Abstract
We report on an Ebola virus disease (EVD) survivor who showed Ebola virus in seminal fluid 531 days after onset of disease. The persisting virus was sexually transmitted in February 2016, about 470 days after onset of symptoms, and caused a new cluster of EVD in Guinea and Liberia.
Keywords: Ebola virus, sexual transmission, seminal fluid, real-time sequencing, Ebola virus persistence
On 29 December 2015, the World Health Organization declared the end of Ebola virus (EBOV) transmission in the Republic of Guinea. On 16 March 2016, the health authorities of N'Zérékoré, Guinea, classified 3 community deaths that had occurred between 27 February and 15 March 2016 in the Koropara subprefecture as probable Ebola virus disease (EVD) cases (Figure 1A, cases 1–3) [1]. A national investigation team was deployed on 17 March in the region. Subsequently, several contacts of probable cases 1, 2, and 3—mostly family members in Koropara and Macenta prefecture—were diagnosed with EVD by EBOV real-time reverse-transcription polymerase chain reaction (RT-PCR) (Figure 1A, cases 4–10). In addition, the disease further spread to Liberia.
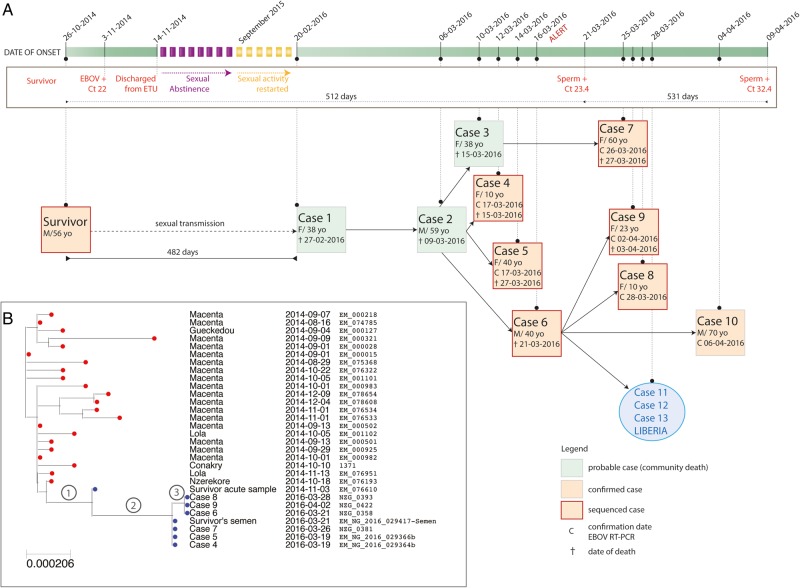
Figure 1.
Chains of transmission within the new Ebola virus disease cluster (A) and maximum likelihood phylogenetic analysis of the sequences from the new EVD cluster in historical and geographical context (B). A, The chart depicts all patients involved in the cluster and the likely chains of transmission as revealed by epidemiological investigation and virus sequencing. The timeline of events is shown on top. The blue circle depicts the 3 additional Liberian cases, which originated from case 6. B, The new sequences were aligned with 1066 historical Ebola virus genome sequences and a maximum likelihood phylogenetic tree was estimated under the general time reversible + Gamma model using RAxML [4]. Only the relevant part of the tree including the new sequences is shown. Abbreviations: +, positive Ebola virus reverse-transcription polymerase chain reaction; Ct, cycle threshold; EBOV, Ebola virus; ETU, Ebola treatment unit; RT-PCR, reverse-transcription polymerase chain reaction.
As of 29 April 2016, Guinea had recorded 3 probable and 7 confirmed cases, of which 6 were admitted at an Ebola treatment unit (ETU). Four patients died in the community and 4 patients in the ETU. Thus, the overall fatality ratio among probable and confirmed cases in this cluster in Guinea was 80% (8/10).
The National Committee of Ethics in Medical Research of Guinea approved the use of diagnostic leftover samples and corresponding patient data for this study (permits N°11/CNERS/14). All necessary consent required by applicable law from the patients whose information is included in the article was obtained in writing.
