Abstract
Epigenetic mechanisms by which cells inherit information are, to a large extent, enabled by DNA methylation and posttranslational modifications of histone proteins. These modifications operate both to influence the structure of chromatin per se and to serve as recognition elements for proteins with motifs dedicated to binding particular modifications. Each of these modifications results from an enzyme that consumes one of several important metabolites during catalysis. Likewise, the removal of these marks often results in the consumption of a different metabolite. Therefore, these so-called epigenetic marks have the capacity to integrate the expression state of chromatin with the metabolic state of the cell. This review focuses on the central roles played by acetyl-CoA, S-adenosyl methionine, NAD+, and a growing list of other acyl-CoA derivatives in epigenetic processes. We also review how metabolites that accumulate as a result of oncogenic mutations are thought to subvert the epigenetic program.
Keywords: chromatin modification, methylation, acetylation, S-adenosyl methionine, folate, oncometabolite
INTRODUCTION
Epigenetics is a field with two rather different manifestations. The classical definition involves heritable phenotypic differences for which no underlying difference in the DNA sequence exists relevant to that phenotype. In more recent times, epigenetics has been used to define the collection of covalent modifications to DNA and posttranslational modifications to histones that influences the expression of genes and the structure of chromatin. The mechanisms behind the heritability of some of the modifications, such as DNA methylation, are clear. For other modifications, it is not clear whether they are heritable or require de novo additions after each cell cycle.
Nevertheless, the dozen or more known posttranslational modifications, and new ones discovered regularly, involve the consumption of a metabolite by the enzyme that catalyzes the modification. As such, changes in the metabolic state of the cell have the potential to affect modifications of chromatin in ways that change gene expression and, in some cases, have heritable consequences.
This review focuses on the metabolism of the cofactors that are used as substrates for DNA and histone modifications and on the emerging evidence for metabolites that inhibit these enzymes. A substantial portion of this review is dedicated to metabolic processes that affect DNA/histone methylation, histone acetylation, and other histone acylations. This is reflective of the current body of literature on the subject, in which studies on methylation and acetylation are the most numerous. We highlight what we believe are critical gaps in the field and offer some suggestions for experiments or measurements that will be particularly enlightening. Multiple excellent reviews address related issues treated lightly here (Ma & Vosseller 2014, Masri & Sassone-Corsi 2014, Venneti & Thompson 2013, Verdin & Ott 2015).
THE CAST OF CHARACTERS
The metabolites of focus in this review are S-adenosyl methionine (SAM), the universal donor for all epigenetic methylation reactions; acetyl-coenzyme A (acetyl-CoA), the donor for histone acetylation; nicotinamide adenine dinucleotide (NAD+), the cofactor consumed by a class of histone deacetylases (HDACs); multiple newly discovered acyl-CoA species; α-ketoglutarate, the cofactor along with FeII for chromatin demethylases; R-2-hydroxyglutarate (R-2HG, also called D-2HG), an oncometabolite and inhibitor of at least some demethylases; and the vitamins folic acid, vitamin C, and niacin.
CHROMATIN METHYLATION
The enzymes responsible for the methylation of DNA and histones all use SAM as the methyl-group donor. SAM, in turn, is made directly from methionine. Although humans and other organisms can synthesize methionine, humans cannot synthesize enough methionine to meet the needs for normal growth and function. Hence, methionine is one of the essential amino acids. The synthesis of methionine depends on acquiring a methyl group, whose origin traces back to dietary sources of folic acid, an essential vitamin (Figure 1).
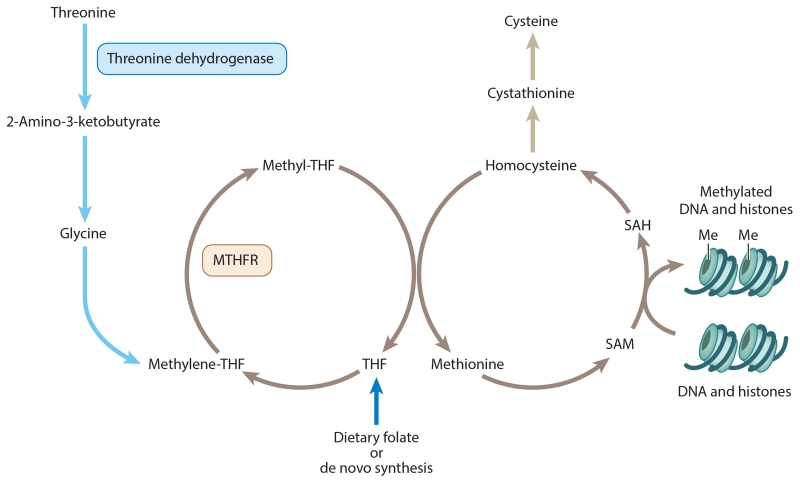
Figure 1.
The one-carbon pathway (brown arrows) is important for the production of methionine and cysteine, as well as the SAM used in DNA and histone methylation reactions. Several inputs into the pathway replenish metabolites consumed during one-carbon metabolism. In cells expressing threonine dehydrogenase, threonine is oxidized to produce glycine, which is converted to methylene-THF (blue arrows). Folate obtained through diet or by de novo synthesis is converted into THF. Abbreviations: Me, methyl; MTHFR, methylenetetrahydrofolate reductase; SAH, S-adenosyl homocysteine; SAM, S-adenosyl methionine; THF, tetrahydrofolate.
Folate: The Link Between Diet, Metabolism, and Epigenetics
Multiple well-controlled genetic studies have established the epigenetic consequences of disrupting the folate-SAM pathway in organisms ranging from yeast (Sadhu et al. 2013), Neurospora (Roberts & Selker 1995), and Arabidopsis (Baubec et al. 2010), to human cells (Friso et al. 2002, Sadhu et al. 2013). A rich epidemiological literature also describes the lasting and heritable consequences of extreme nutritional deprivation in human populations. These health consequences are hypothesized to reflect genes controlling nutrition being set in the greedy mode as a result of the deprived metabolic state, leading to predisposition for diseases such as diabetes (Hales & Barker 2001 and references therein). The potential for such heritable effects of nutrition on human health is important to understand. Indeed, 9% of the human population is homozygous for a hypomorphic mutation involving folate metabolism (MTHFR C677T), whose penetrance depends on the level of dietary folate (Friso et al. 2002, Yamada et al. 2001). In principle, these individuals may have some epigenetic lability when sources of folic acid are limited.
Methyl Donors and Heritable Phenotypes
One remarkable example of a link between diet and epigenetics is the effect of the level of dietary methyl donors on the phenotype caused by a mutation in mice known as agouti viable yellow (Avy). Avy is caused by the insertion of an endogenous retrotransposon near the 5′ end of the agouti gene and creates a coat-color phenotype that varies among individual mutants. The expression level of the Avy allele anticorrelates with the extent of DNA methylation of the retrotransposon; high methylation of CpG sites is associated with low expression and the agouti-colored coat, and low methylation of CpG sites is associated with high expression and a yellow-colored coat. The diet of the mother with respect to methyl-donor levels influences the phenotype of the offspring (Morgan et al. 1999, Wolff et al. 1998). Remarkably, the effect of the mutation on the coat color of a mouse is epigenetically inherited through the mother for several generations after restoration of methyl donors to the diet (Cropley et al. 2006). These observations, along with the heritability of DNA methylation patterns, suggest a simple model for transgenerational effects of diet: high levels of methyl donors in the diet support de novo DNA methylation, which is then maintained by maintenance DNA methyltransferases even when the diet no longer contains high levels of the methyl donors.
However, multiple studies cast doubt on whether DNA methylation is the heritable mark influencing Avy expression (Blewitt et al. 2006, Cropley et al. 2010). Histones also have the potential to carry epigenetic information through methylation, providing an alternative mechanism for dietary-induced epigenetic imprints. Indeed, the methylation levels of histone H4 at lysine (K) 20 vary between animals with different Avy phenotypes (Dolinoy et al. 2010). As appealing as this correlation appears, however, we know of no compelling demonstration of a causal link between histone methylation set by dietary levels of methyl donors and the Avy phenotype.
So what else could be the carrier of transgenerational impacts induced by previous nutritional states? One clue comes from the maternally limited influence of methyl donors on the phenotype of succeeding generations. The most obvious candidate is the microbiome, which is largely inherited from the mother. The potential impact of the maternal microbiome has not yet been adequately controlled for in studies of transgenerational maternal inheritance.
In principle, epigenetic inheritance could be realized through the impact of diet and metabolism on sperm. This possibility is now being explored (reviewed in Rando 2012), but clear evidence has been elusive.
Special Features of Stem Cell Metabolism and Methylation
A recent and surprising discovery of a new input to folate synthesis with potential epigenetic consequences stems from the unusual dependence of mouse embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) on threonine. In contrast to mouse fibroblasts, for example, mouse ESCs contain high levels of mitochondrial threonine dehydrogenase, which oxidizes threonine to drive the synthesis of acetyl-CoA and glycine. Glycine, in turn, can drive one-carbon metabolism by stimulating the synthesis of the methyl donors that ultimately result in elevated SAM levels (Figure 1) (Wang et al. 2009).
