Abstract
A toxicity pathway approach was taken to develop an in vitro assay using human uterine epithelial adenocarcinoma (Ishikawa) cells as a replacement for measuring an in vivo uterotrophic response to estrogens. The Ishikawa cell was determined to be fit for the purpose of recapitulating in vivo uterine response by verifying fidelity of the biological pathway components and the dose-response predictions to women of child-bearing age. Expression of the suite of estrogen receptors that control uterine proliferation (ERα66, ERα46, ERα36, ERβ, G-protein coupled estrogen receptor (GPER)) were confirmed across passages and treatment conditions. Phenotypic responses to ethinyl estradiol (EE) from transcriptional activation of ER-mediated genes, to ALP enzyme induction and cellular proliferation occurred at concentrations consistent with estrogenic activity in adult women (low picomolar). To confirm utility of this model to predict concentration-response for uterine proliferation with xenobiotics, we tested the concentration-response for compounds with known uterine estrogenic activity in humans and compared the results to assays from the ToxCast and Tox21 suite of estrogen assays. The Ishikawa proliferation assay was consistent with in vivo responses and was a more sensitive measure of uterine response. Because this assay was constructed by first mapping the key molecular events for cellular response, and then ensuring that the assay incorporated these events, the resulting cellular assay should be a reliable tool for identifying estrogenic compounds and may provide improved quantitation of chemical concentration response for in vitro-based safety assessments.
Keywords: TT21C, endocrine disruption, human health safety assessment, in vitro and alternative models.
INTRODUCTION
The National Research Council report “Toxicity Testing in the 21st Century” defines the need to develop human cell-based assays for assessing chemical safety (Krewski et al., 2010). However, to use in vitro assays for risk assessment, they must be carefully designed to be fit for the purpose of predicting human response. Validation of these assays should ensure utility for the intended purpose, as well as define any limitations with respect to the in vivo outcome of concern. Existing initiatives, including ToxCast, Tox21 and EDSP integrate high-throughput and tiered testing approaches for chemical prioritization (Attene-Ramos et al., 2013; Browne et al., 2015; Huang et al., 2014; Judson et al., 2010; Knudsen et al., 2011; Reif et al., 2010). These programs use a suite of assays to support a weight-of-evidence approach to prioritize chemicals for further testing, and are likely to include in vivo studies (Attene-Ramos et al., 2013; Browne et al., 2015; Huang et al., 2014; Judson et al., 2010; Knudsen et al., 2011; Reif et al., 2010). Our goal, however, is to develop human cell-based assays sufficient for in vitro-only risk assessments where results can define regions of safety for human exposure without requiring animal testing.
During the female estrous cycle, uterine tissue undergoes synchronized changes in response to alterations in 17β-estradiol (E2) with proliferation of the epithelial lining occurring in response to increased E2 prior to the secretory phase (Bondesson et al., 2014; Maruyama and Yoshimura, 2008). Many chemicals mimic estrogen and induce uterine proliferation in the human and in animal models. In the context of chemical risk assessment, proliferative pressure on the cell is a risk factor for carcinogenicity. Potential for chemical-induced uterotrophic response is currently monitored under OECD guideline 440: rodent uterotrophic assay (OECD, 2003). We developed a human uterine epithelial cell (Ishikawa)-based assay with the intention of replacing the uterotrophic assay for screening compounds and, more importantly, as a means of providing realistic estimates of concentration-response for human uterine effects in vivo.
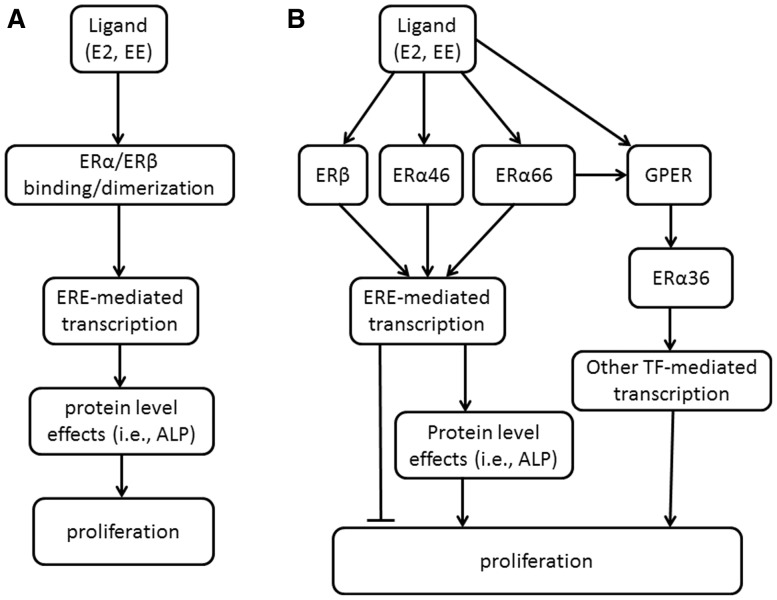
We took a toxicity pathway approach to assay development—first outlining the key events in the uterine proliferative response, then designing a model system that accounts for key signaling events from molecular initiating event (ER binding) to phenotypic outcome, i.e., proliferation (Fig. 1). This stepwise process is similar to defining the molecular and cellular events in an Adverse Outcome Pathway (AOP), but stops short of defining tissue, organism and population effects. Molecular events leading to increased proliferation have traditionally been attributed to transcriptional activity after estrogen binds classical ERs—ERα66 or ERβ—and the majority of current HTS assays for screening focus on measuring ERα66 or ERβ ligand binding and transactivation (Browne et al., 2015; Rotroff et al., 2013, 2014). The toxicity pathway these efforts portray is a significant simplification of E2-mediated signaling (Fig. 1A). A current understanding of estrogen signaling incorporates the more complete regulatory network for estrogen-mediated tissue responses, including feedback and feed-forward interactions between additional ERs (Fig. 1B), that serve as the key determinants of chemical concentration-response.
FIG. 1.
Developing a framework for the estrogen receptor signaling network. A. A simplified schematic for estrogen receptor activity relying on only two of the suite of ERs. B. Regulatory network driving concentration–response with the fuller suite of estrogen receptors.
In addition to ERβ and full-length ERα (ERα66), two shorter isoforms of ERα (ERα46, ERα36) and the G-protein Estrogen Receptor (GPER) are key mediators of estrogen-induced proliferation (Filardo et al., 2000; Penot et al., 2005; Wang et al., 2006). These receptors all bind estrogen, but differences in binding domains lead to large variations in affinity for native ligands and exogenous chemicals (Li et al., 2003; Lin et al., 2013; Tamrazi et al., 2002; Zhao et al., 2009). Current literature supports pro-proliferative effects of ERα66, GPER and ERα36 (Filardo et al., 2000; Tong et al., 2010; Wang et al., 2006), and anti-proliferative effects of ERβ and ERα46 (Klinge et al., 2010; Lin et al., 2007; Omoto et al., 2003). The inherent feedback in the pathway ensures tight regulatory control of estrogen-mediated events. Further, the relative expression of the ER isoforms, differential affinity for ligands, and regulatory feedback signals between the receptors will influence the shape of chemical-specific dose-response curves and, subsequently, estimated points of departure (PoDs). For these reasons, we considered all key ERs and down-stream cellular responses in the toxicity pathway when selecting the appropriate cell model and developing in vitro assays intended for safety assessment applications (Fig. 1B).
Using a toxicity pathway approach to assay design can increase the likelihood that the assay will provide biological- and dose-relevant information. Here, we have used this approach to develop a model of uterine proliferation. The ultimate goal is to recommend an in vitro assay in human cells that moves beyond screening and prioritization to reliably predict concentrations at which estrogenic activity can be expected in humans.
MATERIALS AND METHODS
Ishikawa Cells
The cell line used for assay development (Ishikawa) is of human uterine adenocarcinoma origin. It is commercially available through the European Collection of Authenticated Cell Cultures (ECACC) and has been genotyped for confirmation of cell type.
Chemicals and Reagents
17β-estradiol (E2; CAS #50-28-2) and 17α-ethinyl estradiol (EE; CAS #57-63-6) were purchased from Tocris Bioscience (Bristol, UK); tamoxifen (TAM; CAS #10540-29-1) and 4-Hydroxytamoxifen (OHT; CAS #68392-35-8) from Sigma (St. Louis, MO); HEPES 1 M solution, L-Glutamine 200mM solution, Trypsin 2.5%, Penicillin—Streptomycin solution, phenol red-free DMEM/F-12 (1:1) and DPBS/Modified from Hyclone Laboratories, Inc. (Logan, Utah); MEM NEAA (100x) from Gibco® by Life Technologies (Grand Island, NY). Fetal bovine serum (FBS) and charcoal stripped FBS (CSS) were from Atlanta Biologicals (Flowery Branch, GA).
