Activation of a wheat gene product by a fungal protein leads to cell death in the plant, allowing the pathogen to cause disease.
Keywords: Wheat, fungal disease, pathogens, necrotrophic pathogens, plants, receptor kinase
Abstract
Necrotrophic pathogens live and feed on dying tissue, but their interactions with plants are not well understood compared to biotrophic pathogens. The wheat Snn1 gene confers susceptibility to strains of the necrotrophic pathogen Parastagonospora nodorum that produce the SnTox1 protein. We report the positional cloning of Snn1, a member of the wall-associated kinase class of receptors, which are known to drive pathways for biotrophic pathogen resistance. Recognition of SnTox1 by Snn1 activates programmed cell death, which allows this necrotroph to gain nutrients and sporulate. These results demonstrate that necrotrophic pathogens such as P. nodorum hijack host molecular pathways that are typically involved in resistance to biotrophic pathogens, revealing the complex nature of susceptibility and resistance in necrotrophic and biotrophic pathogen interactions with plants.
INTRODUCTION
Plants have evolved several means of protection against biotrophic pathogens, which include bacteria, viruses, nematodes, insects, and fungi with biotrophic lifestyles. One is the detection of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) (1). PRRs are cell surface receptors in the host, and PAMPs are usually conserved pathogen molecules that serve essential functions. Recognition leads to PAMP-triggered immunity (PTI), which results from the activation of a host response involving an oxidative burst, activation of mitogen-associated and calcium-dependent kinases, transcriptional activation of defense response genes, and, in some cases, localized cell death (2). Pathogens have evolved the ability to evade PTI by secreting effectors, many of which suppress PTI-mediated defense responses. Accordingly, a second layer of immunity termed effector-triggered immunity (ETI) involves the recognition of effectors by products of effector-specific resistance genes, the most common being the so-called nucleotide-binding and leucine-rich repeat domain proteins (NB-LRR class). ETI responses overlap with some of the same responses as PTI, with both leading to the restriction of biotrophic growth (3).
Less is known about how plants combat diseases caused by necrotrophic fungal pathogens, which, as opposed to biotrophs, gain nutrients from dead or dying tissue. In the past, necrotrophs were considered generalists that kill their hosts by secreting a barrage of cell wall–degrading enzymes and/or toxins. However, recent studies have revealed that some peptide toxins of necrotrophic pathogens are necrotrophic effectors (NEs) that, when recognized by the host, induce cell death, allowing the pathogen to feed and ultimately sporulate (4, 5). Molecular cloning of several plant NE sensitivity genes revealed that they harbor NB and LRR domains, features of classic disease “resistance” genes involved in ETI (5–7). Additional studies of the host response upon recognition of the Parastagonospora nodorum–produced NE known as SnToxA by the wheat gene Tsn1 (which contains NB and LRR domains) revealed hallmarks of an ETI response (5, 8, 9). However, because the pathogen is a necrotroph, the end result was susceptibility as opposed to resistance. Therefore, this work demonstrated that the pathogen acquired the ability to hijack the host’s own ETI pathway, resulting in NE-triggered susceptibility (NETS) (10).
Common wheat (Triticum aestivum) is grown worldwide and provides about 20% of the calories consumed by humans. Diseases and pests pose a serious threat to wheat production and can threaten the global supply. P. nodorum is an important necrotrophic fungal pathogen of wheat that infects the leaves and glumes, causing the disease Septoria nodorum blotch (SNB), which can result in substantial yield losses and reduced grain quality. Compatible wheat–P. nodorum interactions rely on the recognition of pathogen-produced NEs by specific host genes. A typical biotroph-induced “defense” response and cell death followed by susceptibility occur upon activation of the wheat Snn1 gene product by the P. nodorum–produced protein SnTox1 (11). Here, we cloned and characterized Snn1 to gain a better understanding of the mechanisms of susceptibility operating in plant–necrotrophic pathogen interactions.
RESULTS
Map-based isolation and identification of an Snn1 candidate gene
The Snn1 gene lies in a region on the short arm of wheat chromosome 1B (Fig. 1A) for which we previously conducted saturation and fine mapping (12). For high-resolution mapping, we developed a population consisting of 17,000 gametes (8500 F2 plants) derived from a cross between the common wheat landrace Chinese Spring (CS) and CS that had a pair of 1B chromosomes from the common wheat variety “Hope” (CS-Hope 1B) substituted for the native pair of 1B chromosomes. The molecular markers Xpsp3000, Xfcp618, Xfcp619, and Xfcp624 (table S1) were used to genotype the F2 population, and phenotypic assessment of the population was conducted by infiltrating the plants with cultures containing the P. nodorum NE SnTox1 and by scoring them as sensitive or insensitive based on the presence or absence of necrosis, respectively. The resulting genetic linkage map spanned 2.73 centiMorgans (cM) with markers Xfcp618 and Xfcp624 flanking Snn1 at distances of 1.9 and 0.16 cM, respectively (Fig. 1B). Screening of the chromosome 1BS bacterial artificial chromosome (BAC)–based minimum tiling path (MTP) clones (13) revealed an approximately 2.5-Mb contig consisting of 44 clones containing markers Xfcp618, Xfcp619, and Xfcp624 and thus encompassing Snn1 (Fig. 1C). Markers derived from BAC end sequences (BESs), or corresponding whole-genome survey sequences (table S1) (14), were used to develop and anchor additional markers to the genetic map. This led to the delineation of Snn1 to a segment of 0.16 cM spanned by four BAC clones (Fig. 1C).
Fig. 1. Map-based cloning of the Snn1 gene.
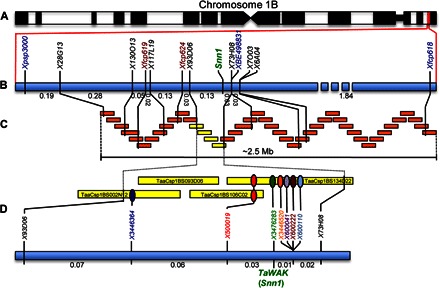
(A) The genomic region containing the Snn1 gene on the short arm of chromosome 1B is shown in red. (B) The genetic linkage map of the Snn1 region. Markers in blue are from Reddy et al. (12), markers in red are from previous unpublished work, and markers in black were developed in this research. (C) BAC-based physical map of the Snn1 region anchored to the genetic linkage map. The four BACs in yellow represent the Snn1 candidate gene region. (D) Genetic linkage mapping of the seven candidate genes identified in the four BACs from the candidate gene region in (C). The green oval represents TaWAK, which cosegregated with Snn1.
Sequencing and bioinformatic analyses of the four BAC clones revealed the presence of seven putative genes (Fig. 1D). The placement of markers representing each of the candidate genes (table S2) onto the linkage map showed that only one of them cosegregated with Snn1, and the other six candidates were separated from Snn1 by recombination events (Fig. 1D).