The induction of pluripotent stem cells alters the levels of many metabolites, but among those most changed are metabolites involved in threonine and SAM metabolism. In mouse iPSCs, the levels of cysteine, threonine, and folate are substantially reduced relative to the levels in noninduced cells. By contrast, SAM and cystathionine levels are relatively high in iPSCs (Shyh-Chang et al. 2013). The increase in cystathionine levels is curious, because cystathionine is one possible metabolite resulting from the salvage pathway for SAM after it donates a methyl group (Figure 1). The simultaneous elevation of SAM and cystathionine suggests both that the SAM flux must be high and that threonine metabolism is a major source of SAM in these cells. Histone H3 appears to be a major beneficiary of the enhanced methylation capacity of iPSCs: The level of H3K4me3 is high in induced pluripotent cells relative to the fibroblasts they are derived from (Shyh-Chang et al. 2013). Moreover, limitation of threonine in the culture medium dramatically reduces the levels of H3K4me3 and H3K4me2. Curiously, threonine limitation has no perceptible impact on the levels of H3K4me1, H3K9me3, H3K27me3, H3K36me3, or H3K79me3. How do reduced SAM pools resulting from threonine limitation have such a specific effect on particular modifications? Multiple mechanisms could account for these observations: the Km for a methyltransferase that produces H3K4me2,3 could be higher than the Km for other methyltransferases, the expression levels of particular methyltransferases could rise during iPSC generation, or other methylated species could turn over far less than the H3K4me2,3 species. Clearly, information on the dynamics of these modifications will be important.
Whether threonine is an important donor to the SAM supply in humans is not clear. The human threonine dehydrogenase gene is a transcribed pseudogene with a splice-site mutation and an in-frame nonsense mutation, as well as additional mutations in some individuals. Humans can catabolize threonine by an L-serine/threonine dehydratase (Edgar 2002). It will be interesting to determine whether that activity, especially in human ESCs, contributes to one-carbon metabolism.
CHROMATIN DEMETHYLATION
Given the significant roles of methylated histone residues and 5-methylcytosine (5mC) in the regulation of gene expression, much excitement arose from the discovery that methyl groups could be enzymatically removed from histones and DNA (He et al. 2011, Ito et al. 2011, Shi et al. 2004, Tahiliani et al. 2009). Interestingly, most of the enzymes that catalyze these reactions are FeII- and α-ketoglutarate–dependent dioxygenases.
Histone Demethylation
The largest class of histone demethylases consists of FeII- and α-ketoglutarate–dependent dioxygenases that contain the conserved Jumonji C (JmjC) protein domain, which was first identified in the Jumonji protein (JARID2) (Cloos et al. 2006, Takeuchi et al. 1995, Tsukada et al. 2006, Whetstine et al. 2006). The neural plates of mice lacking JARID2 display an abnormal cross-like morphology; hence the name Jumonji, which means cruciform in Japanese (Takeuchi et al. 1995). JmjC domain–containing histone demethylases target specific mono-, di-, and tri-methylated lysine residues in histones. It remains unclear whether JmjC domain–containing proteins target any of the methylated arginine (R) residues in histones; JMJD6 was proposed to demethylate H3R2me2 and H4R3me2, although subsequent studies have characterized JMJD6 as a lysyl hydroxylase (Chang et al. 2007, Mantri et al. 2011, Webby et al. 2009).
In the first step toward removal of a methyl group on a methylated lysine, JmjC domain–containing histone demethylases catalyze a hydroxylation reaction using ferrous iron as a cofactor and α-ketoglutarate (also called 2-oxoglutarate) as a cosubstrate (Figure 2a). The reaction begins with the oxidative decarboxylation of α-ketoglutarate to produce CO2 and succinate. This step is thought to form a highly reactive oxyferryl species (FeIV=O) that then hydroxylates the methyl-lysine. Release of the products completes the hydroxylation reaction and reduces the iron back to FeII, thus restoring the enzyme to a catalytically active state. The resulting hydroxymethyl-lysine is unstable and spontaneously releases formaldehyde to complete the process of demethylation.
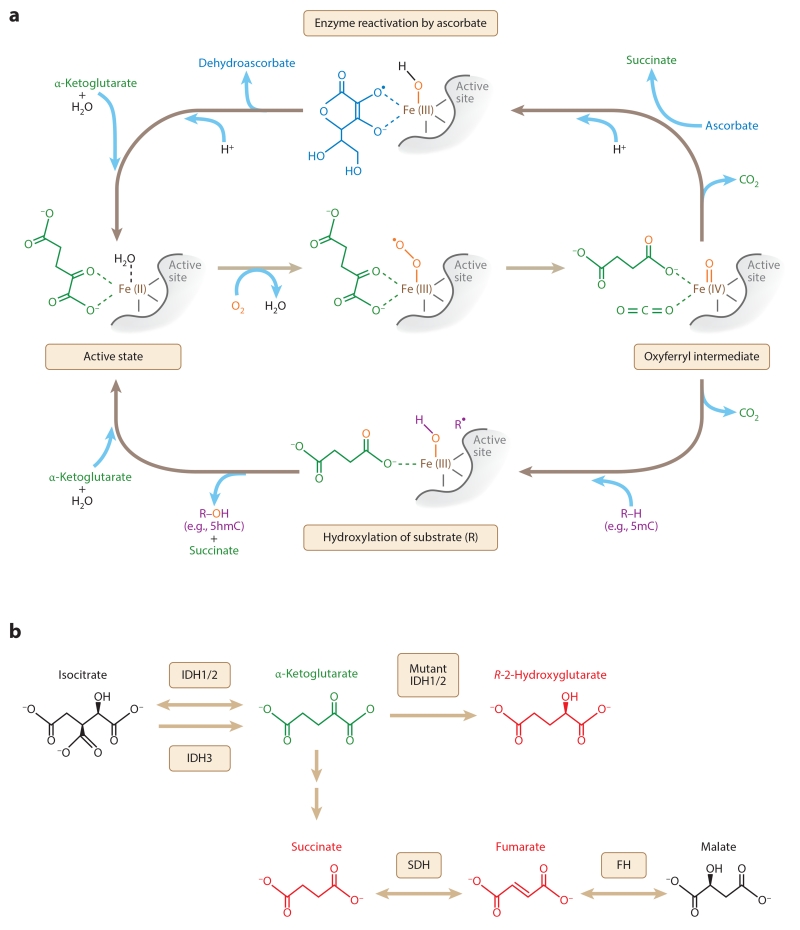
Figure 2.
Potential roles of ascorbate and oncometabolites in regulating the activity of FeII- and α-ketoglutarate–dependent dioxygenases. (a) Following the oxidative decarboxylation of α-ketoglutarate, the dioxygenase can regain enzyme activity either by hydroxylating a substrate such as 5mC or by oxidizing ascorbate. (b) Mutations in IDH, SDH, and FH genes can result in high levels of oncometabolites (red) known to inhibit FeII- and α-ketoglutarate–dependent dioxygenases competitively with respect to α-ketoglutarate (green). Abbreviations: 5hmC, 5-hydroxymethylcytosine; 5mC, 5-methylcytosine; FH, fumarate hydratase; IDH, isocitrate dehydrogenase; SDH, succinate dehydrogenase.
In addition to JmjC domain–containing histone demethylases, the amine oxidases KDM1A and KDM1B also demethylate histones (Shi et al. 2004). Rather than depending on FeII and α-ketoglutarate for enzyme activity, KDM1A/B require flavin adenine dinucleotide as a cofactor. The potential effects of metabolism on KDM1A/B activity remain largely unexplored.
DNA Demethylation
Ten-eleven translocation (TET) enzymes are FeII- and α-ketoglutarate–dependent dioxygenases that catalyze the hydroxylation of 5mC to produce 5-hydroxymethylcytosine (5hmC) (Tahiliani et al. 2009). The hydroxylation reaction is similar to the one described for JmjC domain–containing histone demethylases (Figure 2a). At least two possible mechanisms can account for the removal of the 5hmC intermediate (reviewed in Kohli & Zhang 2013). In the first mechanism, 5hmC is passively removed through DNA replication. In the second mechanism, 5hmC undergoes additional rounds of TET-mediated oxidation to form 5-formylcytosine and 5-carboxylcytosine, which can then be removed through base excision repair. In addition to potentially serving as an intermediate in the demethylation process, 5hmC itself may be a functional modification (reviewed in Hahn et al. 2014).
Ascorbate as a Cofactor for Demethylase Enzymes
Ascorbate, more commonly known as vitamin C, can enhance the activity of JmjC domain–containing histone demethylases, TET oxidases, and various other FeII- and α-ketoglutarate–dependent dioxygenases. The underlying mechanism, which may vary among different dioxygenases, is best understood for prolyl 4-hydroxylase (P4H); therefore, we discuss the details of the mechanism for P4H before turning to the connection between ascorbate and chromatin.
P4H stabilizes the triple-helical structure of collagen and requires ascorbate for maximal activity. Interestingly, primates, guinea pigs, and some bat species have lost the ability to synthesize ascorbate as a result of mutations in the gene encoding L-gulonolactone oxidase and therefore depend solely on dietary ascorbate. Severe ascorbate deficiency in these species is associated with collagen defects that arise largely from the impairment of P4H and other related FeII- and α-ketoglutarate–dependent dioxygenases.
Ascorbate has the capacity to bind P4H directly and reduce the iron center in its oxidized form. Whereas purified P4H does not require ascorbate to catalyze hydroxylation at a maximal rate for the first 15–30 reaction cycles, it does require ascorbate to maintain full activity thereafter (Myllylä et al. 1978). In agreement with the kinetic data, ascorbate functions to reactivate P4H molecules that have catalyzed an alternative reaction, whereby oxidative decarboxylation of α-ketoglutarate occurs without the subsequent hydroxylation of proline and leaves the iron center in an oxidized state (Figure 2a) (de Jong et al. 1982; Myllylä et al. 1978, 1984; Puistola et al. 1980). This uncoupled reaction can build up a pool of inactive enzymes unless ascorbate is present to reduce the iron center back to FeII. Ascorbate likely forms a direct interaction with the enzyme-bound iron (Majamaa et al. 1986), and its role in stimulating P4H is highly specific, as alternative reducing agents do not accelerate hydroxylation activity to a large degree (Myllylä et al. 1978, Tuderman et al. 1977).