Cell Culture
Ishikawa cells were maintained in phenol red-free 1:1 DMEM/F12 medium supplemented with 1% L-Glutamine (100X), 1% penicillin/streptomycin (10,000 U/ml penicillin, 10,000 µg/ml streptomycin), 1% HEPES (1M), 1% non-essential amino acids (100X), 0.1% ITS Premix (insulin, human transferrin, selenium supplement, 10X), and 10% FBS. Prior to gene analysis, protein analysis, enzyme activity or proliferation assays, Ishikawa cells were passaged into 150 mm cell culture plates at a density of 2x106 cells per plate in 10% CSS supplemented media and allowed to grow for 72 hours. Cells were then plated according to assay format requirements in 5% CSS supplemented media. After 48 hours, media was replaced with 5% CSS supplemented media containing various concentrations of EE, E2, TAM, OHT or ethanol (EtOH; vehicle control). EtOH concentrations in media were 0.1% v/v. For the remainder of the experiment, half of the treatment media in each well was replaced daily with fresh 5% CSS media containing appropriate concentrations of EE, E2, TAM, OHT or vehicle (EtOH).
Western Blotting
Ishikawa cells were lysed in ice cold protein lysis buffer (1M Tris-HCL, 0.5M EDTA, 5M NaCl, 10% SDS, 10% sarkosyl diluted in distilled H2O (dH2O)to 100 ml final volume) containing freshly added protease and phosphatase inhibitors (Pierce Biotechnology; Rockford, IL). Samples were stored at -80 °C until further use. To begin western blot analysis, samples were thawed on ice and sonicated 3–5 times for 5 seconds. Protein concentrations were determined via BCA Protein assay (Pierce Biotechnology) and the LDS sample buffer was added directly to lysates at a 1:4 dilution (NuPAGE® LDS Sample Buffer, Life Technologies; Carlsbad, CA). Samples were heated at 90 °C for 3 minutes, cooled on ice, and approximately 20–40 µg of total protein per well was loaded into 10% Novex® Tris-Glycine gels (Life Technologies). SDS-PAGE was run in 1X Tris/Glycine/SDS running buffer (Bio-Rad, Hercules, CA) at 125V constant for 2 hours. Separated protein samples were transferred from gels to PVDF membranes for 7 minutes at 20V (iBlot® Transfer Stack PVDF, Life Technologies) using the iBlot® Gel Transfer Device (Invitrogen, Carlsbad, CA). Membranes were rinsed with dH2O (or d-H2O depending on choice above) and blocked for 2 hours at 25 °C with Odyssey® Blocking buffer (LI-COR Biosciences; Lincoln, NE). Following blocking, primary antibody incubation was performed overnight at 4 °C in Odyssey® blocking buffer with the following anti-human antibodies from Santa Cruz Biotechnology (Dallas, TX): ERα66 and ERα46 (F10), ERα36 (G20), ERβ (H150), or GPER (N15). Anti-human GAPDH (14C10) was used when applicable (Cell Signaling, Beverly, MA). Membranes were subsequently washed 3X in PBS + 0.5% Tween-20 (Sigma-Aldrich, St. Louis, MO) for 15 minutes per wash and secondary fluorescently-tagged antibodies (IRDye® Infrared Dyes, LI-COR Biosciences) were added in PBS + 0.5% Tween-20 for 1 hour at 25 °C protected from light. Membranes were washed again three times in PBS + 0.5% Tween-20 for 15 minutes per wash and imaged on the Odyssey® Classic Infrared Imaging System (LI-COR Biosciences). Densitometry analysis was performed using Image Studio 4.0 software (LI-COR Biosciences).
Real-Time PCR
RNA was extracted using the QIAxtractor® (Qiagen; Maryland, USA) with VX reagent pack according to manufacturer’s instructions. Reverse transcription was performed with 300ng RNA input using a High Capacity cDNA reverse transcription kit as per manufacturer’s instructions (Applied Biosystems® Life Technologies; Grand Island, NY) for a total reaction volume of 20µl. PCR reactions were performed using a 7900HT Sequence Detection System (Applied Biosystems® Life Technologies; Grand Island, NY) in a total reaction volume of 20µl using TaqMan Fast Universal PCR Master Mix (2x). TaqMan probes for PGR (Hs01556702_m1), ALPP (Hs03046558_s1), GREB1 (Hs00536409_m1), and B2M (Hs00984230_m1) were used as per manufacturer’s instructions. For all reactions B2M was used as the internal housekeeping gene and assays were performed in technical and experimental triplicate. Values were calculated using the ΔΔC(t) method and expressed as a ratio of gene of interest to a reference gene normalized to experimental culture condition.
Alkaline Phosphatase (ALP) Activity
Ishikawa cells were cultured as described in the cell culture procedure and plated into 96-well plates at 2x103 cells per well in 200 µl of 5% CSS media. Following treatments, the media was removed and the cells were washed with 100µl of PBS. PBS was aspirated and the plates were incubated for 20–30 minutes at -80 °C to lyse the cells. 100µl of One-Step PNPP reagent (Thermo Fisher Scientific Inc.; Rockford, IL) was added to each well and plates were incubated at room temperature in the dark. Absorbance at 405 nm was measured after 30, 60, or 90 minutes.
MTS Proliferation Assay
Following the outlined culture procedure, cells were plated in 24-well plates at a density of 7×103 cells per well in 0.5 ml of media containing 5% CSS for 48 hours prior to treatment. Medium was then removed and the cells were rinsed twice with 1 ml PBS. 80µl of trypsin (0.25%) was added and the plate was incubated at 37 °C for 5 min. Each well was then treated with 320µl of 5% CSS and 80µl of CellTiter 96® AQueous One Solution (Promega; Madison, WI) and incubated at 37 °C. After 20, 40 and 60 minutes, the plate was read on a spectrophotometer using a wavelength of 490 nm. Results were expressed as relative fluorescent units (RFU) and fold changes were calculated as the change from vehicle control.
CellTrace Proliferation Assay
Cells that had been in 10% CSS for 72 hours were labeled with CellTraceTM Far Red Cell Proliferation Dye according to manufacturer’s instructions (Molecular Probes® Life Technologies, Grand Island, NY) and plated into 24-well plates at 10×106 cells per well in 0.5 ml of 5% CSS supplemented media containing treatment. Following treatment, cells were analyzed on the FACS Canto™ II flow cytometer, BD Biosciences (Mountain View, CA). Proliferation Index was calculated for each sample using ModFit LT (Verity Software House).
Hoechst Proliferation Assay
Following 72 hours of growth in 10% CSS supplemented media, Ishikawa cells were passaged into 96 well plates at a density of 4×103 cells per well in 150µl of 5% CSS supplemented media and allowed to grow for 48 hours. Cells were then treated with various concentrations of EE or vehicle control for 3 days. At the end of the assay, cells were fixed with 4% paraformaldehyde containing 2µg/ml of Hoechst-33342 nuclear stain from Thermo Fisher Scientific Inc. (Rockford, IL). Plates were analyzed on a Molecular Devices FlexStation plate reader using a 9-point scan per well. Total fluorescence was averaged for each well and used to calculate a fold change in cell number as a measure of proliferation.
RESULTS
Confirming Biological Relevance of the In vitro Assay
Based on a comprehensive review of the current understanding of estrogen-mediated cellular responses, we developed an AOP-like (toxicity pathway) diagram to guide assay development (Fig. 1B). This schematic includes key events from estrogen binding to transcriptional activation, enzyme induction and phenotypic response. To ensure that the model incorporated the appropriate biology for predicting uterine response, we evaluated whether the cells contained the appropriate receptors and were capable of recapitulating key responses to estrogenic stimulus, including transcription of ERE-mediated genes, ALP enzyme induction, and proliferation.
Expression of Estrogen Receptors in Ishikawa Cells
Five distinct ERs are expressed in human tissues that bind estrogen and have unique signaling contributions to estrogen-mediated cellular responses: three ERα isoforms (ERα66, ERα46, and ERα36), ERβ, and GPER. We evaluated mRNA and protein expression of each isoform. mRNA profiles are described in the supplementary data (Supplementary Fig. 1). Because ERs are post-transcriptionally regulated (Ishii et al., 2013; Saceda et al., 1991), we performed extensive evaluations of the protein expression of each of these receptors using western blotting. ERα66, ERα46, and ERα36 mRNAs are transcribed from the same open reading frame of the ERα gene and are therefore translated into proteins with homologous amino acid sequences. ERα46 is a truncated version of ERα66, missing the first domain on the N terminus (Flouriot et al., 2000). ERα36 is truncated at both the C and N terminus but contains an additional unique region on the C terminal end (Wang et al., 2005). The majority of commercially available antibodies for ERα are directed at full length ERα66 and it is unknown whether they distinguish between the three ERα isoforms. For this reason, we developed a positive control for protein detection of each isoform by overexpressing GFP-fused ERα isoform constructs in HEK293T cells that do not express ERα (Filardo and Thomas, 2012; Kahlert et al., 2000; Zhao et al., 2009). The specific immunogen used to develop each available antibody as listed on the company product sheet was used to predict isoform recognition.
