary Figure S1B, available at Annals of Oncology online). A list of these additional potential fusion genes is presented in supplementary Table S3, available at Annals of Oncology online, although only a subset of these complex rearrangements may result in actively expressed fusion products.
The gene expression and function of numerous genes were predicted to be altered in this tumor, many with potential cancer driver functions, in addition to the TPM3–ALK fusion protein. In total, 142 genes were directly affected by genomic breakpoints. Of these genes, many have previously been linked to cancer progression, including ABL1, ESR1, ARNT, PAX3, PRRX1, RALGDS and LMNA, which are listed in the COSMIC census cancer driver genes database (http://cancer.sanger.ac.uk/census). Analysis of protein–protein interactions between the rearranged genes [10] revealed an extensive node of interacting proteins (supplementary Figure S4, available at Annals of Oncology online). A complex network of pathways were also potentially affected by somatic variation in these genes, with multiple-hit genes involved in MAPK (MAP4K3, DDR2, NTRK2), PI3K/AKT (RXRA, LAMC3, COL52A2, TNR) and ErbB (ABL1, SRC, TGFA) signaling, as well as cell cycle (ABL1, RBL1) and adhesion (SRC, CTNNA2, PRKCE, TJP2, LAMC3, COL52A2, TNR) pathways. More importantly, other potentially targetable genes were affected in the tumor (supplementary Table S4, available at Annals of Oncology online).
discussion
We present the first case of a patient with an ALK-rearranged IMT who responded to ceritinib after failure of crizotinib. We analyzed this patient's residual disease after a significant partial response and identified a complex pattern of chromosomal rearrangements, consistent with chromoplexy, which involved many additional oncogenes. The timing of our sample acquisition limits our conclusion; however, these data suggest that chromoplexy may be involved with the oncogenesis or progression of IMTs.
Approximately half of the patients with IMT have ALK rearrangements [1, 11, 12], but the rarity of this disease challenges successful completion of large, well-powered clinical trials. Although there is no standard of care for ALK-rearranged IMTs, reports of responses to crizotinib and extrapolations from the experience of treating ALK-rearranged non-small-cell lung cancers (NSCLC) suggest that ALK-targeted therapy is appropriate for this patient population. Very little is known about the mechanisms of resistance to crizotinib in IMT. Accordingly, there is little guidance on how to treat IMT after progression on crizotinib. In NSCLC, approximately one-third of patients who progress on the first-generation ALK inhibitor crizotinib develop mutations in the ALK kinase domain [13], whereas other patients develop amplification of ALK or activation of alternative signaling pathways [14]. In this patient, we only had sequencing base coverage of 3X, limiting our ability to identify a gatekeeper mutation in the ALK domain in the residual tissue specimen obtained at surgery. Instead, we identified a complex pattern of genetic rearrangements involving many oncogenes, consistent with chromoplexy. Chromoplexy was first coined in prostate cancer where a closed chain of multiple, balanced rearrangements associated with the loss-of-function of tumor suppressor genes and gain-of-function of oncogenic fusions was observed in the majority of analyzed specimens [15]. The identification of chromoplectic events in subclones suggested that multiple rounds of chromoplexy may occur, resulting in the ‘punctuated’ evolution of prostate cancer. In contrast, chromothripsis typically involves hundreds of genomic breakpoints on one or two chromosomes [16, 17], and kataegis involves several single-nucleotide mutations clustered in a single locus [18, 19]. Our results suggest that MPSeq is useful for identifying oncogenic rearrangements in patients with IMT, and provides additional insight into chromosomal aberrations that might be missed by standard sequencing approaches. MPSeq is not compatible with formalin-fixed paraffin-embedded specimens but can be used with touch-prep and frozen specimens. Similar to what is seen in prostate cancer, chromoplexy may have a role in the pathogenesis of IMT.