Little is known about the mechanism through which ascorbate stimulates JmjC domain–containing histone demethylases and TET oxidases. Ascorbate may reactivate these enzymes following the uncoupled turnover of α-ketoglutarate, as it does for P4H, but this possibility has not been formally tested. In a different mechanism proposed for TET2 stimulation, ascorbate binds the catalytic domain and thereby induces a structural change that promotes catalytic activity (Yin et al. 2013). In principle, ascorbate could also modulate activity by increasing cellular uptake of iron or by helping to maintain intracellular iron in the reduced state, assuming that FeII availability were limiting for the enzyme. Ultimately, ascorbate may promote dioxygenase activity through multiple mechanisms in vivo.
Ascorbate in Somatic Cell Reprogramming
Whereas the significance of ascorbate in collagen synthesis was established decades ago, evidence linking ascorbate to the epigenetic regulation of gene expression has only recently surfaced. Notably, ascorbate can help reprogram mouse embryonic fibroblasts (MEFs) into iPSCs (Esteban et al. 2010). Other antioxidants do not mediate this effect, suggesting that ascorbate acts primarily through mechanisms other than the scavenging of reactive oxygen species. Given that iPSC generation involves a genome-wide reorganization of chromatin (reviewed in Apostolou & Hochedlinger 2013), it seems likely that ascorbate promotes pluripotency at least in part by enhancing the activity of chromatin modifiers.
Interestingly, ascorbate treatment of MEFs reduces total levels of H3K36 methylation through a mechanism dependent on KDM2A and/or KDM2B, two closely related H3K36 demethylases that contain JmjC domains and require ascorbate for maximal activity in vitro (Tsukada et al. 2006, Wang et al. 2011). Ascorbate treatment depends on KDM2A/B to reduce both H3K36me2 and H3K36me3, even though KDM2A/B specifically prefer the dimethyl form (He et al. 2008, Tsukada et al. 2006, Wang et al. 2011); therefore, either KDM2A/B gain the ability to target H3K36me3 in this context, or the observed effect is indirect. Ascorbate and KDM2B both promote cell proliferation and induce changes in gene expression during reprogramming, although it is unclear whether they act through the same mechanism (Liang et al. 2012, Wang et al. 2011).
Ascorbate also converts partially reprogrammed cells called pre-iPSCs into fully reprogrammed iPSCs (Esteban et al. 2010). In pre-iPSCs, transcription factors fail to bind and activate the expression of genes required for pluripotency (Sridharan et al. 2009). Ascorbate helps transition pre-iPSCs to a pluripotent state in part through the reduction of H3K9 methylation, a repressive modification that occupies pluripotency-associated loci during the early stages of reprogramming (Chen et al. 2013b, Soufi et al. 2012). Individual knockdowns of various JmjC domain–containing H3K9 demethylases impair the ascorbate-mediated conversion of pre-iPSCs to iPSCs, suggesting that ascorbate normally cooperates with these enzymes to remove H3K9 methylation and thereby induce pluripotency (Chen et al. 2013b).
Ascorbate affects the methylation status of DNA as well. Removing ascorbate from serum-free culture medium causes human ESCs to gain DNA methylation near the promoters of almost 2,000 genes (Chung et al. 2010). The mechanism underlying this effect is unknown, although ascorbate has been shown to mediate DNA demethylation in both mouse ESCs and MEFs through the activity of TET enzymes (Blaschke et al. 2013, Dickson et al. 2013, Minor et al. 2013, Yin et al. 2013). Ascorbate treatment of mouse ESCs, which are commonly cultured without ascorbate, leads to a gradual decrease in 5mC and a transient increase in 5hmC at gene promoters, as well as increased expression of germline-associated genes in particular (Blaschke et al. 2013, Yin et al. 2013). Adding ascorbate to the culture medium also increases total levels of 5-formylcytosine and 5-carboxylcytosine, which TET enzymes produce through sequential rounds of 5mC oxidation (Yin et al. 2013). These ascorbate-induced effects require the presence of TET1 and TET2, which are both stimulated by ascorbate in vitro (Blaschke et al. 2013, Yin et al. 2013).
α-Ketoglutarate can induce some of the same effects as ascorbate in mouse ESCs (Carey et al. 2015). Specific kinase inhibitors that help maintain ESCs in a näive pluripotent state rewire glucose and glutamine metabolism in ways that produce a high ratio of α-ketoglutarate/succinate, suggesting that α-ketoglutarate may be important for pluripotency. Indeed, treating mouse ESCs with cell-permeable α-ketoglutarate is sufficient to promote pluripotency in the absence of the kinase inhibitors. Consistent with its role as a cosubstrate for histone demethylases and TET enzymes, α-ketoglutarate reduces total levels of H3K27me3, H4K20me3, and 5mC and induces expression of germline genes in a TET1/2-dependent manner. α-Ketoglutarate may be a limiting factor for TET oxidases in vivo, as injecting mice with various α-ketoglutarate precursors increases 5hmC levels in the liver (Yang et al. 2014).
With respect to iPSC generation, the interplay between ascorbate and TET activity is complex and highlights the need to further investigate the precise mechanisms through which ascorbate alters the epigenetic landscape of chromatin (Chen et al. 2013a). Such a task will require teasing apart the effects of ascorbate on DNA methylation and histone methylation, as these modifications can influence each other (reviewed in Rose & Klose 2014). Case in point, ascorbate affects DNA methylation levels at the Dlk1-Dio3 locus through the maintenance of histone modifications that antagonize the recruitment of DNMT3A, a de novo DNA methyltransferase (Stadtfeld et al. 2012). As an added complication, TET oxidases may regulate gene expression indirectly through recruitment of the histone methyltransferase PRC2 or the O-linked β-N-acetylglucosamine transferase OGT (Chen et al. 2013c, Deplus et al. 2013, Wu et al. 2011).
Importantly, the effects described here are all induced by physiologically relevant concentrations of ascorbate—in humans and mice, ascorbate concentrations in the plasma are approximately 50 μM, whereas brain tissue contains up to millimolar levels (Hornig 1975, Rice & Russo-Menna 1997, Sotiriou et al. 2002). However, the Km values for ascorbate must be measured for JmjC domain–containing histone demethylases and TET oxidases to determine whether it is plausible for ascorbate to directly stimulate these enzymes in vivo. To date, little evidence exists that ascorbate alters the levels of DNA methylation or histone methylation in organisms as opposed to cultured cells (Yin et al. 2013). Therefore, our understanding of ascorbate and its significance in the epigenetic regulation of gene expression will benefit from mechanistic and biochemical studies, as well as studies using organisms deficient in ascorbate synthesis.
Oncometabolites as Inhibitors of Demethylases
In addition to serving as a cosubstrate for more than 60 dioxygenases, α-ketoglutarate plays key roles in amino acid synthesis, nitrogen transport, and the TCA cycle. Mutations that dysregulate these metabolic pathways, particularly in ways that produce high levels of metabolites known to inhibit FeII- and α-ketoglutarate–dependent dioxygenases, are prevalent in certain types of cancers. We discuss the potential roles of FeII- and α-ketoglutarate–dependent dioxygenases in cancer progression as they relate to chromatin.
Loss-of-function mutations in genes encoding subunits of the succinate dehydrogenase (SDH) complex, which converts succinate to fumarate in the TCA cycle and the electron transport chain (Figure 2b), occur frequently in paraganglioma, pheochromocytoma, gastrointestinal stromal tumor (GIST), and renal carcinoma. As a result of SDH inactivity, these tumors accumulate up to millimolar concentrations of succinate, a product of the reactions catalyzed by FeII- and α-ketoglutarate–dependent dioxygenases (Pollard et al. 2005). Studies in vitro and in HEK293T cells show that at such high concentrations, succinate competes with α-ketoglutarate to inhibit the activity of multiple FeII- and α-ketoglutarate–dependent dioxygenases, including TET1, TET2, and the JmjC domain–containing histone demethylases KDM4A, KDM4D, and KDM4DL (Rose et al. 2008, 2011; Smith et al. 2007; Xiao et al. 2012).
Recently, more than 100 pheochromocytoma and paraganglioma samples were clustered into three groups based on genome-wide measurements of DNA methylation; in the one group that exhibited hypermethylation at CpG sites, 16 of the 17 tumors contained mutations in SDH genes, whereas the other two groups were associated with mutations in other known susceptibility genes (Letouzé et al. 2013). Interestingly, the only tumor that lacked any SDH mutations in the hypermethylated group instead carried inactivating mutations in the gene encoding fumarate hydratase (FH), which converts fumarate to malate (Figure 2b). A similar trend was observed in SDH-deficient GISTs, which exhibit widespread hypermethylation at CpG sites in comparison with GISTs driven by alternative mutations (Killian et al. 2013). Thus, aberrantly high levels of DNA methylation in paragangliomas, pheochromocytomas, and GISTs are largely unique to SDH-mutated tumors.
Loss of SDH activity is a likely cause of the hypermethylation phenotype, as disrupting SDH function in cultured cells or in mice increases DNA methylation levels, particularly at genes subject to hypermethylation in SDH-deficient tumors (Letouzé et al. 2013, Xiao et al. 2012). Histone methylation also increases in response to SDH inactivation, and in at least some cases, treatment with α-ketoglutarate reverses this effect (Cervera et al. 2009, Smith et al. 2007, Xiao et al. 2012). Altogether, these observations indicate that succinate accumulation in SDH mutants inhibits the activity of TET oxidases and JmjC domain–containing histone demethylases. Likewise, fumarate has been suggested to competitively inhibit these enzymes in tumors containing loss-of-function mutations in the gene encoding FH (Xiao et al. 2012). It is plausible that these effects promote tumorigenesis, given that TET oxidases and histone demethylases have known roles in tumor suppression (reviewed in Kaelin 2011, Losman & Kaelin 2013).