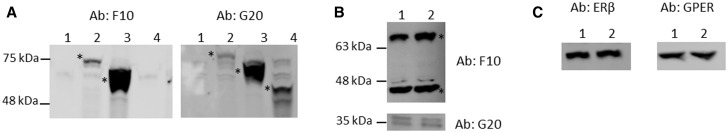
Of the antibodies tested, two from Santa Cruz Biotechnology consistently detected each ERα. ERα66 and ERα46 were detected by clone F10, which targets the C-terminal end of the protein that is present only in these two isoforms and provides specific detection as confirmed with the GFP-fusion controls (Fig. 2A). ERα36 was best detected using the Santa Cruz ERα antibody clone G20 which recognizes the central hinge domain of the protein, an epitope present in all 3 isoforms (Fig. 2A). G20 produced a number of nonspecific bands at higher molecular weights, preventing its use for detecting the two larger isoforms (Supplementary Fig. 2). For each primary antibody, no protein bands were detected in the WT HEK negative control lysate and the appropriate GFP-tagged ERα isoform was detected in the transfected HEK lysate (Fig. 2A). Additional western blots were run with F10 or G20 then stripped and re-probed with anti-GFP to confirm the correct band for fusion proteins (Supplementary Fig. 3). F10 and G20 were then used to probe for each ERα isoform in Ishikawa cells grown in either maintenance media (10% FBS) or in media supplemented with CSS. In all conditions, each of the three isoforms was present (Fig. 2B). Antibodies against ERβ and GPER identified specific bands at expected molecular weights for each protein, confirming the expression of these other estrogen receptors (Fig. 2C).
FIG. 2.
Detection of all five ER proteins by western blot. Protein expression levels of ERα66, ERα46, ERα36, ERβ, and GPER were detected by Western blot. A. Lysates from ERα-deficient HEK cells (Lane 1), ERα66-GFP fusion protein transfected HEK cells (Lane 2), ERα46-GFP fusion protein transfected HEK cells (Lane 3), or ERα36-GFP fusion protein transfected HEK cells (Lane 4) were run using either F10 for detecting 66/46 or G20 for detecting all 3 isoforms. B. Lysates from WT Ishikawa cells in 10% FBS containing media (Lane 1) or WT Ishikawa cells in 10% CSS containing media (Lane 2) were run using F10 for detection of ERα66 and ERα46, and G20 for detection of ERα36. For detection of ERα36, twice as much lysate was used to improve band clarity. C. ERβ and GPER were present in Ishikawa cells in 10% FBS media (Lanes 1) and in 10% CSS media (Lanes 2). *Asterisks denote protein band of interest in each blot, molecular weights determined by protein ladders.
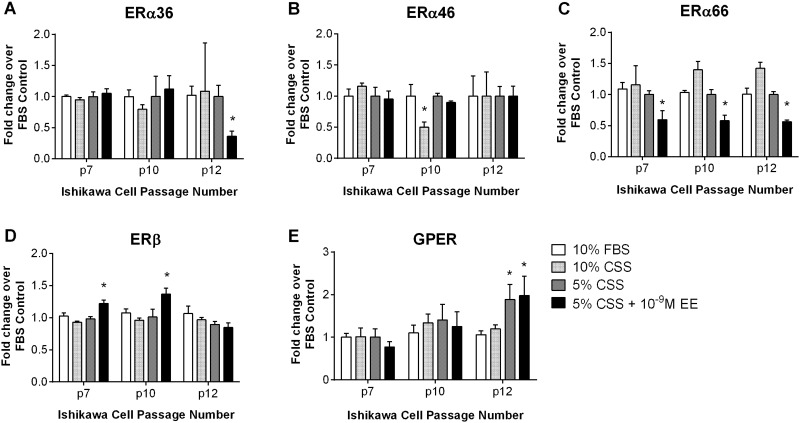
To evaluate the stability of the test system, we examined protein expression in Ishikawa cells under each experimental condition outlined in our cell culture method including growth medium (10% FBS), charcoal-stripped serum supplemented medium (10% CSS), low charcoal-stripped serum medium (5% CSS, 1% v/v EtOH), and cells treated for 3 days with EE (5% CSS + 10 − 9M EE) over several consecutive passages. All of the ERα isoforms were present in the Ishikawa cells under all test conditions (Fig. 3). However, some changes in expression levels occurred across passages and for certain treatment conditions. ERα36 expression displayed a statistically significant decrease in response to EE treatment at passage 12 (p12), but was fairly stable for other treatments and passages tested (Fig. 3A). Likewise, ERα46 expression was steady across passages, with a decrease observed only in 10% CSS media at p10 (Fig. 3B). ERα66 was consistent across all passages in vehicle control samples, but showed a trend towards decreased protein expression following EE treatment (Fig. 3C). ERβ expression increased following EE treatment at p7 and p10, but not at p12 (Fig. 3D). GPER protein expression levels were consistent across treatments through passage 10, but displayed significant differences between treatments at p12 (Fig. 3E). Cell lines reportedly lose estrogen sensitivity through alteration in ER expression at higher passages (Daly and Darbre, 1990), unpublished observation) and fluctuations in expression patterns were observed in Ishikawa cells at the higher passages tested here. Our results indicate that the expression of the suite of ER receptors in these cells remains relatively stable through p10.
FIG. 3.
Expression of ERα isoforms, ERβ, and GPER protein across passages and treatment conditions in Ishikawa cells. Expression of A. ERα36, B. ERα46, C. ERα66, D. ERβ, or E. GPER was measured in Ishikawa cells at passages 7, 10, and 12 by western blotting. Densitometry was performed using GAPDH as a housekeeping gene and expression was normalized as fold change over 10% FBS controls for each experiment. Data shown represents the mean + SEM (A-C, n = 3; D-E, n = 1 for each treatment condition, *p ≤ 0.05 statistical difference from 10% FBS controls, two-way ANOVA).
Regulation of ER-Mediated Transcription in Ishikawa Cells
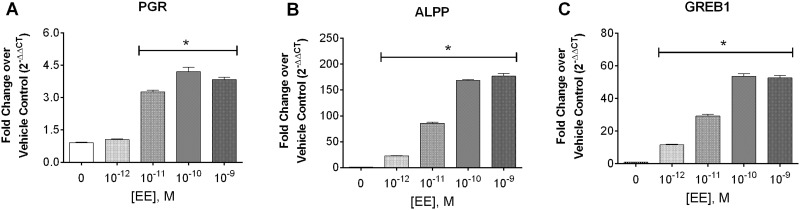
Classical signaling through ERα (ERα66) occurs through dimerization of the cytosolic receptor, translocation to the nucleus and initiation of transcription through binding of the receptor to estrogen response elements (EREs). To determine if Ishikawa cells maintain this classically described estrogen-mediated transcriptional signaling, expression of three estrogen-mediated genes was evaluated using RT-PCR after 3 days of treatment with the potent estrogen derivative, ethinyl estradiol (EE). Progesterone receptor (PGR) and a gene regulated in breast cancer (GREB1) contain EREs in their promoter regions for direct ER dimer binding and subsequent transcriptional regulation (Deschenes et al., 2007; Petz and Nardulli, 2000). Additionally, GREB1 is essential for ER transcriptional regulation (Mohammed et al., 2013). The full ERE has not been described for alkaline phosphatase, placental (ALPP), but increased transcription of ALPP has been specifically associated with estrogen-mediated responses in uterine cells (and not breast cells) in vitro and in vivo (Albert et al., 1990; Naciff et al., 2009). All of these genes demonstrated dose-dependent increases, with significant induction occurring at doses as low as 10 − 11 M for PGR and 10 − 12 M for ALPP or GREB1 (Fig. 4). We also evaluated the full genome transcriptomic profile using gene array with Ishikawa cells following 72-hour treatment with vehicle or 10 − 9M EE (see Supplementary Materials). The genomic data demonstrated regulation of expected estrogen receptor pathways and was consistent with previous reports for this cell type and with intact uterine tissue (Naciff et al., 2009). Transcriptomic results are provided in Supplementary Figure 4.
FIG. 4.
Expression of estrogen-mediated gene transcripts following treatment of Ishikawa cells with EE for 3 days. mRNA transcript levels of A. PGR, B. ALPP, and C. GREB1, in p8 Ishikawa cells treated for 3 days with increasing doses of EE in 5% CSS media were analyzed using qRT-PCR. At each concentration, samples were analyzed in triplicate. Fold change over vehicle control (0 M EE) was calculated by the 2-ΔΔCT method and data shown represents the mean + SEM (n = 4, *≤ 0.05 Mann-Whitney statistical difference from vehicle control).