In this case of a young patient with metastatic IMT, with no known effective chemotherapeutic options, ceritinib was administered on a compassionate use basis, given the known association of IMTs with ALK rearrangements, detectable ALK expression by his tumor, his prior response to crizotinib and the reported responses to ceritinib after crizotinib in ALK-rearranged NSCLC [20]. In our analysis of the treatment-resistant, residual IMT, we identified a complex pattern of genetic rearrangements consistent with chromoplexy. Although it is difficult to know for certain if these chromoplectic rearrangements preceded treatment, their presence suggests that chromoplexy has a role in the oncogenesis of IMTs. Furthermore, this patient's remarkable response suggests that ceritinib should be considered as an option after progression on crizotinib for patients with metastatic or unresectable IMT and ALK mutations.
funding
This work was supported by National Institutes of Health (K12CA090628 to ASM) and Mayo Clinic's Center for Individualized Medicine's Biomarker Discovery Program.
disclosure
The authors have declared no conflicts of interest.
Supplementary Material
acknowledgements
The authors would like to acknowledge our patient, and Bobbi-Ann Jebens for her assistance preparing this manuscript.
references
- 1.Lovly CM, Gupta A, Lipson D et al. . Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov 2014; 4: 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butrynski JE, D'Adamo DR, Hornick JL et al. . Crizotinib in ALK-rearranged inflammatory myofibroblastic tumor. N Engl J Med 2010; 363: 1727–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy SJ, Aubry MC, Harris FR et al. . Identification of independent primary tumors and intrapulmonary metastases using DNA rearrangements in non-small-cell lung cancer. J Clin Oncol 2014; 32: 4050–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drucker TM, Johnson SH, Murphy SJ et al. . BIMA V3: an aligner customized for mate pair library sequencing. Bioinformatics 2014; 30: 1627–1629. [DOI] [PubMed] [Google Scholar]
- 5.Vasmatzis G, Johnson SH, Knudson RA et al. . Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood 2012; 120: 2280–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krzywinski M, Schein J, Birol I et al. . Circos: an information aesthetic for comparative genomics. Genome Res 2009; 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy SJ, Cheville JC, Zarei S et al. . Mate pair sequencing of whole-genome-amplified DNA following laser capture microdissection of prostate cancer. DNA Res 2012; 19: 395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamant L, Dastugue N, Pulford K et al. . A new fusion gene TPM3-ALK in anaplastic large cell lymphoma created by a (1;2)(q25;p23) translocation. Blood 1999; 93: 3088–3095. [PubMed] [Google Scholar]
- 9.Lawrence B, Perez-Atayde A, Hibbard MK et al. . TPM3-ALK and TPM4-ALK oncogenes in inflammatory myofibroblastic tumors. Am J Pathol 2000; 157: 377–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franceschini A, Szklarczyk D, Frankild S et al. . STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013; 41: D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffin CM, Patel A, Perkins S et al. . ALK1 and p80 expression and chromosomal rearrangements involving 2p23 in inflammatory myofibroblastic tumor. Mod Pathol 2001; 14: 569–576. [DOI] [PubMed] [Google Scholar]
- 12.Griffin CA, Hawkins AL, Dvorak C et al. . Recurrent involvement of 2p23 in inflammatory myofibroblastic tumors. Cancer Res 1999; 59: 2776–2780. [PubMed] [Google Scholar]
- 13.Gainor JF, Varghese AM, Ou SH et al. . ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clin Cancer Res 2013; 19: 4273–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katayama R, Lovly CM, Shaw AT. Therapeutic targeting of anaplastic lymphoma kinase in lung cancer: a paradigm for precision cancer medicine. Clin Cancer Res 2015; 21: 2227–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baca SC, Prandi D, Lawrence MS et al. . Punctuated evolution of prostate cancer genomes. Cell 2013; 153: 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen MM. Chromoplexy: a new category of complex rearrangements in the cancer genome. Cancer Cell 2013; 23: 567–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forment JV, Kaidi A, Jackson SP. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat Rev Cancer 2012; 12: 663–670. [DOI] [PubMed] [Google Scholar]
- 18.Nik-Zainal S, Alexandrov LB, Wedge DC et al. . Mutational processes molding the genomes of 21 breast cancers. Cell 2012; 149: 979–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding L, Wendl MC, McMichael JF, Raphael BJ. Expanding the computational toolbox for mining cancer genomes. Nat Rev Genet 2014; 15: 556–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friboulet L, Li N, Katayama R et al. . The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov 2014; 4: 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.