R-2HG is another relevant oncometabolite that accumulates in tumors containing neomorphic mutations in the genes encoding isocitrate dehydrogenase (IDH)1 or IDH2 (Dang et al. 2009, Gross et al. 2010, Ward et al. 2010). IDH1 and IDH2 normally catalyze the interconversion of isocitrate and α-ketoglutarate; however, a single amino acid change at R100 or R132 in the active site of IDH1, or at the analogous residues in IDH2, disfavors the forward reaction by reducing the binding affinity for isocitrate and alters the reverse reaction to produce R-2HG instead of isocitrate (Figure 2b). Mutations that confer these changes frequently occur in gliomas and acute myelogenous leukemia (AML) and produce R-2HG levels ranging from approximately 1 mM to 10 mM (Dang et al. 2009, Gross et al. 2010, Ward et al. 2010). By contrast, the R-2HG concentration in normal cells is less than 100 μM (Losman & Kaelin 2013). Neomorphic IDH1/2 mutations likely promote oncogenesis through R-2HG accumulation, as IDH1 R132H expression and R-2HG treatment are individually sufficient to induce transforming effects in cultured hematopoietic cells (Losman et al. 2013).
At concentrations relevant to IDH1/2-mutated tumors, R-2HG competes with α-ketoglutarate to inhibit the activity of TET oxidases and JmjC domain–containing histone demethylases in vitro (Chowdhury et al. 2011, Lu et al. 2012, Xu et al. 2011). R-2HG may also inhibit these enzymes in vivo, as introducing mutant IDH1/2 alleles into cultured human cells induces a global increase in DNA methylation and histone methylation (Duncan et al. 2012, Figueroa et al. 2010, Lu et al. 2012, Turcan et al. 2012, Xu et al. 2011). Much like SDH-mutated tumors and their counterparts, IDH1/2-mutated gliomas exhibit a distinct signature of DNA hypermethylation in comparison with non-IDH1/2-mutated gliomas (Figueroa et al. 2010, Noushmehr et al. 2010).
The striking observation that loss-of-function TET2 mutations and neomorphic IDH1/2 mutations are common but mutually exclusive in AML suggests that TET2 inactivation and R-2HG accumulation promote oncogenesis through the same mechanism (Figueroa et al. 2010, Gaidzik et al. 2012, Weissmann et al. 2012). However, drug-mediated inhibition of IDH1 R132H attenuates the proliferation of IDH1-mutated glioma cells without largely affecting DNA methylation levels (Rohle et al. 2013). Furthermore, the S enantiomer of 2HG is a potent inhibitor of TET oxidases and JmjC domain–containing histone demethylases (Xu et al. 2011), yet S-2HG treatment of cultured hematopoietic cells is not sufficient to induce transforming effects (Losman et al. 2013). Thus, the role of TET enzymes in R-2HG–mediated oncogenesis remains unclear, partly owing to the complication that R-2HG accumulation also exerts non-epigenetic effects (reviewed in Adam et al. 2014).
CHROMATIN ACETYLATION
Acetyl-CoA is a key metabolic intermediate in both catabolic and anabolic processes. Both glycolysis and β-oxidation of fatty acids break down hydrocarbons to supply the TCA cycle with acetyl-CoA and thereby fuel cellular respiration. Anabolic pathways essential for cellular growth consume acetyl-CoA for the production of lipids, cholesterol, and amino acids. Beyond these metabolic transactions, acetyl-CoA serves as the donor of the acetyl moiety for reactions catalyzed by acetyltransferase enzymes. Protein acetylation is a major posttranslational modification within the cell, comparable in scale to phosphorylation and ubiquitination with respect to the number of substrates (Choudhary et al. 2009). Recent work demonstrates a direct link between acetyl-CoA levels and protein acetylation. This section focuses on the impact of acetyl-CoA metabolism on the acetylation of histone residues.
The first-described example of acetylation as a posttranslational modification was the acetylation of lysine residues on histone tails isolated from calf thymus nuclei (Allfrey et al. 1964). In this landmark study, the authors speculated that acetylation of histone tails could serve as a mechanism to regulate transcription and “allow a means of switching-on or -off RNA synthesis at different times, and at different loci of the chromosomes” (Allfrey et al. 1964, p. 792). This remarkably prescient statement foreshadowed the explosion of research in the field of epigenetics. The conditional acetylation of histones determines, in part, the compaction state of chromatin and organizes chromatin into distinct euchromatic and heterochromatic domains. At physiological pH values, lysine residues within the tail region of histones are positively charged, and acetylation is thought to neutralize the charge of these residues and thereby disrupt the electrostatic interaction between histones and DNA (Hong et al. 1993). Acetylation of histone tails also modulates interactions with neighboring nucleosomes and with certain chromatin-regulating proteins (Hecht et al. 2015, Luger et al. 1997, Wolffe & Hayes 1999). Additionally, histone acetyl marks serve as conditional docking sites for proteins containing bromodomains, which recognize and bind to acetyl-lysines (Dhalluin et al. 1999, Hassan et al. 2002, Jacobson et al. 2000). These acetyl marks are reversible through the activity of three different classes of HDACs, one of which consumes NAD+ in the process (see below), enabling the possibility for spatiotemporal regulation of the interactions between bromodomain-containing proteins and chromatin.
Acetyl-CoA Metabolism Is Linked to Histone Acetylation
A recurring theme within this review is the apparent reorganization of chromatin in response to changes in cellular levels of intermediary metabolites. In principle, changes in chromatin modifications could result through regulation of the enzymes that place the modification or through changes in the level of a metabolite that serves as the donor for that modification. In the instance of histone acetylation, it appears that the acetyl-CoA level fluctuates to the extent that, under certain conditions, it becomes limiting for acetyltransferase activity. Acetyl-CoA is thought to freely diffuse through the nuclear pore complex, and altering the pool of available acetyl-CoA in the cytoplasm or nucleus can cause changes in histone acetylation. The experimental inactivation of acetyl-CoA synthetases, the major source of cytoplasmic and nuclear acetyl-CoA, leads to a drop in bulk histone acetylation in Saccharomyces cerevisiae and cultured mammalian cells (Takahashi et al. 2006, Wellen et al. 2009). By contrast, histone acetylation in S. cerevisiae increases in response to the decreased expression of acetyl-CoA carboxylase (Acc1), the enzyme that converts nucleocytosolic acetyl-CoA to malonyl CoA in the first step of de novo fatty acid synthesis (Galdieri & Vancura 2012). Acc1p competes with histone acetyltransferases for the limited pool of nuclear acetyl-CoA. Thus, a decrease in Acc1p activity results in greater availability of acetyl-CoA for histone acetylation (Galdieri & Vancura 2012). These studies demonstrate that histone acetylation can be regulated by physiologically relevant changes in the level of nucleocytosolic acetyl-CoA. One of the current challenges in the field is to understand the conditions that cause acetyl-CoA levels to fluctuate and whether fluctuations can trigger a regulatory response aimed at adapting to changes in cellular and environmental conditions.
Physiological Fluctuations of Acetyl-CoA Levels
The best evidence of physiological fluctuations in acetyl-CoA levels comes from studies of the yeast metabolic cycle, in which, under glucose-limited conditions, prototrophic yeast cells oscillate through three distinct metabolic phases with regular periodicity (Tu et al. 2005). The yeast metabolic cycle consists of an oxidative-respiratory phase during which high levels of oxygen are consumed in cellular respiration, a reductive-building phase marked by the expression of genes important for mitochondrial function and cell division, and a reductive-charging phase in which genes important for nonrespiratory modes of energy production are upregulated. Acetyl-CoA is among the most dynamic of the tested metabolites across the yeast metabolic cycle (Cai et al. 2011, Tu et al. 2007). Acetyl-CoA levels peak during the oxidative-respiratory phase, and bulk acetylation of specific lysine residues on histones H3 and H4 shows a strong periodicity that coincides with acetyl-CoA levels in the cell (Cai et al. 2011).
Adding various carbon sources to cultures during the reductive phases of the metabolic cycle rapidly breaks its normal periodicity. Cells immediately enter the oxidative-respiratory phase and begin dividing (Cai et al. 2011). This forced entry into the oxidative-respiratory phase of the metabolic cycle is accompanied by an increase in histone acetylation that correlates with acetyl-CoA levels. Similarly, adding glucose to stationary-phase yeast cultures causes a burst of histone acetylation that is dependent on the histone acetyltransferases Gcn5 and Esa1 (Friis et al. 2009). In both cases, acetyl-CoA levels increase two-to fourfold following the addition of different carbon sources, with a corresponding increase in histone acetylation. The picture resulting from these studies is that carbon metabolism drives acetyl-CoA production, which in turn impacts the extent of histone acetylation within the nucleus. The marked difference in acetylation levels from one position in the genome to another appears to result in some cases from competition between acetyltransferase and deacetylase enzymes or from differences in the accessibility of large protein complexes such as histone acetyltransferases to some regions of the genome.