ER-Mediated Induction of Enzyme Activity in Ishikawa Cells
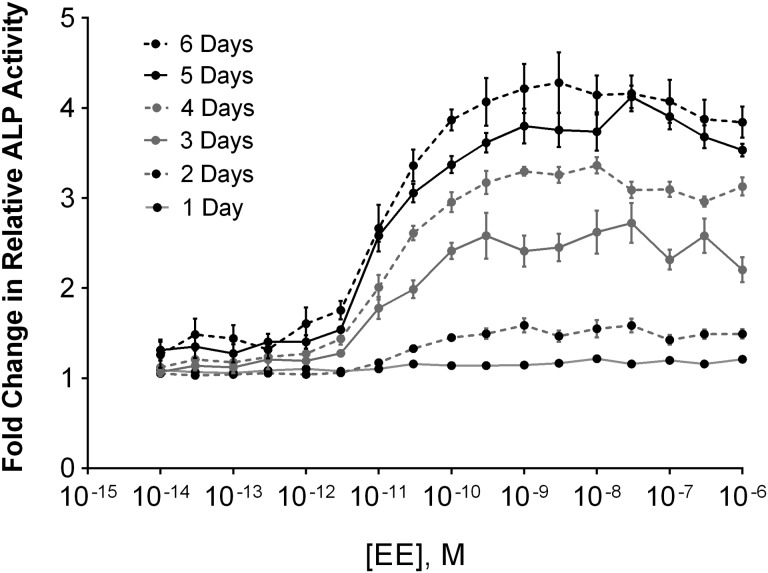
ALP enzyme activity is associated with estrogen-specific signaling in endometrial tissues and plays an important role in physiologic changes in uterine stromal and epithelial cells during implantation and pregnancy (Vatin et al., 2014). Up-regulation of ALP activity in response to estrogen is a useful measure of estrogen-mediated cell signaling (Bansode et al., 1998). ALP enzyme activity was measured across doses of EE and treatment times using PNPP, a substrate for ALP that yields a yellow product after enzymatic cleavage and can be read on a plate reader (absorbance). Ishikawa cells demonstrated both a dose- and time-dependent induction of ALP enzyme activity (Fig. 5). Minimal responses were observed on days 1 or 2 of treatment, but by day 3, ALP activity was significantly increased in the presence of EE at concentrations greater than 10 − 11 M (Mann-Whitney test for significance over vehicle treated).
FIG. 5.
Induction of ALP enzyme activity in Ishikawa cell following EE treatment. ALP enzyme activity was measured in Ishikawa cells across increasing doses of EE daily up to 6 days after treatment. At each time point and concentration, samples were analyzed in triplicate and fold change over vehicle control was calculated. Data shown represents the mean ± SEM (n = 3).
ER-Mediated Proliferation in Ishikawa Cells
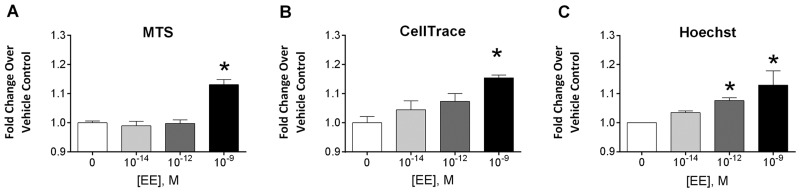
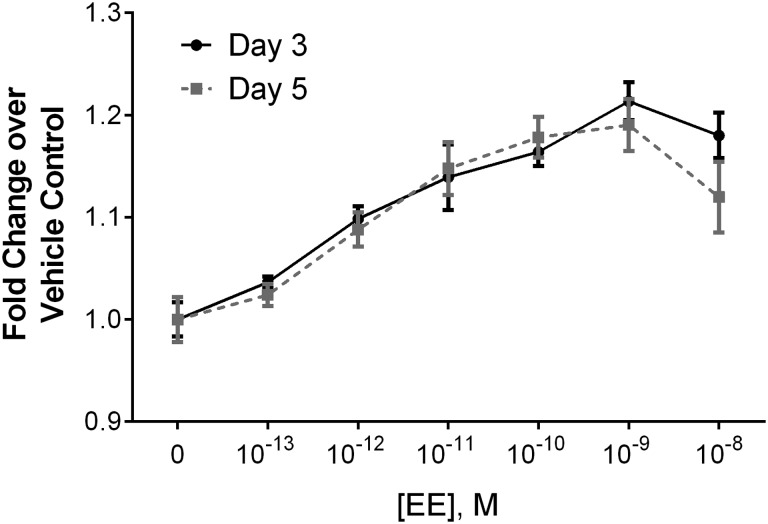
On a cellular level, the characteristic phenotypic response to estrogenic signaling is the induction of proliferation. Multiple methods for measuring proliferation in Ishikawa cells were evaluated, including the same ACEA platform used in the ToxCast T47D assay, MTS, CellTrace, and Hoechst nuclear staining. The ACEA assay measures cellular impedance of an electrical current, which correlates to cellular responses including adhesion, proliferation and cytotoxicity. Our preliminary efforts with the platform revealed that the loss of cell–cell contact associated with estrogen signaling in the uterine cells confounded the proliferative signal in the impedance readout (see Supplementary Fig. 6). The platform was therefore not useful as measure for proliferation with Ishikawa cells. MTS is a widely used enzymatic viability assay with a colorimetric read-out, where viability is used as a direct correlate to cell number. CellTrace is a cytoplasmic dye that uniformly labels the cell population of interest at the start of treatment. With each division, cells become half as bright, allowing for quantitation of the number of divisions and relative percent of proliferative responders by flow cytometry. Hoechst dye labels the cell nucleus and provides a method for quantification of the number of nuclei, and therefore cells, in each well of the assay using a fluorescent measurement. Despite very different assay readouts, all three methods showed similar results, with maximum induction of approximately 20% fold increase in the proliferation over controls following 3 days of 10 − 9 M EE treatment (Fig. 6). Of the assays, the Hoechst assay provided the least amount of inter-experimental variability, which allowed greater sensitivity at low doses. In-depth dose-response studies were then performed using the Hoechst assay at 3 and 5 days of EE treatment, with concentrations ranging from 10 − 13 M to 10 − 8 M EE. Significant increases in proliferation were observed at 10 − 12 M EE and above on both days 3 and 5 (Fig. 7). Additional studies using the MTS assay on days 2–6 showed maximal responses on day 3 with the induction of proliferation at concentrations as low as 3×10 − 12 M EE (Supplementary Fig. 5). Although the dynamic range of the proliferation response was modest, it was highly consistent across experiments. The Z factor (Zhang et al., 1999) for these assays revealed that the Hoechst assay was the most robust of the methods with a Z-factor of 0.610 compared to 0.246 for MTS and 0.219 for CellTrace.
FIG. 6.
Comparison of three methods for evaluating EE-induced proliferation in Ishikawa cells. Several methods for detection of cellular proliferation were compared following EE treatments in Ishikawa cells. A. MTS was performed on day 3 following EE treatment (n = 3). B. Cells were labeled with CellTrace proliferation dye prior to treatment and then analyzed on a flow cytometer for proliferation at day 3 (n = 3). C. Cells were labeled with Hoechst dye and total fluorescence was analyzed using 9 fields of view on a Flex Station plate reader on day 3 following treatment (n = 3). For each assay, fold change over vehicle control (0 M EE) was calculated and the mean + SEM is shown (*≤ 0.05 Mann-Whitney statistical difference from vehicle control).
FIG. 7.
Comparison of 3 and 5 day endpoints for the Hoechst proliferation assay in EE treated Ishikawa cells. Ishikawa cells were treated across multiple doses of EE and Hoechst staining was performed at day 3 or 5 post treatment. Fold change in proliferation over vehicle control (0M EE) was calculated for each time and dose. Days 3 and 5 both represent the maximal proliferative dose response and are shown here as mean ± SEM. For each experiment, samples were run in triplicate (n = 3). For both time points, significance was reached at doses of 10 − 12M EE or higher (*≤ 0.05 Mann-Whitney statistical difference from vehicle control).
Evaluating Predictivity of Concentration-Response
Our goal in evaluating estrogen responses in Ishikawa cells was to provide a fit-for-purpose in vitro assay useful for predicting a surrogate point of departure (PoD) for uterine response to estrogenic stimulus. To accomplish this goal, the surrogate test system should have concentration-response characteristics consistent with those seen in intact tissue. Comparison of these two situations requires concentration-response information in both systems. Toward this end, we first evaluated the concentration-response for estrogens (EE and the endogenous ligand E2), the selective estrogen receptor modulator (SERM) tamoxifen (TAM) and its metabolite 4-hydroxytamoxifen (OHT) in Ishikawa cells (Fig. 8). As EE and E2 are potent agonists for this pathway, the concentration range tested for these compounds was from 10 − 8M through 10 − 13M at 1 log intervals. For TAM and OHT, the dose range was increased to span 10 − 6M through 10 − 11M in order to include the highest nontoxic doses in our analysis. Because EE and TAM are pharmaceuticals, and E2 is a well-studied endogenous estrogen involved in regulating the menstrual cycle, it was possible to compare the active in vitro concentrations to in vivo serum concentrations in women. EE and E2 were used to test the ability of the Ishikawa cells to identify activation via classical estrogenic compounds. TAM, and its potent metabolite OHT, tested the ability to identify a SERM with uterine-specific activity.