Cloning and sequencing of the full-length complementary DNA (cDNA) and subsequent analysis of the deduced amino acid sequence of the candidate gene indicated that it was a member of the wall-associated kinase (WAK) class of plant receptor kinases. The gene, hereafter referred to as TaWAK, has 3045 base pairs (bp) from the start codon to the stop codon with three exons and a coding sequence of 2145 bp with 5′ and 3′ untranslated regions (UTRs) of 164 and 102 bp, respectively (Fig. 2A). The deduced amino acid sequence indicated that the protein contains conserved wall-associated receptor kinase galacturonan binding (GUB_WAK), epidermal growth factor–calcium binding (EGF_CA), transmembrane, and serine/threonine protein kinase (S/TPK) domains (Fig. 2A), with the S/TPK domain predicted to be intracellular and the GUB_WAK and EGF_CA binding domains predicted to be extracellular. Alignment and phylogenetic analysis of TaWAK together with WAK genes from other dicot and monocot members of the plant kingdom indicated that it is a member of a diverse group of monocot WAK genes, and the GUB_WAK domain contains a 22–amino acid insertion specific to monocots (figs. S1 and S2).
Fig. 2. Functional validation of the TaWAK gene by mutagenesis and transgenesis.
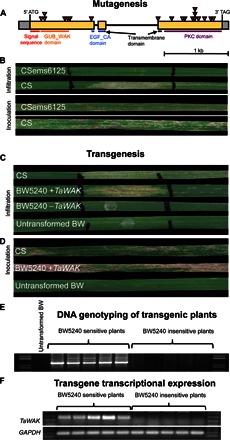
(A) Gene structure of the TaWAK (Snn1) gene with exons in yellow and UTRs in gray. Red arrowheads indicate mutation sites in EMS-induced mutants, all of which were insensitive to SnTox1. PKC, protein kinase C. (B) Infiltration and inoculation reactions on leaves of wild-type CS and the EMS mutant CSems6125 are shown as an example. (C) Reactions to SnTox1 infiltrations of CS (Snn1+), untransformed Bobwhite (BW; Snn1−), and sensitive and insensitive T1 transgenic plants both derived from the same event (BW5240). (D) Transgenic plants that were sensitive to SnTox1 were also susceptible to disease caused by spores of an SnTox1-producing fungal isolate. (E) All SnTox1-sensitive T1 plants derived from event BW5240 had the TaWAK transgene, and all insensitive plants lacked the transgene. (F) Similarly, all SnTox1-sensitive BW5240 T1 plants expressed TaWAK, whereas the insensitive plants did not.
Validation of Snn1 by mutagenesis and transgenesis
We treated seeds of the SnTox1-sensitive wheat line CS with the chemical mutagen ethylmethane sulfonate (EMS), and upon infiltrating ~2000 M2 families with SnTox1, we identified 16 independent SnTox1-insensitive mutants (Fig. 2A, fig. S3, and table S3). Comparative sequence analysis of TaWAK from each of the SnTox1-insensitive mutants with TaWAK from CS indicated that 14 of the mutant lines harbored missense mutations, 3 of which harbored two mutations each (table S3). These mutations occurred in each of the conserved GUB_WAK, EGF_CA, and S/TPK domains, indicating that all three domains are essential to confer SnTox1 sensitivity. In addition to the 14 mutant lines with missense mutations, one SnTox1-insensitive line had a nonsense mutation inducing a stop codon in the first exon, and another was found to have a point mutation in the splice acceptor site of the second intron, which led to the relocation of the splice site and a cDNA fragment that was 60 bp shorter than that in CS (fig. S4). The SnTox1-insensitive mutants were also resistant to SNB caused by an SnTox1-producing isolate of P. nodorum (Fig. 2B).
To further validate the TaWAK gene as Snn1, we transformed the SnTox1-insensitive wheat genotype BW with the full-length TaWAK cDNA driven by the maize ubiquitin promoter. Analysis of the T1 generation revealed families segregating for sensitivity to SnTox1 (Fig. 2C). SnTox1-sensitive transgenic plants had and transcribed the TaWAK transgene, whereas the SnTox1-insensitive plants lacked the gene (Fig. 2, C to F). Furthermore, the TaWAK-possessing transgenic plants were also susceptible to SNB (Fig. 2D).
The mutagenesis and transgenesis experiments together demonstrated that the TaWAK gene was both sufficient and necessary to confer sensitivity to SnTox1 and susceptibility to SnTox1-producing isolates of P. nodorum. Therefore, this provided conclusive evidence that TaWAK was indeed the Snn1 gene.
Snn1 comparative and diversity analysis
DNA gel blot analysis was conducted on the set of CS nullisomic-tetrasomic lines with probe FCG36 derived from a region encompassing the 5′ end of the coding region and the 5′UTR to determine the copy number of Snn1 (fig. S5). Absence of the hybridizing fragment in the nullisomic 1B–tetrasomic 1D line indicated that the gene is located on chromosome 1B as expected, but the lack of hybridizing fragments from chromosomes 1A and 1D suggested that homoeoalleles of Snn1 do not exist.
Bioinformatic analysis of wheat sequences (14) revealed that the homoeologous chromosome arm 1AS contained no significant matches, but 1DS harbored a gene with 91% nucleotide identity across the entire gene with the exception of the first 800 bp, which had no similarity (fig. S6). Evaluation of CS mRNA indicated that the 1D copy is not transcribed and therefore not functional (Fig. 3A). At the amino acid level, matches to predicted proteins from Triticum urartu and Aegilops tauschii, the diploid ancestral donors of the A and D genomes of polyploid wheat, respectively, were identified (table S4 and fig. S2), but the level of identity (70%) suggested that they are not orthologs.
Fig. 3. Transcriptional expression of Snn1.
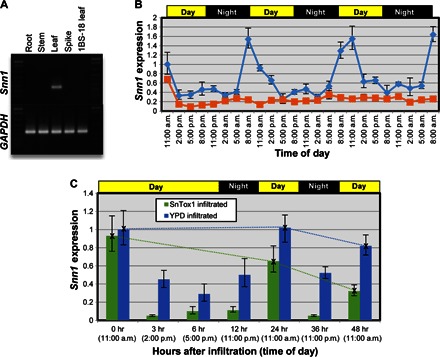
(A) Snn1 expression survey in CS by reverse transcription polymerase chain reaction (RT-PCR) with GAPDH as an endogenous control. The absence of an amplicon in CS 1BS-18 indicates that Snn1 transcription is unique to 1B. (B) Snn1 expression levels in 2-week-old plants entrained with a 12-hour light/dark cycle evaluated every 3 hours over a 72-hour period (blue) and in plants subjected to continuous darkness for the same time points (orange) using relative quantitative PCR (RQ-PCR). (C) RQ-PCR evaluation of Snn1 expression in SnTox1-challenged plants (green bars) and YPD medium–infiltrated plants (blue bars). The dotted lines are shown to compare expression of Snn1 at 11:00 a.m. over 3 days. Note that the expression of Snn1 in the SnTox1-infiltrated plants becomes reduced over time compared to the control.