CHROMATIN DEACETYLATION
Sirtuins Link NAD+ Metabolism to Histone Deacetylation
NAD+ is a cofactor used to carry electrons in numerous metabolic redox reactions. In addition, NAD+ is involved in processes including calcium signaling (Guse 2015), ADP-ribosylation (Honjo et al. 1968, Schreiber et al. 2006), and protein deacetylation (Imai et al. 2000, Landry et al. 2000, Smith et al. 2000). NAD+-dependent protein deacetylation is unique to a group of proteins collectively known as sirtuins, named after the budding yeast protein Sir2 (Rine & Herskowitz 1987). Most of the sirtuins identified to date are capable of removing acetyl marks from histone tails. Other classes of HDACs do not require NAD+ or any other metabolic cofactor. Hence, the NAD+ dependence of sirtuins provides a potentially important link between metabolism and the epigenetic regulation of gene expression. Analogous to the coupling of acetyl-CoA levels and histone acetyltransferase activity, changes in NAD+ metabolism could impact sirtuin-mediated deacetylation of histones and other targets. In this way, the acetylation status of histones could provide a form of molecular memory of the recent physiological state of the cell (Downey et al. 2015). Moreover, this potential form of memory has the capacity to control the expression of genes to maintain a physiological state when favorable, or adjust the state if needed (Downey et al. 2013).
Work on the budding yeast protein Sir2 has been paradigmatic for sirtuin studies. The NAD+-dependent histone deacetylation activity of Sir2 is required for transcriptional silencing at the HML and HMR loci, at rDNA, and at telomeres. In addition to Sir2, budding yeast have four other sirtuins (Hst1, Hst2, Hst3, Hst4) that also have histone deacetylation activity. Sirtuins are conserved across all domains of life, and seven different sirtuins have been identified in mammals. In this section, we consider work that illuminates how different aspects of metabolism can impact sirtuin activity and lead to epigenetic consequences in gene expression.
NAD+ Metabolism and the Function of Sir2
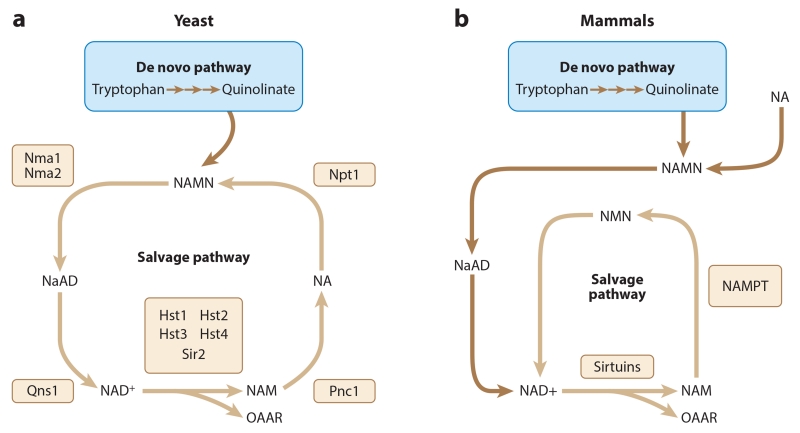
NAD+ is consumed during ADP-ribosylation and deacetylation reactions. The deacetylation reaction performed by Sir2 hydrolyzes one molecule of NAD+ to produce O-acetyl-ADP-ribose (OAAR) and nicotinamide. Nicotinamide is reported to be a noncompetitive inhibitor of Sir2 and human SIRT1 (Avalos et al. 2005, Bitterman et al. 2002). Conversely, under different assay conditions, nicotinamide has also been reported to competitively inhibit the deacetylation activity of SIRT1 and SIRT2 (Marcotte et al. 2004). Eukaryotic cells have two metabolic pathways for NAD+ synthesis (Figure 3). Tryptophan serves as a precursor for de novo synthesis of NAD+. Alternatively, nicotinamide, produced from sirtuin-based deacetylation or ADP-ribosylation, is utilized in a salvage pathway for NAD+ synthesis. In addition, some NAD+ precursors can be taken up from the medium and incorporated into the NAD+ salvage pathway. Studies in budding yeast demonstrate the link between NAD+ metabolism and sirtuin function. Mutation of NPT1, which encodes the NAD+ salvage pathway enzyme nicotinate phosphoribosyltransferase, leads to a two- to threefold drop in NAD+ levels and consequent losses in rDNA and telomeric silencing (Sandmeier et al. 2002, Smith et al. 2000). These effects result from the loss of Sir2 function, which in this case appears to be caused by the limitation of available NAD+ in npt1 mutants. By contrast, mutations in the de novo NAD+ synthesis pathway do not impair silencing, nor do they lower NAD+ levels. Apparently, at least under laboratory conditions, the NAD+ salvage pathway generates sufficient NAD+ to support Sir2 function. Consistent with this model, Npt1 and Nma2 of the NAD+ salvage pathway show strong nuclear localization, whereas proteins involved in the de novo pathway are evenly distributed throughout the cell (Anderson et al. 2002, Sandmeier et al. 2002).
Figure 3.
Metabolic pathways used for synthesis of NAD+ in yeast and mammals. (a) The de novo NAD+ synthesis pathway converts tryptophan to quinolinate (blue box), which is then converted into NAMN, a precursor to NAD+. The NAD+ salvage pathway (light brown arrows) converts NAM, one of the by-products of sirtuin-mediated deacetylation, back into NAD+. Yeast converts NAM into NAD+ through the generation of NA. (b) Mammals are able to process NA in the same manner as yeast to generate NAD+. However, the mammalian salvage pathway does not generate NA; instead, NAM is converted into NMN and then directly back into NAD+. NAD+ is consumed during the deacetylation of histones catalyzed by sirtuins. Abbreviations: NA, nicotinic acid; NaAD, nicotinic acid adenine dinucleotide; NAD+, nicotinamide adenine dinucleotide; NAM, nicotinamide; NAMN, nicotinic acid mononucleotide; NAMPT, nicotinamide phosphoribosyltransferase; NMN, nicotinamide mononucleotide; OAAR, O-acetyl ADP-ribose.
NAD+ Salvage Pathway Flux and Nicotinamide Metabolism
Though conditions that lower NAD+ availability clearly impact Sir2 function, a simple model in which Sir2 function reflects overall NAD+ levels is inadequate to explain several additional observations. Overexpression of Npt1, Nma1, and Nma2, all proteins involved in the NAD+ salvage pathway (Figure 3a), increases Sir2-dependent silencing of rDNA and extends cellular lifespan but does not impact overall NAD+ levels in the cell (Anderson et al. 2002). These observations can be reconciled if Sir2 consumes the additional NAD+ produced by overexpression of the salvage pathway, as implied by the increased gene silencing. Thus, both absolute NAD+ levels and flux through the NAD+ salvage pathway may impact Sir2 function. Additionally, as with npt1 mutants, mutation of the NAD+ salvage pathway gene PNC1 results in impaired rDNA and telomeric silencing (Sandmeier et al. 2002). However, pnc1 mutants are distinct from npt1 mutants in that they do not exhibit decreased NAD+ levels (Sandmeier et al. 2002). So how do pnc1 mutants reduce gene silencing? Pnc1 functions one step before Npt1 in the NAD+ salvage pathway, preventing accumulation of nicotinamide by converting it to nicotinic acid (Anderson et al. 2003, Gallo et al. 2004). The effects of a pnc1 mutation can be explained by simple feedback inhibition of Sir2 when nicotinamide accumulates. Because sirtuins both consume NAD+ and produce nicotinamide, Pnc1 has dual importance for Sir2 function: (a) promoting continuous flux through the NAD+ salvage pathway to maintain NAD+ levels and (b) preventing the accumulation of nicotinamide to concentrations capable of inhibiting Sir2 activity. The deamidation of nicotinamide by Pnc1 is conserved across bacteria, yeast, plants, and invertebrates and may be equally as important in modulating sirtuin function in these other species (Balan et al. 2008, Ghislain et al. 2002, Vrablik et al. 2009, Wang & Pichersky 2007).
Mammalian Sirtuins and NAD+ Metabolism
The diversity of mammalian sirtuins, and specifics regarding the enormous range of biology in which they are involved, is beyond the scope of this review. However, the themes established for NAD+ and Sir2 function in yeast have been highly informative for mammalian sirtuin biology. For example, nicotinamide is also scavenged and utilized in the mammalian NAD+ salvage pathway through conversion into nicotinamide mononucleotide by the enzyme nicotinamide phosphoribosyltransferase (NAMPT) (Revollo et al. 2004). As in yeast, the turnover of nicotinamide by mammalian NAMPT is important for the function of sirtuins (Fulco et al. 2008, van der Veer et al. 2007). From this large body of work emerges a theme in which sirtuins link changes in a cell’s environment, as mediated through metabolites, to transcriptional regulation, sometimes through epigenetic mechanisms. A recent example demonstrates the relationship between the natural oscillation of NAD+ during a typical circadian cycle and fluctuations in SIRT1 histone deacetylase activity (Asher et al. 2015, Nakahata et al. 2009, Ramsey et al. 2009, Trickey et al. 2008). An interesting link between SIRT1 function and diseases that impair DNA repair pathways has also been uncovered. Both xeroderma pigmentosum and Cockayne syndrome are disorders caused by mutations in nucleotide excision repair genes that lead to premature aging, increased risk for cancer, and reduced NAD+ levels as a result of the constitutive activation of poly-ADP-ribose polymerase (PARP). In both cases, diminished NAD+ levels result in decreased SIRT1 function and reduced transcription of genes controlled by SIRT1 (Fang et al. 2014, Scheibye-Knudsen et al. 2014).
In many studies of mammalian sirtuins, the precise mechanistic underpinnings of sirtuin-dependent effects on cellular physiology are not adequately understood. Even in yeast, the full spectrum of substrates for sirtuins is just now coming into focus (Downey et al. 2013, 2015). Understanding how target specificity is achieved and integrated with NAD+ levels, competing acetyltransferases, and other metabolites will require an ever-wider range of approaches and continued exploitation of the power offered by model organisms.