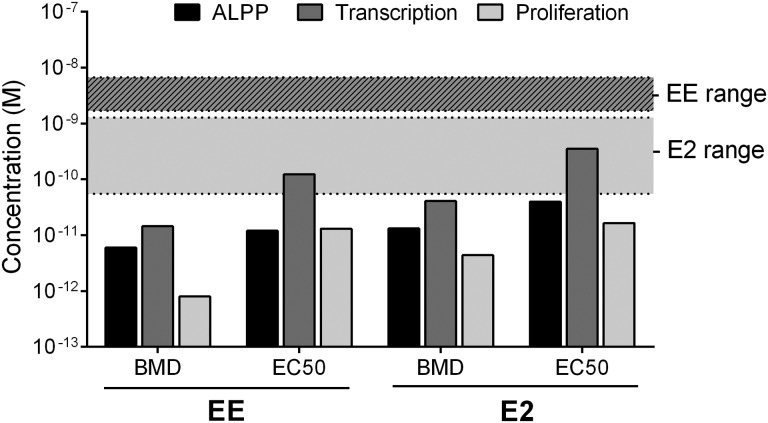
FIG. 8.
Assay sensitivity and physiologic relevance. The BMD and EC50 were calculated for each of the three end points assessed in Ishikawa cells, including ALPP enzyme activity (PNPP assay), transcriptional responses (PCR for PGR upregulation), and proliferation (Hoechst assay) following 3 day treatment with various concentrations of either EE or E2. Average circulating levels of E2 in healthy female adults and average circulating levels of EE in females taking standard oral EE at contraceptive doses from available publications are displayed as shaded ranges.
Comparing Active Estrogen Concentrations In vitro to In vivo Serum Concentrations
We compared dose-response parameters for enzyme activity (ALP), gene transcription (PGR), and proliferation endpoints (Hoechst) following estrogen treatment in Ishikawa cells to active concentrations in serum of healthy women of reproductive age. Biological activity may be evaluated in several different ways, including benchmark dose (BMD), concentration with 50% maximal activity (EC50), the lowest observed effect (LOEL), etc. Figure 8 shows a comparison of the BMD and EC50 for each in vitro endpoint following 3 days of EE or E2 treatment. In general, responses occurred in the low to mid-picomolar range. Reference ranges for steady state serum concentrations in women using EE as a contraceptive are 0.5 to 2 ng/ml with a range of 1.69 to 6.75 nM (Brody et al., 1989). The in vitro concentrations of EE required to initiate response in the Ishikawa cells were lower than these reported blood concentrations (Fig. 8). Because EE is used as a primary pharmacologically active component in oral contraceptives, it is reasonable that in vivo serum concentrations would be well above the lowest activating dose. These results provide evidence that the Ishikawa cell assay is sufficiently sensitive to identify estrogenic response in the human population.
To better compare the assay to normal response in vivo, in vitro concentrations of EE and E2 were also compared to published values for circulating estrogen (E2) in normal, premenopausal women (Fig. 8) (Mayo Clinic physician reference ranges). E2 and EE have similar potency for ER-mediated effects (Fig. 9), although they have different pharmacokinetic characteristics in vivo, due to a decreased clearance of EE (Dickson and Eisenfeld, 1981; Van den Belt et al., 2004). Estrogenic responses in the Ishikawa cells stimulated with EE or E2 occurred at or below active concentrations in healthy premenopausal women, further supporting the suitability and sensitivity of these Ishikawa cells for predicting the appropriate dose range of human responses to estrogens (MacNaughton et al., 1992; Reyes et al., 1977).
FIG. 9.
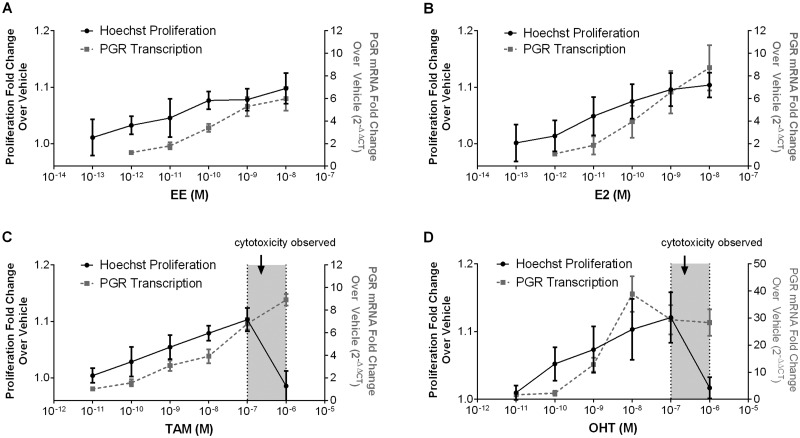
Estrogenic activity of pharmaceutical estrogen modulators in the Ishikawa proliferation assay. A-D. Ishikawa cells were treated with EE (A), E2 (B), Tamoxifen (C, TAM) or 4-Hydroxytamoxifen (D, OHT) at increasing doses. After 3 days, the Hoechst assay to determine changes in proliferation and RT-PCR for upregulation of PGR were performed. Fold change over control cells was calculated (n = 3). For TAM and OHT, cytotoxicity was determined by propidium iodide labeling and is shown as shaded regions along the x axis.
Using Ishikawa Cells to Derive In vitro-Based Points of Departure for Pharmaceutical Chemicals
To test whether the assay could provide accurate dose-response profiles for exogenous compounds with known SERM activity, we assessed TAM, a breast cancer therapeutic, and its active metabolite OHT in our Ishikawa assay. TAM and OHT inhibit ERα-mediated proliferation in breast tissue, but induce proliferation in uterine tissues (Barakat, 1995; Diel, 2002; Gallicchio et al., 2004). As pharmaceuticals, these chemicals have been subject to extensive clinical review and data on circulating levels is readily accessible, making them ideal candidates for comparisons of in vitro activity levels to in vivo effective doses. We ran a full dose response analysis for proliferation (Hoechst) and transcriptional responses (PGR) for EE, E2, TAM, and OHT, and monitored potential cytotoxic effects of compounds at high doses with PI labeling experiments (Fig. 9). Data are also available for the activity of these compounds in a suite of in vitro assays for endocrine disruptor screening under the EPA’s ToxCast initiative. With these results we can also compare results from the Ishikawa cells with those arising for the HTS endocrine disruptor screening efforts.
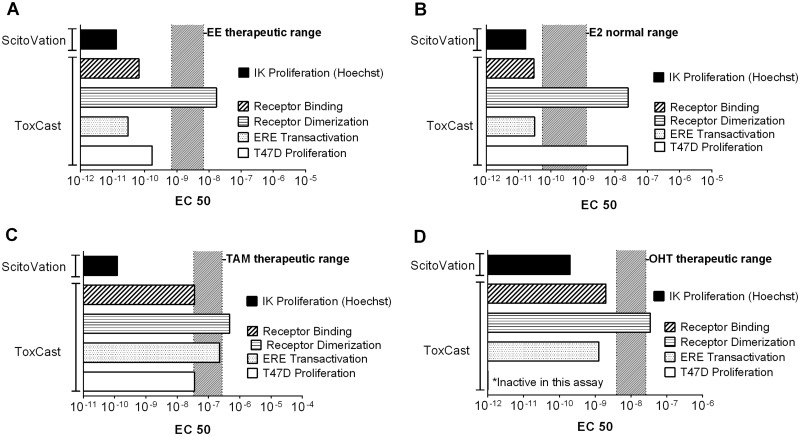
The proliferation data sets were used to calculate EC50 values that we then compared to the EC50 for four different ToxCast assays (Fig. 10). The ToxCast assays selected were the most sensitive assay (as determined by lowest EC50 values) from each of four categories; those measuring (1) ER binding, (2) ER dimer formation, (3) ERE-mediated transcriptional activity, and (4) cell proliferation in a breast cancer cell line (T47D). All data was taken from the latest version of the iCSS ToxCast Dashboard available at the time of publication (v2; https://www.epa.gov/chemical-research/toxicity-forecasting). The therapeutic range of serum concentrations for each compound is also overlaid onto the appropriate chemical doses. The range for EE is based on the measured circulating levels of serum EE in women taking a standard daily oral dose for contraceptive purposes (Brody et al., 1989). The range for E2 is derived from the normal serum levels for premenopausal adult females (Mayo Clinic Physician References). The ranges for TAM and OHT are serum levels measured in women taking the recommended therapeutic dose of 20 mg/day (Decensi et al., 1999; Gallicchio et al., 2004). OHT, the active metabolite of TAM, was positive in the Ishikawa proliferation assay, but not in the ToxCast breast cell proliferation assay. This discrepancy is consistent with its tissue–specific SERM activity. However, in general, the breast cell proliferation assay and other ToxCast assays were less sensitive than the Ishikawa proliferation assay for these chemicals, based on the EC50 values obtained from the iCSS Dashboard (v2). The EC50 values for the Ishikawa proliferation assay were below therapeutic ranges for all pharmaceutical compounds (Fig. 10) whereas the EC50 values for the four ToxCast assays were all higher, with several assay EC50s overlapping with the therapeutic ranges.
FIG. 10.