We evaluated 826 accessions of wheat and its ancestral species for the presence of Snn1 alleles and for sensitivity to SnTox1 (table S5). This included 123 accessions of Aegilops speltoides, the closest living relative of the diploid B genome progenitor of polyploid wheat. On the basis of DNA analysis, many accessions were positive for harboring an allele of Snn1, but none of the primitive diploid (Ae. speltoides) or tetraploid wild emmer (Triticum turgidum ssp. dicoccoides) accessions were sensitive to SnTox1. However, three accessions of tetraploid T. turgidum ssp. dicoccum were sensitive, along with 73 and 16% of domesticated durum and common wheat varieties, respectively.
Sequencing and phylogenetic analysis of Snn1 from 24 accessions including 12 SnTox1-insensitive and 12 SnTox1-sensitive lines (fig. S7) indicated that Snn1 alleles from Ae. speltoides and wild emmer (T. turgidum ssp. dicoccoides) were more divergent relative to cultivated emmer (T. turgidum ssp. dicoccum), durum (T. turgidum ssp. durum), and common wheat (T. aestivum ssp. aestivum), which agrees with what is known about the evolutionary history of wheat and its relatives (15). This analysis also revealed two single-nucleotide polymorphisms that caused nonsynonymous mutations at amino acid positions 347 and 429 and gave rise to SnTox1-insensitive snn1 alleles (figs. S3 and S7). The mutation at position 347 caused a Gly-to-Arg amino acid substitution within the EGF_CA binding domain, and the mutation at position 429 caused a Lys-to-Asn substitution between the S/TPK and transmembrane domains.
The durum variety Lebsock was insensitive to SnTox1, but DNA sequence analysis indicated that the Snn1 sequence of Lebsock was identical to several other SnTox1-sensitive genotypes (fig. S7). Transcriptional analysis revealed that the gene was not expressed, thus providing an explanation for the lack of SnTox1 sensitivity in Lebsock (fig. S8).
Transcriptional regulation of Snn1
Evaluation of Snn1 transcriptional expression in different plant tissues of CS indicated that it is transcribed in the leaves, but not in the roots, stems, or spikes (Fig. 3A). Evaluation of Snn1 transcription levels in leaves under a 12-hour light/dark regime every 3 hours for a 72-hour time period indicated that Snn1 transcription levels decrease throughout the daylight hours and increase throughout the subjective nighttime hours, peaking at subjective dawn (Fig. 3B). Evaluation of Snn1 transcription levels at the same time points of plants placed under continuous darkness since the first time point indicated that expression levels were reduced during the hours of the first day and then remained low through the remainder of the 72-hour evaluation period.
To determine whether SnTox1 affects Snn1 transcription levels, we evaluated expression in plant leaves infiltrated with SnTox1 and included leaves infiltrated with yeast potato dextrose (YPD) medium as controls (Fig. 3C). Expression levels of Snn1 transcript in the medium-infiltrated leaves agreed with what was observed in the previous experiments (Fig. 3B). However, Snn1 expression in SnTox1-infiltrated plants increased to about 62% of the level of the control plants at 24 hours after infiltration and to less than half the level of control plants at 48 hours. This indicated that Snn1 is not induced or up-regulated by SnTox1 but is, instead, down-regulated over time.
Snn1-SnTox1 induction of mitogen-activated protein kinase genes
The induction of mitogen-activated protein kinase (MAPK) genes MAPK3 and MAPK6 leading to transcriptional reprogramming is a hallmark of a defense response to a biotroph (16). The expression levels of the wheat MAPK genes TaMAKP3 and TaMAPK6 were evaluated in 2-week-old plants inoculated with the SnTox1 protein. Transcription of TaMAPK3, but not of TaMAPK6, was up-regulated within 15 min after SnTox1 treatment in a compatible Snn1-SnTox1 interaction, and activation was transient, returning to the initial levels by 3 hours after inoculation (fig. S9).
Yeast two-hybrid analysis of Snn1-SnTox1 interactions
Related research has shown that SnTox1 does not enter the plant cell upon fungal infection but remains extracellular (17). Here, we used yeast two-hybrid (Y2H) analysis of the extracellular fragments of the Snn1 protein to determine whether the Snn1 and SnTox1 proteins interacted directly in yeast. Snn1 and SnTox1 were expressed in yeast as the prey and the bait proteins, respectively, and the Snn1 prey protein was expressed as full-length (minus signal peptide) or partial (with specific deletion) proteins as illustrated in fig. S10. The results indicated that the SnTox1 protein likely interacts directly with Snn1 within a region of 140–amino acid residues between the extracellular GUB_WAK and EGF_CA domains.
DISCUSSION
Here, we report the cloning and characterization of the wheat gene Snn1, which confers susceptibility to the disease SNB caused by SnTox1-producing strains of P. nodorum. Snn1 is a member of the WAK family of receptor kinases. Plant WAK proteins have been implicated in cell expansion, biotrophic pathogen resistance, and general perception of the extracellular environment (18). WAKs are known to serve as PRRs for pectin fragments known as oligogalacturonides (OGs) (19), which act as damage-associated molecular patterns released by the plant cell upon pathogen attack or wounding. Recognition of OGs by the WAK PRRs is followed by the activation of a classic biotrophic defense response (20). Recent research has implicated WAK genes in conferring resistance to the maize diseases head smut, caused by the soil-borne biotrophic fungus Sporisorium reilianum (21), and northern corn leaf blight, caused by the hemibiotrophic fungus Exserohilum turcicum (22). The mechanisms of pathogen detection for these WAK resistance genes are unknown. Perhaps they perceive OGs generated by pathogen-induced perturbations in the cell wall or perhaps they directly interact with pathogen-produced effectors, as appears to be the case for Snn1. Nevertheless, the maize WAK genes operate to confer resistance through recognition and activation of a defense response to limit growth of the pathogen. Snn1 induces a similar response upon activation. However, the end result is susceptibility as opposed to resistance.
There are currently three known examples of plant NB-LRRs conferring sensitivity to necrotrophic pathogen NEs leading to susceptibility (5–7), which can be considered as examples of the pathogen hijacking the ETI pathway, leading to sporulation. Snn1 is the first case of a receptor kinase–driven pathway being hijacked by a pathogen and resulting in disease. Receptor kinases are usually PRRs that drive PTI pathways (2). Although the ETI and PTI transcriptomes may significantly overlap, there are some common differences, such as activation patterns of MAPK genes. Studies indicate that MAPK activation in PTI occurs very early and is transient, whereas it is more prolonged in ETI (16). Activation of TaMAPK3 upon the recognition of SnTox1 by Snn1 agrees with the former in that it was activated within 15 min of inoculation and lasted only a short time.