O-Acetyl-ADP-Ribose Metabolism and Histone Deacetylation
Since the discovery that sirtuins produce OAAR from NAD+, much speculation has focused on a possible role for OAAR in either gene silencing or cellular signaling (Jackson & Denu 2002, Tanner et al. 2000). For example, microinjection of OAAR into starfish oocytes leads to inhibited development, indicating that accumulation of OAAR can interfere with normal cellular development (Borra et al. 2002), though the millimolar concentrations used in this study would be substantially outside the likely physiological range. In addition, the identification of enzymes capable of metabolizing OAAR suggests that OAAR levels are actively regulated within the cell (Ono et al. 2006). To date, the evidence for a role of OAAR in gene silencing comes from two studies. The inclusion of pure OAAR in an in vitro assembly reaction for the Sir2-Sir3-Sir4 complex increases the ratio of Sir3 relative to Sir2-Sir4 and leads to structural changes in the complex, as assessed by electron microscopy (Liou et al. 2005). Additionally, the affinity of the purified Sir protein complex, and of Sir3 alone, toward a recombinant chromatin template increases with the addition of OAAR (Martino et al. 2009). Together, these studies suggest that OAAR has the potential to affect the Sir protein complex’s ability to interact with chromatin. Whether OAAR-induced changes to the complex translate to an effect on gene silencing in vivo remains unresolved. Specifically, an elegant genetic study using cleverly designed fusion proteins has established that OAAR is not essential for silencing the HML and HMR loci in budding yeast (Chou et al. 2008). However, whether OAAR might influence the in vivo function of any sirtuin remains an open question.
AN EXPANDING RANGE OF HISTONE ACYLATIONS
Most, if not all, posttranslational modifications discovered in the proteome are eventually found on histones. Hence, the discovery of succinylation and malonylation of proteins has led to the rapid recognition of specific sites of succinylation and malonylation on histones in species ranging from yeast to human (Xie et al. 2012). These modifications involve succinyl-CoA or malonyl-CoA donors for modification of lysine residues on all four histone species. The most conserved positions are on H3 and H4; we focus on these positions because of their greater potential for epigenetic contributions. Succinylation and malonylation are found on some of the same lysines known to be substrates for other modifications. For example, H3K56, which is acetylated in S phase, is also succinylated and malonylated depending on the species. H3K79 can also be succinylated, methylated, or acetylated (Xie et al. 2012).
Mutational strategies to test the significance of succinylated lysines in yeast have typically used a glutamic acid (E) substitution to mimic the constitutively succinylated state and alanine (A) or arginine substitutions to block succinylation. It is difficult to deconvolute the phenotypes of mutations at positions where several different marks are possible. Which modification’s loss is responsible for the phenotype? However, uniquely succinylated positions exist, such as H4K77 and H2AK21. The H4K77E mutation has a unique silencing phenotype not shared with the H4K77R or H4K77A mutations. Likewise, H2AK21E confers DNA damage sensitivity not shared with the H2AK21R or H2A21A mutations. A glutamic acid substitution is thematically (but not sterically) similar in structure to succinylated lysine, sharing the distal carboxyl moiety. Hence, the phenotypes of these mutations should be interpreted with caution. Nevertheless, the clear inference is that the failure to remove the succinyl group could have important consequences.
Sir2 and its four yeast paralogs are classified as NAD+-dependent deacetylases (reviewed in Imai & Guarente 2010), though the number of possible substrates they have been tested on is limited. From work on mammalian cells, it is now clear that some sirtuins are better thought of as deacylases because of their ability to remove multiple acyl modifications of lysine. Indeed, the SIRT5 enzyme, which exists both in the mitochondrion and in the cytoplasm and nucleus, can remove a succinyl group from multiple substrates, including succinyl dehydrogenase, providing another potential route for metabolism to impact epigenetic processes (Papanicolaou et al. 2014).
Malonylation is a less abundant histone modification than succinylation, but it is found in histones from yeast to humans. The malonyl-CoA donor comes from fatty acid synthesis, but the function of malonylation is as yet unclear. Mutation of one of the two sites of malonylation on yeast histones (H2AK199) produces no detectable phenotype. The other site of malonylation, H3K56, is also a site of acetylation and, as described above, a site of succinylation. Hence the reduced viability of the H3K56E mutation has an unresolved cause (Xie et al. 2012).
Additional classes of acylation have been described on mammalian histones and illuminate other potential links to metabolism. Fatty acid oxidation produces propionyl-CoA and butyryl-CoA, both of which are donors for cognate posttranslational modifications of specific lysines in histones H3 and H4. In the absence of genetic evidence, the abundance of modifications can provide some measure of their potential importance. Propionylation occurs on 7% of H3K23 and butyrylation occurs on 31% of H3K115 in some leukemia cells; both levels may be too high to be accounted for as accidental acylations. Crotonyl-CoA, which can be derived from either butyryl-CoA or amino acid metabolism, modifies H4K12 as well as multiple positions on H2A and H2B. SIRT1 and SIRT2, cytoplasmic members of the sirtuin family, can remove these acyl modifications in a presumably NAD+-dependent manner (reviewed in Papanicolaou et al. 2014). Obviously, CoA substrates for these modifications and the NAD+ dependence of their removal could tie aspects of metabolism to epigenetic processes in abundant ways, but so far the field has only begun evaluating their significance. These modifications could reflect fundamental links between metabolism and epigenetics. Alternatively, they may have no regulatory function and could instead reflect the spurious and potentially detrimental accumulation of errors in acylation, perhaps better thought of as chromatin damage, analogous to DNA damage that accumulates over time.
CONCLUDING THOUGHTS
Metabolism and epigenetics is a moving target for any would-be reviewers, with many rapid discoveries and an expansive literature. We have omitted several modifications of potential importance, including O-GlcNAcylation, a modification found on histones, RNA polymerase, and several chromatin-modifying enzymes (reviewed in Lewis & Hanover 2014). Likewise, we have ignored citrulline, a noncanonical amino acid found in histones and other proteins. Citrulline results from the deamination of arginine or the demethylimination of methyl-arginine posttranslationally. The enzymes responsible for this, known as peptidylarginine deiminases, require calcium and a reducing agent. So far, citrullination has one clear epigenetic impact: Citrullination of arginine 8 on histone H3 causes loss of HP1 binding on H3K9me3 and subsequent loss of chromatin condensation (Sharma et al. 2012), similar in spirit to how phosphorylation of H3S10 affects the binding of HP1 to H3K9me3 (Fischle et al. 2005). Ah yes, phosphorylation, yet another link between metabolism and epigenetics. These omissions reflect no shortage of interest, merely the limits of length (and our endurance).
SUMMARY POINTS.
Fluctuations in acetyl-CoA levels are directly coupled to the level of histone acetylation at certain loci; hence, changes in this epigenetic mark are a direct readout of this aspect of metabolism.
Changes in the level of dietary methyl donors can cause heritable changes in the phenotype of the Avy mouse mutation and correlated changes in DNA and histone methylation.
Demethylation of DNA and histones involves enzymes that require FeII and α-ketoglutarate.
Vitamin C may aid somatic cell reprogramming through the stimulation of demethylases.
Mouse embryonic stem cells can use threonine as a secondary source of S-adenosyl-methionine and acetyl-CoA.
Oncometabolites can inhibit both DNA and histone demethylases.
Individual lysine positions on histones can be modified by covalent attachment to several different metabolites.
The NAD+ salvage pathway impacts the deacetylation function of sirtuins.
FUTURE ISSUES.
Progress would be vastly enhanced by the availability of genetically encoded reporters that could provide real-time and localized measurements of metabolite levels.
For positions in histones that are subject to multiple different modifications, does each different modification confer a unique function?
In such examples, modification-site mutations must be paired with mutation or perturbation of the enzymes responsible for those modifications to infer specific roles for a particular modification.
Analyses of the Km of chromatin modifiers for their substrates and of the dynamics of each modification will enable useful modeling.
Genetic variation in the human population, such as the common mutation affecting folate metabolism, should be evaluated for its impact on potential epigenetic lability as a function of nutritional status.
As intriguing as the link is between mutations in IDH1/2 and the inhibition of TET and Jumonji C domain demethylases, does this mechanism drive cancer?
To what extent does a modification of one site on a nucleosome affect the probability or consequences of other modifications?
Careful consideration should be given to the potential role of the microbiome’s response to diet as a possible source of transgenerational inheritance.