Comparison of EC50 values for estrogenic compounds across ToxCast assays or Ishikawa proliferation assay. A-D. For each compound, EC50s for our proliferation data (IK proliferation) were calculated and are shown for comparison with published EC50 values from the ToxCast suite of assays (version 2, http://actor.epa.gov/dashboard/). Therapeutic levels for each compound are shown as a hatched overlay. ToxCast assay identifications: T47D Proliferation (ACEA_T47D_80hr_Positive), ERE Transcription (OT_ERa_EREGFP_0120), Receptor Binding (NVS_NR_hER), Receptor Dimerization (OT_ER_ERaERa_0480).
DISCUSSION
Using an AOP Driven Approach to Select a Cell Model and Appropriate Endpoints for In vitro Assay Development
One of the goals of development of an Adverse Outcome Pathway or toxicity pathway is establishing a well-supported framework for informing in vitro assay design to increase the accuracy of animal-free risk assessment strategies. AOPs are not in themselves quantitative; they do not account for dose-response. However, if the AOP is sufficiently well-described as to include key events mediating the dose-response, an assay built on this framework should inherently account for the dose-dependence of cellular events. This paper describes our effort to use an AOP-type approach for developing an in vitro assay to serve as a surrogate for uterine proliferation (specifically, responses of the uterine epithelium), an outcome associated with prolonged exposure to estrogenic compounds and a potential precursor to uterine cancer.
The process described in this paper embodies a biology-driven approach, starting with curation of the current toxicology, biology, and medical literature to identify the key events in estrogenic signaling in the uterus—starting from initiating event (ligand–receptor binding) to downstream response (epithelial proliferation). Because the primary goal of this effort was fit-for-purpose in vitro assay development, we did not focus on individual or population level effects, but instead on defining key events driving cell type-specific cellular response. It is now clear that a suite of at least 5 receptors have specific roles in E2-mediated proliferative signaling and the coordinated interactions among these signaling pathways serve both to initiate and to limit proliferative responses in the uterus. In designing our assay, the markers measured in the various assays accounted for initial molecular interactions (ligand-receptor binding), transcriptional events, protein level response, the coordination of signaling among the various estrogen-responsive receptors and, ultimately, cellular response (Fig. 1). In organizing this cellular AOP for E2-mediated responses in vitro, we concluded that in order to most accurately recapitulate the key biology of this system, the cell model should: (1) represent the tissue type of interest, (2) express each of the key receptors and (3) initiate these key molecular events at physiologic concentrations of estrogens. The Ishikawa uterine epithelial cell line was chosen for consistency with these criteria.
Most other intact cell models used for estrogen screening are breast cell models. In ToxCast, the ACEA_T47D_80hr_Positive assay provides intact cell proliferation measurements using the T47D breast cancer cell line. The National Center for Toxicological Research (NCTR) of the Food and Drug Administration (FDA) has made potency data available for chemical endocrine activity through a database called the Endocrine Disruptor Knowledge Base (EDKB) (Ding et al., 2010). This resource includes such information as receptor binding, in vivo uterotrophic activity (rodent), and in vitro cellular responses. 160 relative potency calculations based on cell proliferation (RPP, relative proliferative potency) are documented in this system based on E-SCREEN, which measures estrogen-mediated proliferation in MCF-7 breast cells (Fang et al., 2000; Soto et al., 1995). In addition to the EDKB, the FDA has curated data from publically available data sets to create a larger database, the Estrogen Activity Database or EADB. This data is derived from 444 publications covering a wide range of chemical space. Of these, 107 studies reported cell proliferation in response to chemical treatment. The overwhelming majority of these experiments were performed using either MCF-7 cells or another estrogen-responsive breast cell line. Four of these studies reported results using Ishikawa cells but three were ALP induction (not direct proliferation measurements) and one reported inhibition of estradiol-induced proliferation by a specific pharmaceutical antagonist (Shen et al., 2013). Thus, we undertook an extensive validation of the Ishikawa as a model for uterine response.
Ensuring the In vitro Model Contains Appropriate Biology
The use of the Ishikawa cell line in testing for estrogenic activity has several advantages. It is commercially available; it has been genotyped for authenticity; it is the cell-type of interest (uterine epithelium); and it retains estrogen responsiveness across several passages. Our first task was to ensure that the cell line retained key biological responses associated with the estrogen signaling pathway. Importantly, these Ishikawa cells express all five endogenous estrogen receptors known to regulate proliferation (ERα66, ERα46, ERα36, ERβ, and GPER) and their expression is stable across several passages in vitro. The fidelity of the signaling network was evaluated by examining the effect of EE on several key events in the estrogen response pathway, including regulation of estrogen-mediated gene expression, induction of uterine-specific ALP enzyme activity, and changes in proliferation. Although some uncertainty exists with the use of a cancer cell line to predict normal tissue response, all of the tested endpoints displayed reproducible responses at doses in the picomolar to nanomolar range, which is consistent with in vivo measures of circulating estrogens in healthy premenopausal women. Further, each of key events in the proposed estrogen pathway were recapitulated by this cell model, indicating that Ishikawa cells are a reasonable model of uterine epithelium signaling. These findings support the use of Ishikawa cells to recapitulate endogenous estrogenic responses in vitro, though studies with normal human primary uterine cells would be a valuable next step in evaluating the system.
Evaluating Utility of the In vitro Model to Predict Concentration-Dependence of Chemical Response
In the effort to move towards in vitro-based chemical safety assessments, in vitro fit-for-purpose assays should recapitulate not only the phenotypic response of interest, but also the concentration-dependence of this response. Although current HTS efforts incorporate dose-response evaluations into the screening process, there is rarely an effort to ground-truth the estimates of chemical activity against behavior in the intact human. In this regard, there has been a large scale effort to test the specificity and selectivity of the EPA ToxCast assays to predict in vivo activity of estrogenic chemicals, comparing positive and negative responses in vitro to the OECD guideline in vivo studies, i.e., the rat uterotrophic assay (OECD, 2003). One challenge in this comparison is species-differences—the HTS tests use human cells and cell-free preparations, although the guideline standard is a rodent in vivo. A computational approach to use in vitro human cell-based and in vitro cell-free assay data to predict rodent in vivo responses has been undertaken (Browne et al., 2015). These studies were able to demonstrate a high degree of predictivity for classifying estrogenic vs. non-estrogenic compounds using reference chemicals and are able to rank compounds as strong or weak activators. The ability of these assays to quantitatively predict in vivo human concentration response has not yet been assessed, however. It is unclear whether any of these in vitro assays, either by themselves or in combination, are capable of providing quantitative chemical potency estimates that are consistent with those seen in the human population.
Here, we evaluated in vitro responses to (E2) and two pharmaceutical compounds (TAM/OHT, EE) where human in vivo data are available to compare in vitro and in vivo dose-response. Although the number of chemicals tested is small, the advantage of being able to compare results to the species and population of interest will be essential to development of reliable assays for use in safety assessment. There are many potential methods of in vitro in vivo extrapolation with varying degrees of complexity, each with associated advantages and limitations (Teeguarden et al., 2005; Yoon et al., 2015). In this case, we compared media concentrations to measured human serum concentrations to estimate consistency between the in vitro and in vivo concentration response. By evaluating responses to compounds with known estrogenic effects in the human and comparing both the concentration-response data from the Ishikawa assay and those from the publicly available ToxCast data to in vivo serum concentrations, we drew two important conclusions. First, the Ishikawa assay responded to estrogen at concentrations that are consistent with in vivo dosimetry. Second, our fit-for-purpose assay, incorporating the key molecular events for proliferation, had lower EC50s than those from the high throughput ToxCast assays. In fact, the EC50s derived from the ToxCast assays often overlapped with therapeutic serum concentrations. Our Ishikawa cell assay, with all the tested compounds (E2, EE, TAM and OHT), were well below concentration ranges representing their therapeutic levels.
There are some limitations in the currently available data that confound direct comparison of the ToxCast assays to our model, including limited dose ranges in the ToxCast data, which undermines confidence in EC50 (a.k.a. AC50) values and the small number of chemicals tested in our study (4 chemicals), which limits the number of chemicals for comparison. For a large scale effort such as ToxCast, it is not feasible to tailor dosing to each individual chemical’s potency, as it is in a study such as that presented here. Conversely, it is not feasible to perform the in depth analysis described here for thousands of compounds. Our current efforts are expanding the chemical repertoire of the Ishikawa assay to further test comparisons made here as that used for the ToxCast assays. Nonetheless, we contend that there is value to the broader perspective provided by ToxCast and to the deeper perspective presented here, and that insights gained from both efforts can help refine future testing strategies.