The long gap between the new cluster and previous cases in Guinea and the location of N'Zérékoré in the forest region of Guinea near the presumed source of the epidemic raised the possibility of a new spillover event from an animal reservoir. Alternative hypotheses included transmission from an EVD survivor or an undetected unbroken chain of human-to-human transmission. Virus genome sequencing can determine which of these scenarios are most likely. Therefore, blood samples from cases 4 and 5 were collected on 18 March and sent to the sequencing facility of the European Mobile Laboratory in Nongo, Conakry. Real-time sequencing was performed using MinION technology (Oxford Nanopore) as described by Quick et al [2], with results available on 20 March 2016. Validation sequencing was performed by the University of Makeni laboratory, Sierra Leone, on the Ion Torrent platform. A database of 1066 previously sequenced EBOV genomes was used to place the sequences into geographical and historical context. Viruses from cases 4 and 5 were genetically indistinguishable by both methods of sequencing. They were closely related to viruses from the 2014–2016 outbreak, ruling out a new introduction from an animal reservoir. Notably, the viruses did not belong to the GN1 or SL3 lineages that were circulating in Guinea in the second half of 2015 [2], but instead to a cluster of cases from N'Zérékoré and Macenta that was last detected in 2014 (Figure 1B; Supplementary Figure 1) [3, 4]. The most closely related sequence in the database (EM_076610; accession number KR817153) had been obtained from a male patient on admission to the ETU in Guéckédou in November 2014 (he survived) (Figure 1B) [3].
Independently and in parallel, epidemiological investigation identified the same survivor as the likely source of the new EVD cluster. On 19 March, the investigation revealed that case 1 had sexual intercourse with a male EVD survivor from the village of Koropara Centre (Figure 1A, “survivor”). He had been treated in the ETU of Guéckédou from 3 to 14 November 2014. On admission, the cycle threshold (Ct) value in the RealStar Filovirus Screen RT-PCR (Altona Diagnostics) was 22 (the sequence of that sample [EM_076610; accession number KR817153] was available in the database). He reported sexual abstinence from discharge in November 2014 through August 2015 as recommended by current protocols. From September 2015, he had occasional sexual intercourse with various partners, including case 1 at the end of January 2016. Except for episodes of asthenia, he reported no post-EVD complications. In March 2016, a rapid diagnostic test for human immunodeficiency virus types 1 and 2 (Alere Determine HIV-1/2) was negative. A seminal fluid sample obtained on 21 March was positive for EBOV with a Ct value of 23.4 in the RealStar Zaire Ebolavirus RT-PCR (Altona Diagnostics). On 9 April 2016, seminal fluid still tested positive with a Ct value of 32.4.
EBOV in seminal fluid from 21 March was sequenced in Guinea and in Sierra Leone as described above. On 23 March, the results revealed that the virus was identical to that of acute cases 4 and 5 (Figure 1B), supporting the epidemiological evidence of sexual transmission. Additional sequencing and phylogenetic analysis of 7 acute cases confirmed that the entire cluster is linked with the survivor. The sequences are either identical to or only differ by up to 1 nucleotide from that of the survivor's semen (Figure 1B; Supplementary Table 1).
The PCR and sequencing data strongly suggest that EBOV persisted in the survivor from 26 October 2014 (date of onset) to 9 April 2016 (last EBOV RNA–positive semen sample)—that is, for 531 days. Remarkably, the virus in blood from 3 November 2014 differed from that in semen from 21 March 2016 by just 5 novel mutations (Supplementary Table 1) despite being collected 504 days prior to the collection of the semen sample. This equates to an evolutionary rate of 0.19 × 10−3 substitutions per site per year, roughly 6 times slower than the average evolutionary rate seen during human-to-human transmission in this outbreak with reported mutation rates of 1.19 × 10−3 and 1.42 × 10−3, respectively (Supplementary Figure 2) [2, 3, 5]. The predicted effects on proteins encoded by the EBOV genome are shown in Supplementary Table 1. Notably, one mutation is predicted to encode a premature stop codon in the VP30 gene, truncating the expected gene product by 7 amino acids.
We report on an EVD survivor in whom EBOV RNA was detected in seminal fluid 531 days after onset of disease, the longest reported period of filovirus persistence in an individual. Furthermore, we provide evidence that the persisting virus remains viable and infectious during the long-term persistence. Evidence for sexual transmission of the persisting EBOV in February 2016, about 470 days after onset of symptoms in the survivor, is compelling. The absence of other cases in Guinea around this time, the epidemiological link between the survivor and case 1, and the identity of strains in the survivor's seminal fluid and the acute cases strongly argue for the survivor being the source of the new infections. EBOV persistence in seminal fluid is a known phenomenon, and previous reports have also indicated that persisting virus may be sexually transmitted [6–8]. However, so far the longest reported period of virus RNA persistence in semen was 284 days [6], and reported likely transmission events have been observed a maximum of 179 days after onset of acute illness [7].