The results of this study indicate that the SnTox1 and Snn1 proteins interact directly, at least in yeast. This is perhaps not surprising, given that Snn1 spans the plasma membrane and contains extracellular binding domains and that SnTox1 does not enter the plant cell (17). This is in contrast to the other well-characterized wheat–P. nodorum interaction involving the NB-LRR gene Tsn1 and the P. nodorum–produced NE SnToxA, which do not interact directly in yeast (5). In this case, Tsn1 is an intracellular protein perhaps operating as a guard for another membrane-spanning protein, and SnToxA is likely internalized within the plant cell in Tsn1-containing lines (23).
Together, these findings provide evidence that Snn1 and Tsn1 activate PTI and ETI pathways upon recognition of SnTox1 and SnToxA, respectively (Fig. 4). Therefore, P. nodorum (and likely other necrotrophic pathogens) has acquired the ability to essentially “trick” the plant into inducing a response that the plant has evolved to defend itself against biotrophic pathogens. The response includes cell death, which allows this pathogen—being a necrotroph—to feed, proliferate, and sporulate. Plants carrying both Snn1 and Tsn1 experience significantly higher levels of disease than plants having only one of the genes (Fig. 4) (24). Therefore, the secretion of NEs that result in the hijacking of both PTI and ETI to induce NETS benefits the pathogen in terms of survival and propagation.
Fig. 4. Overview of the Snn1-SnTox1 and Tsn1-SnToxA interactions and known downstream events that result in NETS in the wheat–P. nodorum pathosystem.
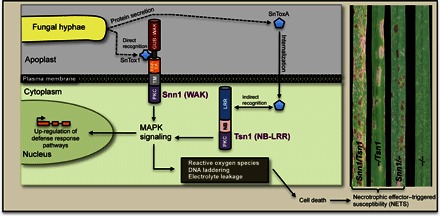
The SnTox1 and SnToxA proteins are secreted by the fungus. SnToxA is internalized into the cell (23), but SnTox1 is not (11). Upon recognition of SnTox1 and SnToxA by the Snn1 and Tsn1 proteins, respectively, signaling leading to up-regulation of defense response pathways and events resulting in programmed cell death (9, 11) ultimately provide a means for the pathogen to gain nutrients and reproduce. Plants with either Tsn1 or Snn1 are susceptible, and plants with both genes experience even higher levels of disease (24). Elimination of both genes renders the plant resistant.
Common wheat (T. aestivum) is a disomic allohexaploid (2n = 6x = 42; AABBDD genomes), which arose through the convergence of three diploid ancestors [T. urartu, 2n = 2x = 14 (AA genome); an extinct close relative of Ae. speltoides, 2n = 2x = 14 (SS genome), which donated the B genome; and Ae. tauschii, 2n = 2x = 14 (DD genome)] through two separate hybridization events (15). The first event involved the hybridization of the diploid A and B genome progenitors about half a million years ago to form the first tetraploid wheat, which was wild emmer (T. turgidum ssp. dicoccoides) with AABB genome constitution. Several mutations in wild emmer that made it more amenable to harvest by early farmers led to the formation of cultivated emmer wheat (T. turgidum ssp. dicoccum), which was later fully domesticated in the form of durum (macaroni) wheat (T. turgidum ssp. durum). Hexaploid common (bread) wheat (T. aestivum) arose under cultivation as a result of hybridization between a T. turgidum AB tetraploid and Ae. tauschii. Here, we found Snn1 alleles to exist at all three ploidy levels of the B genome lineage. However, Snn1 alleles that conferred SnTox1 sensitivity were not identified in the diploid (Ae. speltoides) or wild emmer (T. turgidum ssp. dicoccoides) accessions. A few accessions of cultivated emmer were sensitive to SnTox1, which suggests that the first functional Snn1 alleles in terms of having the ability to recognize SnTox1 originated in cultivated emmer wheat and were then passed from there to our modern durum and bread wheat varieties.
Note the disparity in the fraction of durum varieties (73%) that were sensitive to SnTox1 compared to that of common wheat varieties (16%). There are two possible explanations for this. First, it is possible that a functional Snn1 allele was not present in the AB tetraploid involved in the hybridization event of Ae. tauschii to give rise to the first ABD hexaploid and that the few hexaploid genotypes that do have a functional Snn1 allele acquired it through secondary hybridization events leading to gene flow. The second possibility is that the AB tetraploid giving rise to the hexaploid did have a functional Snn1 allele and passed it to early hexaploids, but the allele was eliminated from most current wheat varieties through natural or artificial selection. The second scenario would suggest that a functional Snn1 may serve a beneficial purpose in a tetraploid background, thus causing it to be retained in most varieties, whereas it may not contribute the same benefit in the hexaploid background, possibly because of additional redundancy conferred by the presence of the D genome, and therefore has been lost from most common wheat varieties.
Evaluation of Snn1 transcription showed that it exhibited a diurnal rhythm tightly regulated by light signals. Although light signals serve as primary cues for circadian clock entrainment, Snn1 is not circadian-controlled because the rhythmic oscillations dampened rapidly upon transfer of plants to continuous darkness. Although Tsn1 is a member of the NB-LRR class of genes as opposed to a WAK, it is regulated in a manner very similar to Snn1 (5). Because both Tsn1 and Snn1 belong to classes of genes that typically confer resistance to biotrophs, their peak expression at subjective dawn may be due to mechanisms that evolved to allow the plant to “anticipate” pathogen infection, which often occurs at that time (25). Another study has demonstrated that plant defense responses are influenced by light and circadian rhythms (26). It would be interesting to study the effects of light and the circadian clock on genes that confer resistance to biotrophs and hemibiotrophs as well as other necrotrophs.
The results of this work, together with previous studies, provide strong evidence that common signaling pathways associated with biotroph resistance, that is, PTI and ETI, are hijacked by necrotrophs, leading to susceptibility. Therefore, whether pathway induction resulting in cell death leads to resistance or susceptibility depends on the invading pathogen, because host cell death is detrimental or lethal to biotrophs, but highly beneficial to necrotrophs. Given this model, it should be possible that a single gene product could simultaneously confer resistance to a biotroph and susceptibility to a necrotroph, which would obviously pose a challenge for plant breeders. Indeed, the Victoria blight susceptibility/crown rust resistance locus in oat appears to fit this dual-function description (6). Nevertheless, breeders should embrace a paradigm shift; that is, disease susceptibility genes that serve no beneficial purpose should be selected against and removed from germplasm, and genes that condition resistance to biotrophs should be retained and/or introgressed, all while being diligently aware of the possibility of necrotrophic hijacking.
MATERIALS AND METHODS
Plant materials
A high-resolution mapping population was developed from a cross between CS and CS-Hope 1B. The population consisted of 8500 F2 plants (17,000 gametes) and was used to map Snn1 and anchor the BAC contig to the genetic linkage map. CS was also used for mutagenesis to generate CSems lines. The spring wheat variety BW was used for transformation experiments. A total of 826 tetraploid and hexaploid Triticum accessions and 123 Ae. speltoides accessions were used to determine the prevalence of Snn1 alleles (table S5).