As the toxicity testing field moves forward with the transition to alternative testing strategies, there are several challenges that must be addressed. For instance, due to the large numbers of unregulated chemicals, higher throughput assays are necessary to evaluate likely biological responses across diverse toxicity pathways, such as estrogenic signaling. In addition, continued development of in vitro kinetics and consideration of metabolism in chemical activity estimates will be necessary to extrapolate from the cellular systems to expected dosimetry in the intact human. In our opinion, more extensive work will be needed to develop pathway-specific AOP frameworks and subsequent fit-for-purpose assays to provide sufficient coverage of biological space. Development of fit-for-purpose assays requires a systematic approach to ensure that the assay recapitulates specific characteristics of the pertinent biology. These assays may or may not be amenable to high throughput strategies. The goal is not high throughput by itself, but relevance of the measured endpoints for safety. Certainly any mature safety assessment strategy using only in vitro results will require a tiered approach with consideration of the necessary information for each tier, beginning with broader based screens with cell-based genomic evaluations and HTS assays for prioritization and identifying likely AOPs, and then moving towards more accurate fit-for-purpose approaches for determining points of departure. The case study described here shows the value in using AOP-structured approaches to increase the utility and predictivity of in vitro assays.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
ACKNOWLEDGEMENTS
The authors would like to thank Drs Kim Boekelheide and Marguerite Vantangoli of Brown University, Dr Gary Minsavage of ExxonMobil, and Dr Chad Deisenroth of ScitoVation for their scientific advice and valuable discussions.
FUNDING
This work was supported by the Long-Range Research Initiative (LRI) of the American Chemistry Council and was also performed as part of the Hamner Institutes’ “TT21C Consortium” with funding from Agilent, CropLife America, The Dow Chemical Company, Dow Corning, and ExxonMobil Biomedical Sciences Foundation.
REFERENCES
- Albert J. L., Sundstrom S. A., Lyttle C. R. (1990). Estrogen regulation of placental alkaline phosphatase gene expression in a human endometrial adenocarcinoma cell line. Cancer Res 50, 3306–3310. [PubMed] [Google Scholar]
- Attene-Ramos M. S., Miller N., Huang R., Michael S., Itkin M., Kavlock R. J., Austin C. P., Shinn P., Simeonov A., Tice R. R., et al. (2013). The Tox21 robotic platform for assessment of environmental chemicals - from vision to reality. Drug Discovery Today 18, 716–723. 10.1016/j.drudis.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansode F. W., Chauhan S. C., Makker A., Singh M. M. (1998). Uterine luminal epithelial alkaline phosphatase activity and pinopod development in relation to endometrial sensitivity in the rat. Contraception 58, 61–68. [DOI] [PubMed] [Google Scholar]
- Barakat R. R. (1995). The effect of tamoxifen on the endometrium. Oncology (Williston Park) 9, 129–134. discussion 139-40, 142. [PubMed] [Google Scholar]
- Bondesson M., Hao R., Lin C. Y., Williams C., Gustafsson J. A. (2014). Estrogen receptor signaling during vertebrate development. Biochim Biophys Acta 10.1016/j.bbagrm.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody S. A., Turkes A., Goldzieher J. W. (1989). Pharmacokinetics of three bioequivalent norethindrone/mestranol-50 micrograms and three norethindrone/ethinyl estradiol-35 micrograms OC formulations: are “low-dose” pills really lower? Contraception 40, 269–284. [DOI] [PubMed] [Google Scholar]
- Browne P., Judson R. S., Casey W. M., Kleinstreuer N. C., Thomas R. S. (2015). Screening chemicals for estrogen receptor bioactivity using a computational model. Environ. Sci. Technol.49, 8804–8814. 10.1021/acs.est.5b02641. [DOI] [PubMed] [Google Scholar]
- Daly R. J., Darbre P. D. (1990). Cellular and molecular events in loss of estrogen sensitivity in ZR-75-1 and T-47-D human breast cancer cells. Cancer Res. 50, 5868–5875. [PubMed] [Google Scholar]
- Decensi A., Gandini S., Guerrieri-Gonzaga A., Johansson H., Manetti L., Bonanni B., Sandri M. T., Barreca A., Costa A., Robertson C., et al. (1999). Effect of blood tamoxifen concentrations on surrogate biomarkers in a trial of dose reduction in healthy women. J. Clin. Oncol. 17, 2633–2638. [DOI] [PubMed] [Google Scholar]
- Deschenes J., Bourdeau V., White J. H., Mader S. (2007). Regulation of GREB1 transcription by estrogen receptor alpha through a multipartite enhancer spread over 20 kb of upstream flanking sequences. J. Biol. Chem. 282, 17335–17339. 10.1074/jbc.C700030200. [DOI] [PubMed] [Google Scholar]
- Dickson R. B., Eisenfeld A. J. (1981). 17 Alpha-ethinyl estradiol is more potent than estradiol in receptor interactions with isolated hepatic parenchymal cells. Endocrinology 108, 1511–1518. 10.1210/endo-108-4-1511. [DOI] [PubMed] [Google Scholar]
- Diel P. (2002). Tissue-specific estrogenic response and molecular mechanisms. Toxicol. Lett. 127, 217–224. [DOI] [PubMed] [Google Scholar]
- Ding D., Xu L., Fang H., Hong H., Perkins R., Harris S., Bearden E. D., Shi L., Tong W. (2010). The EDKB: an established knowledge base for endocrine disrupting chemicals. BMC Bioinformatics 11 Suppl 6, S5. 10.1186/1471-2105-11-s6-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H., Tong W., Perkins R., Soto A. M., Prechtl N. V., Sheehan D. M. (2000). Quantitative comparisons of in vitro assays for estrogenic activities. Environ. Health Perspect. 108, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filardo E. J., Quinn J. A., Bland K. I., Frackelton A. R., Jr. (2000). Estrogen-induced activation of Erk-1 and Erk-2 requires the G protein-coupled receptor homolog, GPR30, and occurs via trans-activation of the epidermal growth factor receptor through release of HB-EGF. Mol. Endocrinol. (Baltimore, Md.) 14, 1649–1660. 10.1210/mend.14.10.0532. [DOI] [PubMed] [Google Scholar]
- Filardo E. J., Thomas P. (2012). Minireview: G Protein-Coupled Estrogen Receptor-1, GPER-1: Its Mechanism of Action and Role in Female Reproductive Cancer, Renal and Vascular Physiology. Endocrinology 153, 2953–2962. 10.1210/en.2012-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flouriot G., Brand H., Denger S., Metivier R., Kos M., Reid G., Sonntag-Buck V., Gannon F. (2000). Identification of a new isoform of the human estrogen receptor-alpha (hER-α) that is encoded by distinct transcripts and that is able to repress hER-α activation function 1. EMBO J. 19, 4688–4700. 10.1093/emboj/19.17.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallicchio L., Lord G., Tkaczuk K., Danton M., Lewis L. M., Lim C. K., Flaws J. A. (2004). Association of tamoxifen (TAM) and TAM metabolite concentrations with self-reported side effects of TAM in women with breast cancer. Breast Cancer Res. Treat. 85, 89–97. 10.1023/B:BREA.0000021050.92539.b0. [DOI] [PubMed] [Google Scholar]
- Huang R., Sakamuru S., Martin M. T., Reif D. M., Judson R. S., Houck K. A., Casey W., Hsieh J. H., Shockley K. R., Ceger P., et al. (2014). Profiling of the Tox21 10K compound library for agonists and antagonists of the estrogen receptor alpha signaling pathway. Sci. Rep. 4, 5664. 10.1038/srep05664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H., Kobayashi M., Munetomo A., Miyamoto T., Sakuma Y. (2013). Novel splicing events and post-transcriptional regulation of human estrogen receptor alpha E isoforms. J. Steroid Biochem. Mol. Biol. 133, 120–128. 10.1016/j.jsbmb.2012.09.027. [DOI] [PubMed] [Google Scholar]
- Judson R. S., Houck K. A., Kavlock R. J., Knudsen T. B., Martin M. T., Mortensen H. M., Reif D. M., Rotroff D. M., Shah I., Richard A. M., et al. (2010). In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ. Health Perspect. 118, 485–492. 10.1289/ehp.0901392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlert S., Nuedling S., van Eickels M., Vetter H., Meyer R., Grohé C. (2000). Estrogen receptor α rapidly activates the IGF-1 receptor pathway. J. Biol. Chem. 275, 18447–18453. 10.1074/jbc.M910345199. [DOI] [PubMed] [Google Scholar]