The reduced evolutionary rate of the virus during persistence is plausible, given the presumed low level of replication in immune-privileged sites such as the testes in the absence of detectable viremia. The few mutations that accumulated during the 500 days are of interest and deserve further functional investigation. Nevertheless, the data presented here indicate that this strain retained its pathogenic properties during persistence, as evidenced by transmissibility, replication capacity (Ct values around 20 in the acute cases of the cluster), and fatality ratio of 80%. We cannot rule out the possibility that the survivor was silently reinfected with Ebola after recovery. However this seems unlikely as there have been no laboratory-confirmed cases of Ebola virus reinfection [9]. Additionally, no cases of EVD had been detected in the N'Zérékoré region since January 2015 until the cluster of cases reported here. In the event of reinfection, the lower-than-expected number of mutations seen would make it unlikely that this case was the result of an undetected continuation of a chain of human-to-human transmission among acute cases.
The case is also a great example demonstrating the yield of enhanced surveillance activities in the affected countries, cross-border cooperation, and improved capabilities to rapidly diagnose and manage EVD patients. In addition, the report underlines the value for outbreak response of combining field epidemiology with real-time molecular epidemiology as well as the availability of an up-to-date virus genome repository for the outbreak.
Supplementary Data
Supplementary materials are available at http://cid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank the field teams and the Guinean health authorities for their commitment and excellent cooperation. The EMLab is a technical partner of the World Health Organization (WHO) Emerging and Dangerous Pathogens Laboratory Network and the Global Outbreak Alert and Response Network (GOARN) and the deployments in West Africa have been coordinated and supported by the GOARN Operational Support Team at WHO headquarters.
Author contributions. B. D., D. S., H. B., M. C. W., I. S. C., R. S., S. F., A. L., J. K. K., N. A. E., and B. A. D. performed epidemiological investigations and patient management. H. A. B., S. H., N. M., and S. D. performed laboratory testing. N. J. L., I. G., L. W. M., M. C., U. J., R. E. G. W., and S. D. performed sequence analysis. A. R. performed phylogenetic analysis. P. R., D. M., X. A., M. W. C., R. B. A., M. H. D., A. D., P. F., S. K., S. G., and S. D. coordinated case management, laboratory investigations, and fieldwork. B. D., D. S., N. J. L., S. G., A. R., and S. D. wrote the manuscript. All authors reviewed the final draft. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention. The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Financial support. This work was carried out in the context of the projects EVIDENT (Ebola virus disease: correlates of protection, determinants of outcome, and clinical management) and REACTION! that received funding from the European Union's Horizon 2020 research and innovation program under grant agreements 666100 (to S. G.) and 666092 (to D. M.), respectively, and in the context of service contract IFS/2011/272-372 (to S. G.) funded by the Directorate-General for International Cooperation and Development. The study was further supported by grant C15-101 from the French Institute of Health and Medical Research (INSERM) to D. S. Work at the University of Makeni was supported by funding from the Wellcome Trust to I. G. (097997/Z/11/A).
Potential conflicts of interest. N. J. L. is on the speakers’ bureau of and has received travel fees from Oxford Nanopore Technologies. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization Ebola Response Team. Ebola virus disease in West Africa—the first 9 months of the epidemic and forward projections. N Engl J Med 2014; 371:1481–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quick J, Loman NJ, Duraffour S et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 2016; 530:228–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carroll MW, Matthews DA, Hiscox JA et al. Temporal and spatial analysis of the 2014–2015 Ebola virus outbreak in West Africa. Nature 2015; 524:97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evolution 2016; doi:10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deen GF, Knust B, Broutet N et al. Ebola RNA persistence in semen of Ebola virus disease survivors—preliminary report. N Engl J Med 2015; doi:10.1056/NEJMoa1511410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mate SE, Kugelman JR, Nyenswah TG et al. Molecular evidence of sexual transmission of Ebola virus. N Engl J Med 2015; 373:2448–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez LL, De Roo A, Guimard Y et al. Persistence and genetic stability of Ebola virus during the outbreak in Kikwit, Democratic Republic of the Congo, 1995. J Infect Dis 1999; 179(suppl 1):S170–6. [DOI] [PubMed] [Google Scholar]
- 9.MacIntyre CR, Chughtai AA. Recurrence and reinfection—a new paradigm for the management of Ebola virus disease. Int J Infect Dis 2016; 43:58–61. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.