SnTox1 culture production, infiltrations, and inoculations
SnTox1 cultures were obtained from SnTox1-expressing yeast cultures as previously described (11). SnTox1 culture filtrates were infiltrated into fully expanded secondary leaves of wheat plants using a 1-ml syringe with the needle removed. Immediately after infiltration, the boundaries of the infiltrated sites were marked with a nontoxic felt pen. Reactions were evaluated 3 to 5 days after infiltration and scored as either insensitive (no necrosis) or sensitive (necrosis).
Conidial spores of the SnTox1-producing strain Sn2000 were inoculated on CS, CSems lines, and transgenic lines as previously described (11). The inoculum was prepared by diluting the spore suspensions to 1,000,000 spores/ml and adding two drops of Tween 20 per 100 ml of inoculum. After applying conidial suspensions (via air-spray) to plants at the two-leaf stage until runoff, plants were then placed in a mist chamber at 100% relative humidity at 21°C for 24 hours, followed by 6 days of incubation in the growth chamber under a 12-hour photoperiod. Disease reactions were evaluated 7 days after inoculation.
Spray applications of SnTox1 were carried out by diluting the SnTox1 yeast culture five times with distilled water and adding two drops of Tween 20 per 100 ml of solution. Spray inoculation was performed by air-spray on 2-week-old plants until runoff. Then, plants were kept in the growth chamber under a 12-hour photoperiod and sampled at specific time points as outlined below.
Genetic linkage and physical mapping
The population of 8500 F2 plants was screened with the PCR-based markers Xpsp3000 and Xfcp618, which were previously shown to delineate Snn1 to a 0.9-cM interval in a different population (12). The Snn1 phenotypic marker was placed on the linkage map relative to these markers, and the PCR-based markers XBE498831 (12), Xfcp619, and Xfcp624 (table S1) were evaluated on the plants with recombination events between Xpsp3000 and Xfcp618 using standard PCR conditions and visualization methods as previously described (5). Linkage distances were calculated manually by dividing the number of recombinants by the total number of gametes analyzed (17,000) multiplied by 100 and expressed as map units. A population of this size has a map resolution of 0.006 map units.
Primer sets for markers Xfcp618 and Xfcp624 were used to screen the MTP for the CS chromosome arm 1BS BAC library as described by Raats et al. (13), which led to the identification of a contig consisting of 44 BACs spanning approximately 2.5 Mb. A total of 40 primer pairs for BESs from 19 MTP BACs generated by Raats et al. (13) were tested for polymorphism between CS and CS-Hope 1B, but only 1 primer pair revealed polymorphism. This marker was mapped as X28G13 (table S1). For the remaining six markers that were used to anchor the BAC contig to the linkage map, we used BESs as queries for searches against the wheat survey sequences (http://wheat-urgi.versailles.inra.fr/) of the chromosome arm 1BS to identify the corresponding survey sequence. The survey sequences were evaluated for the presence of simple sequence repeats, which were targeted for primer design and marker development (table S1). This led to the development and subsequent mapping of markers X6A04, X7O03, X73H08, X93D06, X117L19, and X130O13.
Identification of candidate genes
The four BACs (TaaCsp1BS002N12, TaaCsp1BS093D06, TaaCsp1BS106C02, and TaaCsp1BS134D22) that defined the Snn1 candidate gene region were sequenced to 154× coverage using an Ion Torrent Personal Genome Machine (PGM) next-generation sequencer (Life Technologies). The library construction was performed following the manufacturer’s standard procedures using the Xpress Plus Fragment Library Kit (Life Technologies). Templates were prepared using the Ion PGM Template OT2 200 Kit and the Ion OneTouch 2 System. Sequencing was conducted using the Ion PGM with an Ion 314 chip and the Ion PGM 200 Sequencing Kit v2 according to the manufacturer’s instructions. The Torrent Suite 3.4.2 was used for base calling. The DNASTAR SeqMan NGen 11.1.0 software set to the de novo genome assembly setting was used for the sequence assembly. The sequences were then subjected to BLASTX searches of the National Center for Biotechnology Information (NCBI) nonredundant (nr) database to identify putative protein coding sequences (table S2). Sequences with similarity to putative proteins were targeted for primer design and marker development (table S2). A single marker representing each candidate gene was polymorphic and placed on the genetic linkage map. The gene-based markers were also tested on the four BACs by standard PCR amplification followed by agarose gel electrophoresis.
Mutagenesis
Seeds of CS were treated with EMS as previously described (5), and M2 generation plants were infiltrated with SnTox1 and scored for the presence/absence of necrosis as described above. M2 plants showing insensitivity to SnTox1 were self-pollinated to obtain M3 plants, which were screened with SnTox1 for confirmation. Sixteen SnTox1-insensitive mutants were identified (table S3).
We obtained the full-length genomic sequences of the TaWAK gene from each of the 16 mutants using the primers listed in table S6. The gene was amplified in four overlapping fragments, and three independent PCRs for each fragment were sequenced to eliminate PCR errors. Sequences of the mutants were compared to the wild-type TaWAK gene sequence of CS using the software Sequencher v4.8 (Gene Codes Corporation).
Comparative sequence analysis indicated that all but one mutant (CSems-6141) had either missense or nonsense mutations (table S3). CSems-6141 had a point mutation in the splice acceptor site of intron 2. To determine whether the mutation affected splicing, total RNA was isolated from CSems-6141 and CS and was used to make cDNA as previously described (5). RT-PCR was conducted using primers 3ExonF3 and 600015R (table S7).
Snn1 characterization
Total RNA was isolated from leaf tissue of CS and was used for cDNA synthesis as previously described (5). The full-length cDNA was amplified in three overlapping fragments using the primer pairs in table S7, sequenced, and compared with the genomic sequence to identify the splicing junctions. The 5′ and 3′ rapid amplification of cDNA ends (RACE) was performed using the SMART RACE cDNA Amplification Kit (Clontech) to determine the 5′ and 3′UTRs. The first-round PCR of the 5′ RACE reaction was performed using the primer UPM provided in the kit combined with the gene-specific primer 5′ RACE_R4 (table S8). The second-round PCR of the 5′ RACE was carried out using the primer NUP provided in the kit and the gene-specific primer 5′ RACE_R3 (table S8). The first-round PCR of the 3′ RACE reaction was performed using the primer UPM provided in the kit combined with the gene-specific primer 3′ RACE_F3 (table S8). The second-round PCR of the 3′ RACE was carried out using the primer NUP provided in the kit and the gene-specific primer 3′ RACE_F2 (table S8). PCR products of RACE reactions were purified, cloned, and sequenced using the Sanger method.