- Klinge C. M., Riggs K. A., Wickramasinghe N. S., Emberts C. G., McConda D. B., Barry P. N., Magnusen J. E. (2010). Estrogen receptor alpha 46 is reduced in tamoxifen resistant breast cancer cells and re-expression inhibits cell proliferation and estrogen receptor alpha 66-regulated target gene transcription. Mol. Cell Endocrinol. 323, 268–276. 10.1016/j.mce.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen T. B., Houck K. A., Sipes N. S., Singh A. V., Judson R. S., Martin M. T., Weissman A., Kleinstreuer N. C., Mortensen H. M., Reif D. M., et al. (2011). Activity profiles of 309 ToxCast chemicals evaluated across 292 biochemical targets. Toxicology 282, 1–15, 10.1016/j. tox.2010.12.010. [DOI] [PubMed] [Google Scholar]
- Krewski D., Acosta D., Jr, Andersen M., Anderson H., Bailar J. C., 3rd, Boekelheide K., Brent R., Charnley G., Cheung V. G., Green S., Jr., et al. (2010). Toxicity testing in the 21st century: a vision and a strategy. J. Toxicol. Environ. Health B, Crit. Rev. 13, 51–138. 10.1080/10937404.2010.483176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Haynes M. P., Bender J. R. (2003). Plasma membrane localization and function of the estrogen receptor alpha variant (ER46) in human endothelial cells. Proc. Natl Acad. Sci. U S A. 100, 4807–4812. 10.1073/pnas.0831079100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A. H. Y., Li R. W. S., Ho E. Y. W., Leung G. P. H., Leung S. W. S., Vanhoutte P. M., Man R. Y. K. (2013). Differential ligand binding affinities of human estrogen receptor-α isoforms. PloS One 8, e63199. 10.1371/journal.pone.0063199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C. Y., Strom A., Li Kong S., Kietz S., Thomsen J. S., Tee J. B., Vega V. B., Miller L. D., Smeds J., Bergh J., et al. (2007). Inhibitory effects of estrogen receptor beta on specific hormone-responsive gene expression and association with disease outcome in primary breast cancer. Breast Cancer Res. 9, R25. 10.1186/bcr1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNaughton J., Banah M., McCloud P., Hee J., Burger H. (1992). Age related changes in follicle stimulating hormone, luteinizing hormone, oestradiol and immunoreactive inhibin in women of reproductive age. Clin. Endocrinol. (Oxf) 36, 339–345. [DOI] [PubMed] [Google Scholar]
- Maruyama T., Yoshimura Y. (2008). Molecular and cellular mechanisms for differentiation and regeneration of the uterine endometrium. Endocr. J. 55, 795–810. [DOI] [PubMed] [Google Scholar]
- Mohammed H., D'Santos C., Serandour A. A., Ali H. R., Brown G. D., Atkins A., Rueda O. M., Holmes K. A., Theodorou V., Robinson J. L., et al. (2013). Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 3, 342–349. 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naciff J. M., Khambatta Z. S., Thomason R. G., Carr G. J., Tiesman J. P., Singleton D. W., Khan S. A., Daston G. P. (2009). The genomic response of a human uterine endometrial adenocarcinoma cell line to 17alpha-ethynyl estradiol. Toxicol. Sci. 107, 40–55. 10.1093/toxsci/kfn219. [DOI] [PubMed] [Google Scholar]
- OECD (2003). Detailed Background Review of the Uterotrophic Bioassay: Summary of the Available Literature in Support of the Project of the OECD Task Force on Endocrine Disruptors Testing and Assessment (EDTA) to Standardise and Validate the Uterotrophic Bioassay. OECD 38, [Google Scholar]
- Omoto Y., Eguchi H., Yamamoto-Yamaguchi Y., Hayashi S. (2003). Estrogen receptor (ER) beta1 and ERbetacx/beta2 inhibit ERalpha function differently in breast cancer cell line MCF7. Oncogene 22, 5011–5020. 10.1038/sj.onc.1206787. [DOI] [PubMed] [Google Scholar]
- Penot G., Le Peron C., Merot Y., Grimaud-Fanouillere E., Ferriere F., Boujrad N., Kah O., Saligaut C., Ducouret B., Metivier R., et al. (2005). The human estrogen receptor-alpha isoform hERalpha46 antagonizes the proliferative influence of hERalpha66 in MCF7 breast cancer cells. Endocrinology 146, 5474–5484. 10.1210/en.2005-0866. [DOI] [PubMed] [Google Scholar]
- Petz L. N., Nardulli A. M. (2000). Sp1 binding sites and an estrogen response element half-site are involved in regulation of the human progesterone receptor A promoter. Mol. Endocrinol. (Baltimore, Md.) 14, 972–985. 10.1210/mend.14.7.0493. [DOI] [PubMed] [Google Scholar]
- Reif D. M., Martin M. T., Tan S. W., Houck K. A., Judson R. S., Richard A. M., Knudsen T. B., Dix D. J., Kavlock R. J. (2010). Endocrine profiling and prioritization of environmental chemicals using ToxCast data. Environ. Health Perspect. 118, 1714–1720. 10.1289/ehp.1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes F. I., Winter J. S., Faiman C. (1977). Pituitary-ovarian relationships preceding the menopause. I. A cross-sectional study of serum follice-stimulating hormone, luteinizing hormone, prolactin, estradiol, and progesterone levels. Am. J. Obstet. Gynecol. 129, 557–564. [PubMed] [Google Scholar]
- Rotroff D. M., Dix D. J., Houck K. A., Knudsen T. B., Martin M. T., McLaurin K. W., Reif D. M., Crofton K. M., Singh A. V., Xia M., et al. (2013). Using in vitro high throughput screening assays to identify potential endocrine-disrupting chemicals. Environ. Health Perspect. 121, 7–14. 10.1289/ehp.1205065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotroff D. M., Martin M. T., Dix D. J., Filer D. L., Houck K. A., Knudsen T. B., Sipes N. S., Reif D. M., Xia M., Huang R., et al. (2014). Predictive endocrine testing in the 21st century using in vitro assays of estrogen receptor signaling responses. Environ. Sci. Technol. 48, 8706–8716. 10.1021/es502676e. [DOI] [PubMed] [Google Scholar]
- Saceda M., Knabbe C., Dickson R. B., Lippman M. E., Bronzert D., Lindsey R. K., Gottardis M. M., Martin M. B. (1991). Post-transcriptional destabilization of estrogen receptor mRNA in MCF-7 cells by 12-O-tetradecanoylphorbol-13-acetate. J. Biol. Chem. 266, 17809–17814. [PubMed] [Google Scholar]
- Shen J., Xu L., Fang H., Richard A. M., Bray J. D., Judson R. S., Zhou G., Colatsky T. J., Aungst J. L., Teng C., et al. (2013). EADB: An Estrogenic Activity Database for Assessing Potential Endocrine Activity. Toxicol. Sci. 135, 277–291. 10.1093/toxsci/kft164. [DOI] [PubMed] [Google Scholar]
- Soto A. M., Sonnenschein C., Chung K. L., Fernandez M. F., Olea N., Serrano F. O. (1995). The E-SCREEN assay as a tool to identify estrogens: an update on estrogenic environmental pollutants. Environ. Health Perspect. 103, 113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamrazi A., Carlson K. E., Daniels J. R., Hurth K. M., Katzenellenbogen J. A. (2002). Estrogen receptor dimerization: ligand binding regulates dimer affinity and dimer dissociation rate. Mol. Endocrinol. (Baltimore, Md.) 16, 2706–2719. 10.1210/me.2002-0250. [DOI] [PubMed] [Google Scholar]
- Tong J. S., Zhang Q. H., Wang Z. B., Li S., Yang C. R., Fu X. Q., Hou Y., Wang Z. Y., Sheng J., Sun Q. Y. (2010). ER-alpha36, a novel variant of ER-alpha, mediates estrogen-stimulated proliferation of endometrial carcinoma cells via the PKCdelta/ERK pathway. PloS One 5, e15408. 10.1371/journal.pone.0015408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Belt K., Berckmans P., Vangenechten C., Verheyen R., Witters H. (2004). Comparative study on the in vitro/in vivo estrogenic potencies of 17beta-estradiol, estrone, 17alpha-ethynylestradiol and nonylphenol. Aquat. Toxicol. 66, 183–195. 10.1016/j.aquatox.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Vatin M., Bouvier S., Bellazi L., Montagutelli X., Laissue P., Ziyyat A., Serres C., De Mazancourt P., Dieudonne M. N., Mornet E., et al. (2014). Polymorphisms of human placental alkaline phosphatase are associated with in vitro fertilization success and recurrent pregnancy loss. Am. J. Pathol. 184, 362–368. 10.1016/j.ajpath.2013.10.024. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhang X., Shen P., Loggie B. W., Chang Y., Deuel T. F. (2005). Identification, cloning, and expression of human estrogen receptor-α36, a novel variant of human estrogen receptor-α66. Biochem. Biophys. Res. Commun. 336, 1023–1027. http://dx.doi.org/10.1016/j.bbrc.2005.08.226. [DOI] [PubMed] [Google Scholar]
- Wang Z., Zhang X., Shen P., Loggie B. W., Chang Y., Deuel T. F. (2006). A variant of estrogen receptor-{alpha}, hER-{alpha}36: transduction of estrogen- and antiestrogen-dependent membrane-initiated mitogenic signaling. Proc. Natl Acad. Sci. U S A. 103, 9063–9068. 10.1073/pnas.0603339103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. H., Chung T. D., Oldenburg K. R. (1999). A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J. Biomol. Screen 4, 67–73. [DOI] [PubMed] [Google Scholar]
- Zhao C., Putnik M., Gustafsson J. A., Dahlman-Wright K. (2009). Microarray analysis of altered gene expression in ERbeta-overexpressing HEK293 cells. Endocrine 36, 224–232. 10.1007/s12020-009-9233-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.