ACKNOWLEDGMENTS
We thank Kripa Asrani for help in preparation of the figures. Work in the authors’ lab has been supported continuously by a grant from the National Institutes of Health (GM-31105). Additional support was provided by a National Science Foundation Predoctoral Fellowship (to A.E.D.). We regret the inability to cite a wider selection of the published research relevant to this rapidly expanding topic.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Adam J, Yang M, Soga T, Pollard PJ. Rare insights into cancer biology. Oncogene. 2014;33(20):2547–56. doi: 10.1038/onc.2013.222. [DOI] [PubMed] [Google Scholar]
- Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. PNAS. 1964;51(5):786–94. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, et al. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 2002;277(21):18881–90. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423(6936):181–85. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502(7472):462–71. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, et al. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2015;134(2):317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD+ cosubstrate specificity of a Sir2 enzyme. Mol. Cell. 2005;17(6):855–68. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Balan V, Miller GS, Kaplun L, Balan K, Chong Z-Z, et al. Life span extension and neuronal cell protection by Drosophila nicotinamidase. J. Biol. Chem. 2008;283(41):27810–19. doi: 10.1074/jbc.M804681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baubec T, Dinh HQ, Pecinka A, Rakic B, Rozhon W, et al. Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic states in Arabidopsis. Plant Cell. 2010;22:34–47. doi: 10.1105/tpc.109.072819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast Sir2 and human SIRT1. J. Biol. Chem. 2002;277(47):45099–107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500(7461):222–26. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLOS Genet. 2006;2:e49. doi: 10.1371/journal.pgen.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borra MT, O’Neill FJ, Jackson MD, Marshall B, Verdin E, et al. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J. Biol. Chem. 2002;277(15):12632–41. doi: 10.1074/jbc.M111830200. [DOI] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell. 2011;42(4):426–37. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Finley LWS, Cross JR, Allis CD, Thompson CB. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–16. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervera AM, Bayley J-P, Devilee P, McCreath KJ. Inhibition of succinate dehydrogenase dysregulates histone modification in mammalian cells. Mol. Cancer. 2009;8:89. doi: 10.1186/1476-4598-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B, Chen Y, Zhao Y, Bruick RK. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–47. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- Chen J, Guo L, Zhang L, Wu HH, Yang J, et al. Vitamin C modulates TET1 function during somatic cell reprogramming. Nat. Genet. 2013a;45(12):1504–9. doi: 10.1038/ng.2807. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Liu H, Liu J, Qi J, Wei B, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat. Genet. 2013b;45(1):34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature. 2013c;493:561–64. doi: 10.1038/nature11742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou C-C, Li Y-C, Gartenberg MR. Bypassing Sir2 and O-acetyl-ADP-ribose in transcriptional silencing. Mol. Cell. 2008;31(5):650–59. doi: 10.1016/j.molcel.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Chowdhury R, Yeoh KK, Tian Y-M, Hillringhaus L, Bagg EA, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–69. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung TL, Brena RM, Kolle G, Grimmond SM, Berman BP, et al. Vitamin C promotes widespread yet specific DNA demethylation of the epigenome in human embryonic stem cells. Stem Cells. 2010;28:1848–55. doi: 10.1002/stem.493. [DOI] [PubMed] [Google Scholar]
- Cloos PAC, Christensen J, Agger K, Maiolica A, Rappsilber J, et al. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–11. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Cropley JE, Suter CM, Beckman KB, Martin DI. Germ-line epigenetic modification of the murine Avy allele by nutritional supplementation. PNAS. 2006;1103:17308–12. doi: 10.1073/pnas.0607090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cropley JE, Suter CM, Beckman KB, Martin DI. CpG methylation of a silent controlling element in the murine Avy allele is incomplete and unresponsive to methyl donor supplementation. PLOS ONE. 2010;5(2):e9055. doi: 10.1371/journal.pone.0009055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–44. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong L, Albracht SP, Kemp A. Prolyl 4-hydroxylase activity in relation to the oxidation state of enzyme-bound iron. The role of ascorbate in peptidyl proline hydroxylation. Biochim. Biophys. Acta. 1982;704:326–32. doi: 10.1016/0167-4838(82)90162-5. [DOI] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, et al. TET2 and TET3 regulate GlcNA-cylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–55. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, et al. Structure and ligand of a histone acetyl-transferase bromodomain. Nature. 1999;399(6735):491–96. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Dickson KM, Gustafson CB, Young JI, Züchner S, Wang G. Ascorbate-induced generation of 5-hydroxymethylcytosine is unaffected by varying levels of iron and 2-oxoglutarate. Biochem. Biophys. Res. Commun. 2013;439:522–27. doi: 10.1016/j.bbrc.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinoy DC, Weinhouse C, Jones TR, Rozek LS, Jirtle RL. Variable histone modifications at the A(vy) metastable epiallele. Epigenetics. 2010;5:637–44. doi: 10.4161/epi.5.7.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Johnson JR, Davey NE, Newton BW, Johnson TL, et al. Acetylome profiling reveals overlap in the regulation of diverse processes by sirtuins, Gcn5, and Esa1. Mol. Cell. Proteomics. 2015;14(1):162–76. doi: 10.1074/mcp.M114.043141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey M, Knight B, Vashisht AA, Seller CA, Wohlschlegel JA, et al. Gcn5 and sirtuins regulate acetylation of the ribosomal protein transcription factor Ifh1. Curr. Biol. 2013;23(17):1638–48. doi: 10.1016/j.cub.2013.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan CG, Barwick BG, Jin G, Rago C, Kapoor-Vazirani P, et al. A heterozygous IDH1R132H/WT mutation induces genome-wide alterations in DNA methylation. Genome Res. 2012;22:2339–55. doi: 10.1101/gr.132738.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar AJ. The human L-threonine 3-dehydrogenase gene is an expressed pseudogene. BMC Genet. 2002;3:18. doi: 10.1186/1471-2156-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban MA, Wang T, Qin B, Yang J, Qin D, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Fang EF, Scheibye-Knudsen M, Brace LE, Kassahun H, SenGupta T, et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD+/SIRT1 reduction. Cell. 2014;157(4):882–96. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438(7071):1116–22. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Friis RMN, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res. 2009;37(12):3969–80. doi: 10.1093/nar/gkp270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. PNAS. 2002;99:5606–11. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev. Cell. 2008;14(5):661–73. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidzik VI, Paschka P, Spath D, Häbdank M, Köhne CH, et al. TET2 mutations in acute myeloid leukemia (AML): results from a comprehensive genetic and clinical analysis of the AML study group. J. Clin. Oncol. 2012;30:1350–57. doi: 10.1200/JCO.2011.39.2886. [DOI] [PubMed] [Google Scholar]
- Galdieri L, Vancura A. Acetyl-CoA carboxylase regulates global histone acetylation. J. Biol. Chem. 2012;287(28):23865–76. doi: 10.1074/jbc.M112.380519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo CM, Smith DL, Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol. Cell. Biol. 2004;24(3):1301–12. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Talla E, François JM. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast. 2002;19(3):215–24. doi: 10.1002/yea.810. [DOI] [PubMed] [Google Scholar]
- Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 2010;207:339–44. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse AH. Calcium mobilizing second messengers derived from NAD. Biochim. Biophys. Acta. 2015;1854(9):1132–37. doi: 10.1016/j.bbapap.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Hahn MA, Szabó PE, Pfeifer GP. 5-Hydroxymethylcytosine: a stable or transient DNA modification? Genomics. 2014;104(5):314–23. doi: 10.1016/j.ygeno.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales CN, Barker DJ. The thrifty phenotype hypothesis. Br. Med. Bull. 2001;60:5–20. doi: 10.1093/bmb/60.1.5. [DOI] [PubMed] [Google Scholar]
- Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111(3):369–79. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- He J, Kallin EM, Tsukada Y-I, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b) Nat. Struct. Mol. Biol. 2008;15(11):1169–75. doi: 10.1038/nsmb.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y-F, Li B-Z, Li Z, Liu P, Wang Y, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–7. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 2015;80(4):583–92. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hong L, Schroth GP, Matthews HR, Yau P, Bradbury EM. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 “tail” to DNA. J. Biol. Chem. 1993;268(1):305–14. [PubMed] [Google Scholar]
- Honjo T, Nishizuka Y, Hayaishi O, Kato I. Diphtheria toxin-dependent adenosine diphosphate ribo-sylation of aminoacyl transferase II and inhibition of protein synthesis. J. Biol. Chem. 1968;243(12):3553–55. [PubMed] [Google Scholar]
- Hornig D. Distribution of ascorbic acid, metabolites and analogues in man and animals. Ann. N. Y. Acad. Sci. 1975;258:103–18. doi: 10.1111/j.1749-6632.1975.tb29271.x. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci. 2010;31:212–20. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–3. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MD, Denu JM. Structural identification of 2′ - and 3′ -O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of β-NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 2002;277(21):18535–44. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288(5470):1422–25. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Kaelin WG. Cancer and altered metabolism: potential importance of hypoxia-inducible factor and 2-oxoglutarate-dependent dioxygenases. Cold Spring Harb. Symp. Quant. Biol. 2011;76:335–45. doi: 10.1101/sqb.2011.76.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian JK, Kim SY, Miettinen M, Smith C, Merino M, et al. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov. 2013;3(6):648–57. doi: 10.1158/2159-8290.CD-13-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502:472–79. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. PNAS. 2000;97(11):5807–11. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letouzé E, Martinelli C, Loriot C, Burnichon N, Abermil N, et al. SDH mutations establish a hyper-methylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–52. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Lewis BA, Hanover JA. O-GlcNAc and the epigenetic regulation of gene expression. J. Biol. Chem. 2014;289:3440–8. doi: 10.1074/jbc.R114.595439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, He J, Zhang Y. Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nat. Cell Biol. 2012;14(5):457–66. doi: 10.1038/ncb2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou G-G, Tanny JC, Kruger RG, Walz T, Moazed D. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell. 2005;121(4):515–27. doi: 10.1016/j.cell.2005.03.035. [DOI] [PubMed] [Google Scholar]