DNA blot analysis was conducted on the set of CS nullisomic-tetrasomic lines, where a pair of missing chromosomes is compensated for by a pair of homoeologous chromosomes (27). Restriction digestion and hybridization were performed according to Reddy et al. (12). DNA was digested with Bam HI and probed with FCG36 derived from the 5′UTR and coding region of the Snn1. Probe FCG36 was amplified from BAC TaaCsp1BS134D22 using primers indicated in table S6 and fig. S5.
Coding and deduced amino acid sequences were used in BLAST searches of the NCBI database to identify sequences homologous to the Snn1 gene. Major domains of the Snn1 gene were annotated using numerous tools available at the ExPASy Bioinformatics Resource Portal (www.expasy.org).
Phylogenetic analysis
The full-length genomic sequence of Snn1 was obtained from 24 accessions (shown in boldface in table S5) of different ploidy levels. Four overlapping fragments representing the complete gene were amplified using the primers in table S6. Three independent PCRs for each fragment were sequenced to eliminate PCR errors. Sequences were assembled using the software Sequencher v4.8. Deduced amino acid sequences were aligned using MUSCLE (28), and the phylogenetic tree was constructed using the neighbor-joining method and the p-distance model in MEGA (29).
Transcriptional expression
CS was used for Snn1 transcriptional analysis, and the wheat GAPDH gene was used as an internal control as previously described (5). Plants were grown in a growth chamber at 21°C with a 12-hour light/dark cycle (8:00 a.m./8:00 p.m.), except for the continuous dark treatment, which was performed under the same conditions without light. All transcriptional experiments consisted of at least six biological replicates.
To study the tissue-specific expression of Snn1, samples were collected from leaves, stems, and roots at the seedling stage and immature spikes at Feekes wheat growth stage 8. Total RNA was extracted from plant tissues using the RNeasy Plant Mini Kit (Qiagen). First-strand cDNA was synthesized from 2 μg of total RNA using TaqMan Reverse Transcription Reagents (Applied Biosystems). RT-PCR was carried out using primers 600015R and Snn1_RT_R4 for Snn1 and GAPDH.F152 and GAPDH.R338 for GAPDH (table S9) on the cDNAs from the different plant tissues.
Snn1 transcription was investigated on plants grown under a 12-hour light/dark cycle and on plants placed under continuous darkness beginning at the time of collection of the first sample. Samples were collected from 2-week-old seedlings every 3 hours for three consecutive days, as previously described (5).
The effects of SnTox1 on Snn1 transcription were evaluated by infiltration of 2-week-old plants. Treatments included SnTox1- or YPD medium–infiltrated plants. Plants were grown in a growth chamber under a 12-hour light/dark cycle, and infiltrations were performed as previously described. Samples were collected from infiltrated regions of SnTox1- and YPD medium–infiltrated plants at 0-, 3-, 6-, 12-, 24-, 36-, and 48-hour time points for RNA isolation.
To avoid wounding effects on the induction of MAPK genes, the transcription of TaMAPK3 and TaMAPK6 was studied by spray inoculation of SnTox1 cultures on 2-week-old plants as described by Liu et al. (11), with water-sprayed and nonsprayed plants as controls. RNA samples were collected at 0-, 0.25-, 0.5-, 1-, 3-, 6-, 12-, 24-, 36-, and 48-hour time points. Fragments of TaMAPK genes were amplified using the primers in table S9, which were taken from Rudd et al. (30).
RQ-PCR was performed on a 7500 Real-Time PCR System (Applied Biosystems). Each experiment was conducted using six biological replicates, and all PCRs were done in triplicate. The 20-μl PCRs contained 1× SYBR PCR MasterMix (Applied Biosystems), 0.25 μM of each primer, and 5 μl of 10-fold diluted cDNA. The thermocycler procedure was as follows: 10 min of preincubation at 95°C, followed by 40 cycles for 15 s at 95°C and for 1 min at 60°C. The CS deletion line 1BS-18, which lacks the terminal portion of the chromosome arm 1BS containing Snn1, was used as negative control.
Efficiencies of the different primer combinations were evaluated using serial dilutions of CS cDNA (1:5, 1:10, 1:20, and 1:40), and only primers with efficiencies higher than 95% were used for the RQ-PCR. Transcript levels were expressed as the ratio between the initial numbers of molecules in the target and the internal control using the 2−ΔCT method.
Transformation
The full-length Snn1 cDNA was custom-synthesized (Aldevron) and cloned into the pUC57 vector. The Snn1 gene was amplified using primers Snn1.CACC.start.F1 (5′-CACCATGAGCACCCCAAATTCCCAATTCC-3′) and Snn1.stop.R1 (5′-CTACGCTTTACGAGACCGGCCTAG-3′). The PCR product was cloned into a Gateway-compatible entry vector using the pENTR/D-TOPO Cloning Kit (Thermo Fisher Scientific) according to the manufacturer’s directions. The Gateway destination vector pUNos_C1 was a gift from J. Glazebrook and F. Katagiri (Addgene plasmid #33297). The recombination reaction between the entry clone and the destination vector was performed using the Gateway LR clonase II Enzyme mix (Thermo Fisher Scientific).
The plasmids pUNOS-CL-Snn1 and pHAC20 (containing the herbicide resistance gene bar) (31) were cobombarded into the spring wheat variety BW using a particle inflow gun (32). Genetic transformation and plant recovery were performed as described by Cruz et al. (33) and Anand et al. (34). Briefly, immature embryos were isolated 10 to 14 days after anthesis and cultured on embryo induction medium for 5 to 7 days. Embryogenic calli were then cobombarded with the plasmids above. Sixteen hours after transformation, the calli were placed on a selection medium containing glufosinate (5 mg/liter) for 10 days and a shoot-production medium for 2 weeks and then transferred to an elongation and rooting medium for 2 to 4 weeks. Once roots formed, the plants were transferred to soil. Recovered plants were initially screened for the presence of the bar gene by applying a freshly prepared aqueous solution of 0.2% Liberty (AgEvo) to the midlamina portion (~2.5 cm long) of the second or third youngest leaf. The painted area was marked, and damage observations were recorded 5 to 7 days after application. Resistant events were then subjected to PCR screening using construct-specific primer pairs and self-pollinated to obtain T1 seeds.
T1 plants were tested for reaction to SnTox1 by infiltration as described above. Two T1 families, BW5152 and BW5240, were identified to be segregating for reactions to SnTox1. The sensitive and insensitive plants were genotyped by PCR using primers pUNOS.AscI.F1 (GCCCTGCCTTCATACGCTATT) and 2ExonF (ATGCGGGAGCTTGCATTCAT). To test Snn1 transcriptional gene expression in transgenic lines, RNA samples were obtained, and cDNA was synthesized using the method described above. RT-PCR was conducted using primers 600015R and Snn1_RT_R4 for Snn1 and GAPDH.F152 and GAPDH.R338 for the endogenous control gene GAPDH.
Y2H analysis
The coding region (minus putative signal peptide) of SnTox1 (35) and Snn1 (full-length or individual domains) was cloned in-frame with the GAL4 DNA binding domain of the bait vector pGBKT7 and the GAL4 activation domain of the prey vector pGADT7, respectively. The SnTox1 sequence was PCR-amplified from a cDNA clone with primers SnTox1-52F and SnTox1-tagR (table S10). Snn1 sequences were PCR-amplified from the corresponding cDNA clone (GenBank accession number KP091701). For the prey construct expressing full-length mature Snn1 protein, primers Snn1–Nde I–88F and Snn1–Xho I–tagR (table S10) were used; PCR products were subcloned into the Nde I–Xho I sites instead of the routinely used Eco RI and Bam HI sites (both of which are present in Snn1 at multiple locations). For the prey constructs, the primers in table S10 were used to amplify fragments expressing individual domains of Snn1. The identity of all bait and prey constructs was confirmed by DNA sequencing. For testing potential interactions, the bait and prey constructs were cotransformed into yeast strain AH109 following the standard procedures as previously described (36). Positive (pGBKT7-53 and pGADT7-RecT) and negative (pGBKT7-Lam and pGADT7-RecT) controls (Clontech) were included in all experiments. Yeast transformants were selected on SD/-Leu/-Trp, SD/-Leu/-Trp/-Ade/-His, and SD/-LTAH plus X-α-Gal agar plates to detect the activation of reporter genes HIS3, ADE2, and MEL1 (for α-galactosidase activity). The strengths of interactions were compared using serially diluted yeast cells in α-galactosidase activity assays, as described previously (36). SnToxA (GenBank accession number DQ423483), which interacts with the wheat PR-1-5 protein in Y2H assays (36), was used as a negative bait control against Snn1 prey constructs that showed a positive interaction with SnTox1 under the same conditions.
Supplementary Material
Acknowledgments
We thank B. Oldenburg, M. Stoley, M. Overlander, W. Sargent, and D. Holmes for technical assistance. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture (USDA). USDA is an equal opportunity provider and employer. No human or animal subjects were used in this research. Funding: This research was supported by the USDA Agricultural Research Service Current Research Information System project 5442-22000-037-00D and by the European Community’s Seventh Framework Programme TriticeaeGenome (grant agreement number FP7-212019). Author contributions: G.S. and J.D.F designed the experiments; G.S., T.L.F., D.R., W.C., R.S.B., Z.Z., H.N.T., Z.L., Z.F., J.B.R., T.F., S.S.X., and J.D.F. conducted the experiments and collected the data; G.S., T.L.F., S.L., and J.D.F. analyzed and interpreted the data; and G.S. and J.D.F. wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Sequences have been deposited in GenBank under accession numbers KP091701 and KP085710 through KP085749. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/10/e1600822/DC1
fig. S1. Conserved domains and active sites identified in the deduced Snn1 protein.
fig. S2. An unrooted phylogenetic tree showing relationships between Snn1 and other plant wall–associated receptor kinase (WAK) proteins.
fig. S3. Deduced amino acid sequence alignment of mutants and informative lines.
fig. S4. Transcription analysis of the splice site mutant CSems-6141.
fig. S5. DNA blot analysis.
fig. S6. DNA alignment of Snn1 from chromosome 1B and its putative homoeoallele from chromosome 1D.
fig. S7. Phylogenetic tree of 24 genotypes based on deduced amino acid sequences of the Snn1 gene.
fig. S8. Transcription analysis of Snn1 in the durum wheat variety Lebsock.
fig. S9. TaMAPK3 transcription analysis after SnTox1 spray inoculation.
fig. S10. Y2H assays showing that Snn1 interacts with SnTox1 in a sequence-specific manner.
table S1. PCR-based molecular markers used to anchor the CS chromosome 1BS BAC contig to the genetic linkage map containing the Snn1 gene.
table S2. PCR-based molecular markers developed for the seven candidate genes.
table S3. Descriptions of induced mutations identified within the Snn1 gene-coding region.
table S4. Top five BLASTP hits in the NCBI nr database using the Snn1 deduced amino acid sequence as a query.
table S5. The 826 Triticum accessions and 123 Ae. speltoides accessions evaluated for the presence of the Snn1 DNA sequence and/or for reaction to SnTox1.
table S6. Primers used for sequencing of Snn1 from genomic DNA.
table S7. PCR primers used to amplify the Snn1 cDNA fragments for sequencing.
table S8. PCR primers used to amplify the Snn1 cDNA 5′ and 3′ ends.
table S9. PCR primers used for RQ-PCR analysis.
table S10. PCR primers used for Y2H analysis.
REFERENCES AND NOTES
- 1.Jones J. D. G., Dangl J. L., The plant immune system. Nature 444, 323–329 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Zipfel C., Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 12, 414–420 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Dodds P. N., Rathjen J. P., Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Oliver R. P., Solomon P. S., New developments in pathogenicity and virulence of necrotrophs. Curr. Opin. Plant Biol. 13, 415–419 (2010). [PubMed] [Google Scholar]
- 5.Faris J. D., Zhang Z., Lu H., Lu S., Reddy L., Cloutier S., Fellers J. P., Meinhardt S. W., Rasmussen J. B., Xu S. S., Oliver R. P., Simons K. J., Friesen T. L., A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. U.S.A. 107, 13544–13549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorang J. M., Sweat T. A., Wolpert T. J., Plant disease susceptibility conferred by a “resistance” gene. Proc. Natl. Acad. Sci. U.S.A. 104, 14861–14866 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagy E. D., Bennetzen J. L., Pathogen corruption and site-directed recombination at a plant disease resistance gene cluster. Genome Res. 18, 1918–1923 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adhikari T. B., Bai J., Meinhardt S. W., Gurung S. J., Myrfield M., Patel J., Ali S., Gudmestad N. C., Rasmussen J. B., Tsn1-mediated host responses to ToxA from Pyrenophora tritici-repentis. Mol. Plant Microbe Interact. 22, 1056–1068 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Pandelova I., Betts M. F., Manning V. A., Wilhelm L. J., Mockler T. C., Ciuffetti L. M., Analysis of transcriptome changes induced by Ptr ToxA in wheat provides insights into the mechanisms of plant susceptibility. Mol. Plant 2, 1067–1083 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Friesen T. L., Faris J. D., Characterization of the wheat-Stagonospora nodorum disease system: What is the molecular basis of this quantitative necrotrophic disease interaction?. Can. J. Plant Pathol. 32, 20–28 (2010). [Google Scholar]
- 11.Liu Z. H., Zhang Z., Faris J. D., Oliver R. P., Syme R., McDonald M. C., McDonald B. A., Solomon P. S., Lu S., Shelver W. L., Xu S., Friesen T. L., The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLOS Pathog. 8, e1002467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddy L., Friesen T. L., Meinhardt S. W., Chao S., Faris J. D., Genomic analysis of the Snn1 locus on wheat chromosome arm 1BS and the identification of candidate genes. Plant Genome 1, 55–66 (2008). [Google Scholar]