- Losman J-A, Kaelin WG. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–52. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman J-A, Looper RE, Koivunen P, Lee S, Schneider RK, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339(6127):1621–5. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–78. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8Å resolution. Nature. 1997;389(6648):251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Ma Z, Vosseller K. Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. J. Biol. Chem. 2014;289:34457–65. doi: 10.1074/jbc.R114.577718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majamaa K, Gunzler V, Hanauske-Abel HM, Myllylä R, Kivirikko KI. Partial identity of the 2-oxoglutarate and ascorbate binding sites of prolyl 4-hydroxylase. J. Biol. Chem. 1986;261:7819–23. [PubMed] [Google Scholar]
- Mantri M, Loik ND, Hamed RB, Claridge TDW, McCullagh JSO, Schofield CJ. The 2-oxoglutarate-dependent oxygenase JMJD6 catalyses oxidation of lysine residues to give 5S-hydroxylysine residues. ChemBioChem. 2011;12:531–34. doi: 10.1002/cbic.201000641. [DOI] [PubMed] [Google Scholar]
- Marcotte PA, Richardson PR, Guo J, Barrett LW, Xu N, et al. Fluorescence assay of SIRT protein deacetylases using an acetylated peptide substrate and a secondary trypsin reaction. Anal. Biochem. 2004;332(1):90–99. doi: 10.1016/j.ab.2004.05.039. [DOI] [PubMed] [Google Scholar]
- Martino F, Kueng S, Robinson P, Tsai-Pflugfelder M, van Leeuwen F, et al. Reconstitution of yeast silent chromatin: multiple contact sites and O-AADPR binding load SIR complexes onto nucleosomes in vitro. Mol. Cell. 2009;33(3):323–34. doi: 10.1016/j.molcel.2009.01.009. [DOI] [PubMed] [Google Scholar]
- Masri S, Sassone-Corsi P. Sirtuins and the circadian clock: bridging chromatin and metabolism. Sci. Signal. 2014;7(342):re6. doi: 10.1126/scisignal.2005685. [DOI] [PubMed] [Google Scholar]
- Minor EA, Court BL, Young JI, Wang G. Ascorbate induces ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated generation of 5-hydroxymethylcytosine. J. Biol. Chem. 2013;288:13669–74. doi: 10.1074/jbc.C113.464800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 1999;23:314–18. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Myllylä R, Kuutti-Savolainen ER, Kivirikko KI. The role of ascorbate in the prolyl hydroxylase reaction. Biochem. Biophys. Res. Commun. 1978;83(2):441–48. doi: 10.1016/0006-291x(78)91010-0. [DOI] [PubMed] [Google Scholar]
- Myllylä R, Majamaa K, Günzler V, Hanauske-Abel HM, Kivirikko KI. Ascorbate is consumed stoichiometrically in the uncoupled reactions catalyzed by prolyl 4-hydroxylase and lysyl hydroxylase. J. Biol. Chem. 1984;259:5403–5. [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P. Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science. 2009;324(5927):654–57. doi: 10.1126/science.1170803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Kasamatsu A, Oka S, Moss J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. PNAS. 2006;103(45):16687–91. doi: 10.1073/pnas.0607911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanicolaou KN, O’Rourke B, Foster DB. Metabolism leaves its mark on the powerhouse: recent progress in post-translational modifications of lysine in mitochondria. Front. Physiol. 2014;5:301. doi: 10.3389/fphys.2014.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard PJ, Briére JJ, Alam NA, Barwell J, Barclay E, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1α in tumours which result from germline FH and SDH mutations. Hum. Mol. Genet. 2005;14(15):2231–39. doi: 10.1093/hmg/ddi227. [DOI] [PubMed] [Google Scholar]
- Puistola U, Turpeenniemi-Hujanen TM, Myllylä R, Kivirikko KI. Studies on the lysyl hydroxylase reaction. II. Inhibition kinetics and the reaction mechanism. Biochim. Biophys. Acta. 1980;611:51–60. doi: 10.1016/0005-2744(80)90041-8. [DOI] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–54. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ. Daddy issues: paternal effects on phenotype. Cell. 2012;151:702–8. doi: 10.1016/j.cell.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279(49):50754–63. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Rice ME, Russo-Menna I. Differential compartmentalization of brain ascorbate and glutathione between neurons and glia. Neuroscience. 1997;82:1213–23. doi: 10.1016/s0306-4522(97)00347-3. [DOI] [PubMed] [Google Scholar]
- Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116(1):9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CJ, Selker EU. Mutations affecting the biosynthesis of S-adenosylmethionine cause reduction of DNA methylation in Neurospora crassa. Nucleic Acids Res. 1995;23:4818–26. doi: 10.1093/nar/23.23.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–30. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, Klose RJ. Understanding the relationship between DNA and histone lysine methylation. BBA Gene Regul. Mech. 2014;1839:1362–72. doi: 10.1016/j.bbagrm.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, McDonough MA, King ON, Kawamura A, Schofield CJ. Inhibition of 2-oxoglutarate dependent oxygenases. Chem. Soc. Rev. 2011;40(8):4364–97. doi: 10.1039/c0cs00203h. [DOI] [PubMed] [Google Scholar]
- Rose NR, Ng SS, Mecinović J, Liénard BMR, Bello SH, et al. Inhibitor scaffolds for 2-oxoglutarate-dependent histone lysine demethylases. J. Med. Chem. 2008;51:7053–56. doi: 10.1021/jm800936s. [DOI] [PubMed] [Google Scholar]
- Sadhu MJ, Guan Q, Li F, Sales-Lee J, Iavarone AT, et al. Nutritional control of epigenetic processes in yeast and human cells. Genetics. 2013;195:831–44. doi: 10.1534/genetics.113.153981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier JJ, Celic I, Boeke JD, Smith JS. Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD+ salvage pathway. Genetics. 2002;160(3):877–89. doi: 10.1093/genetics/160.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Mitchell SJ, Fang EF, Iyama T, Ward T, et al. A high-fat diet and NAD+ activate Sirt1 to rescue premature aging in Cockayne syndrome. Cell Metab. 2014;20(5):840–55. doi: 10.1016/j.cmet.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame J-C, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7(7):517–28. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Sharma P, Azebi S, England P, Christensen T, Møller-Larsen A, et al. Citrullination of histone H3 interferes with HP1-mediated transcriptional repression. PLOS Genet. 2012;8(9):e1002934. doi: 10.1371/journal.pgen.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339:222–6. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EH, Janknecht R, Maher LJ. Succinate inhibition of α-ketoglutarate-dependent enzymes in a yeast model of paraganglioma. Hum. Mol. Genet. 2007;16(24):3136–48. doi: 10.1093/hmg/ddm275. [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, et al. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. PNAS. 2000;97(12):6658–63. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiriou S, Gispert S, Cheng J, Wang Y, Chen A, et al. Ascorbic-acid transporter Slc23a1 is essential for vitamin C transport into the brain and for perinatal survival. Nat. Med. 2002;8:514–17. doi: 10.1038/0502-514. [DOI] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret K. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151(5):994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, et al. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–77. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Ferrari F, Choi J, Walsh RM, et al. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat. Genet. 2012;44(4):398–405. doi: 10.1038/ng.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–35. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme A synthetase is required for histone acetylation and global transcription. Mol. Cell. 2006;23(2):207–17. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, et al. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9:1211–22. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. PNAS. 2000;97(26):14178–82. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trickey M, Grimaldi M, Yamano H. The anaphase-promoting complex/cyclosome controls repair and recombination by ubiquitylating Rhp54 in fission yeast. Mol. Cell. Biol. 2008;28(12):3905–16. doi: 10.1128/MCB.02116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–16. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310(5751):1152–58. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, et al. Cyclic changes in metabolic state during the life of a yeast cell. PNAS. 2007;104(43):16886–91. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuderman L, Myllylä R, Kivirikko KI. Mechanism of the prolyl hydroxylase reaction. Eur. J. Biochem. 1977;348:341–48. doi: 10.1111/j.1432-1033.1977.tb11888.x. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–83. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Veer E, Ho C, O’Neil C, Barbosa N, Scott R, et al. Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J. Biol. Chem. 2007;282(15):10841–45. doi: 10.1074/jbc.C700018200. [DOI] [PubMed] [Google Scholar]
- Venneti S, Thompson CB. Metabolic modulation of epigenetics in gliomas. Brain Pathol. 2013;23:217–21. doi: 10.1111/bpa.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2015;16:258–64. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- Vrablik TL, Huang L, Lange SE, Hanna-Rose W. Nicotinamidase modulation of NAD+ biosynthesis and nicotinamide levels separately affect reproductive development and cell survival in C. elegans. Development. 2009;136(21):3637–46. doi: 10.1242/dev.028431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cell on threonine catabolism. Science. 2009;325:435–39. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Pichersky E. Nicotinamidase participates in the salvage pathway of NAD biosynthesis in Arabidopsis. Plant J. 2007;49(6):1020–29. doi: 10.1111/j.1365-313X.2006.03013.x. [DOI] [PubMed] [Google Scholar]
- Wang T, Chen K, Zeng X, Yang J, Wu Y, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell. 2011;9(6):575–87. doi: 10.1016/j.stem.2011.10.005. [DOI] [PubMed] [Google Scholar]
- Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webby CJ, Wolf A, Gromak N, Dreger M, Kramer H, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325(5936):90–93. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- Weissmann S, Alpermann T, Grossmann V, Kowarsch A, Nadarajah N, et al. Landscape of TET2 mutations in acute myeloid leukemia. Leukemia. 2012;26(5):934–42. doi: 10.1038/leu.2011.326. [DOI] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076–80. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–81. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–57. [PubMed] [Google Scholar]
- Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res. 1999;27(3):711–20. doi: 10.1093/nar/27.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–93. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, et al. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26(12):1326–38. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Dai J, Dai L, Tan M, Cheng Z, et al. Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics. 2012;11:100–7. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang PP, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Chen Z, Rozen R, Matthews RG. Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase. PNAS. 2001;98(26):14853–58. doi: 10.1073/pnas.261469998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Lin H, Xu H, Zhang L, Cheng L, et al. TET-catalyzed 5-methylcytosine hydroxylation is dynamically regulated by metabolites. Cell Res. 2014;24:1017–20. doi: 10.1038/cr.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R, Mao SQ, Zhao B, Chong Z, Yang Y, et al. Ascorbic acid enhances Tet-mediated 5-methylcytosine oxidation and promotes DNA demethylation in mammals. J. Am. Chem. Soc. 2013;135:10396–403. doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]