- 13.Raats D., Frenkel Z., Krugman T., Dodek I., Sela H., Šimková H., Magni F., Cattonaro F., Vautrin S., Bergès H., Wicker T., Keller B., Leroy P., Philippe R., Paux E., Doležel J., Feuillet C., Korol A., Fahima T., The physical map of wheat chromosome 1BS provides insights into its gene space organization and evolution. Genome Biol. 14, R138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.International Wheat Genome Sequencing Consortium (IWGSC) , A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788 (2014). [DOI] [PubMed] [Google Scholar]
- 15.J. D. Faris, Wheat domestication: Key to agricultural revolutions past and future, in Genomics of Plant Genetic Resources, Vol. 1. Managing, Sequencing and Mining Genetic Resources, R. Tuberosa, A. Graner, E. Frison, Eds. (Springer, 2014), pp. 439–464. [Google Scholar]
- 16.Tsuda K., Katagiri F., Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Liu Z., Gao Y., Kim Y. M., Faris J. D., Shelver W. L., de Wit P. J.G. M., Xu S. S., Friesen T. L., SnTox1, a Parastagonospora nodorum necrotrophic effector, is a dual-function protein that facilitates infection while protecting from wheat-produced chitinases. New Phytol. 211, 1052–1064 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Wagner T. A., Kohorn B. D., Wall-associated kinases are expressed throughout plant development and are required for cell expansion. Plant Cell 13, 303–318 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brutus A., Sicilia F., Macone A., Cervone F., De Lorenzo G., A domain swap approach reveals a role of the plant wall-associated kinase 1(WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107, 9452–9457 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Lorenzo G., Brutus A., Savatin D. V., Sicilia F., Cervone F., Engineering plant resistance by constructing chimeric receptors that recognize damage-associated molecular patterns (DAMPs). FEBS Lett. 585, 1521–1528 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Zuo W., Chao Q., Zhang N., Ye J., Tan G., Li B., Xing Y., Zhang B., Liu H., Fengler K. A., Zhao J., Zhao X., Chen Y., Lai J., Yan J., Xu M., A maize wall-associated kinase confers quantitative resistance to head smut. Nat. Genet. 47, 151–157 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Hurni S., Scheuermann D., Krattinger S. G., Kessel B., Wicker T., Herren G., Fitze M. N., Breen J., Presterl T., Ouzunova M., Keller B., The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. U.S.A. 112, 8780–8785 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manning V. A., Ciuffetti L. M., Localization of Ptr ToxA produced by Pyrenophora tritici-repentis reveals protein import into wheat mesophyll cells. Plant Cell 17, 3203–3212 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu C.-G., Faris J. D., Xu S. S., Friesen T. L., Genetic analysis of disease susceptibility contributed by the compatible Tsn1-SnToxA and Snn1-SnTox1 interactions in the wheat-Stagonospora nodorum pathosystem. Theor. Appl. Genet. 120, 1451–1459 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Bhardwaj V., Meier S., Petersen L. N., Ingle R. A., Roden L. C., Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLOS ONE 6, e26968 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roden L. C., Ingle R. A., Lights, rhythms, infection: The role of light and the circadian clock in determining the outcome of plant–pathogen interactions. Plant Cell 21, 2546–2552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sears E. R., The aneuploids of common wheat. Mo. Agric. Exp. Stn. Res. Bull. 572, 1–58 (1954). [Google Scholar]
- 28.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudd J. J., Keon J., Hammond-Kosack K. E., The wheat mitogen-activated protein kinases TaMPK3 and TaMPK6 are differentially regulated at multiple levels during compatible disease interactions with Mycosphaerella graminicola. Plant Physiol. 147, 802–815 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christensen A. H., Quail P. H., Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5, 213–218 (1996). [DOI] [PubMed] [Google Scholar]
- 32.Finer J. J., Vain P., Jones M. W., McMullen M. D., Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep. 11, 323–328 (1992). [DOI] [PubMed] [Google Scholar]
- 33.Cruz L., Rupp J. L. S., Trick H. N., Fellers J. P., Stable resistance to Wheat streak mosaic virus in wheat mediated by RNAi. In Vitro Cell. Dev. Biol. 50, 665–672 (2014). [Google Scholar]
- 34.Anand A., Zhou T., Trick H. N., Gill B. S., Bockus W. W., Muthukrishnan S., Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-like protein, chitinase and glucanase against Fusarium graminearum. J. Exp. Bot. 54, 1101–1111 (2003). [DOI] [PubMed] [Google Scholar]
- 35.S. Lu, Use of the yeast two-hybrid system to identify targets of fungal effectors, in Plant Pathogen Protocols, Methods in Molecular Biology, M. Bolton, B. Thomma, Eds. (Humana Press Inc., 2012), vol. 835, pp. 165–189. [DOI] [PubMed] [Google Scholar]
- 36.Lu S., Faris J. D., Sherwood R., Friesen T. L., Edwards M. C., A dimeric PR-1-type pathogenesis-related protein interacts with ToxA and potentially mediates ToxA-induced necrosis in sensitive wheat. Mol. Plant Pathol. 15, 650–663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/2/10/e1600822/DC1
fig. S1. Conserved domains and active sites identified in the deduced Snn1 protein.
fig. S2. An unrooted phylogenetic tree showing relationships between Snn1 and other plant wall–associated receptor kinase (WAK) proteins.
fig. S3. Deduced amino acid sequence alignment of mutants and informative lines.
fig. S4. Transcription analysis of the splice site mutant CSems-6141.
fig. S5. DNA blot analysis.
fig. S6. DNA alignment of Snn1 from chromosome 1B and its putative homoeoallele from chromosome 1D.
fig. S7. Phylogenetic tree of 24 genotypes based on deduced amino acid sequences of the Snn1 gene.
fig. S8. Transcription analysis of Snn1 in the durum wheat variety Lebsock.
fig. S9. TaMAPK3 transcription analysis after SnTox1 spray inoculation.
fig. S10. Y2H assays showing that Snn1 interacts with SnTox1 in a sequence-specific manner.
table S1. PCR-based molecular markers used to anchor the CS chromosome 1BS BAC contig to the genetic linkage map containing the Snn1 gene.
table S2. PCR-based molecular markers developed for the seven candidate genes.
table S3. Descriptions of induced mutations identified within the Snn1 gene-coding region.
table S4. Top five BLASTP hits in the NCBI nr database using the Snn1 deduced amino acid sequence as a query.
table S5. The 826 Triticum accessions and 123 Ae. speltoides accessions evaluated for the presence of the Snn1 DNA sequence and/or for reaction to SnTox1.
table S6. Primers used for sequencing of Snn1 from genomic DNA.
table S7. PCR primers used to amplify the Snn1 cDNA fragments for sequencing.
table S8. PCR primers used to amplify the Snn1 cDNA 5′ and 3′ ends.
table S9. PCR primers used for RQ-PCR analysis.
table S10. PCR primers used for Y2H analysis.


