Abstract
The Base Excision Repair (BER) pathway removes the vast majority of damages produced by ionizing radiation, including the plethora of radiation-damaged purines and pyrimidines. The first enzymes in the BER pathway are DNA glycosylases, which are responsible for finding and removing the damaged base. Although much is known about the biochemistry of DNA glycosylases, how these enzymes locate their specific damage substrates among an excess of undamaged bases has long remained a mystery. Here we describe the use of single molecule fluorescence to observe the bacterial DNA glycosylases, Nth, Fpg and Nei, scanning along undamaged and damaged DNA. We show that all three enzymes randomly diffuse on the DNA molecule and employ a wedge residue to search for and locate damage. The search behavior of the Escherichia coli DNA glycosylases likely provides a paradigm for their homologous mammalian counterparts.
Keywords: Base excision repair, DNA glycosylases, Single molecule fluorescence, Search for radiation damage, Glycosylase diffusion, Wedge residue
Introduction
Base excision repair (BER) (Figure 1) is a multistep pathway involving a number of repair enzymes, which are conserved from bacteria to humans. BER evolved to protect cellular DNA against the deleterious effects of endogenous metabolic processes, as well as damage produced by exogenous agents such as ionizing radiation (David et al., 2007; Duclos et al., 2012; Kim and Wilson, 2012; Prakash et al., 2012; Wallace, 2013; Wilson et al., 2003). In fact, the great majority of damages produced by ionizing radiation are repaired by BER (Wallace, 1998). The BER pathway can be entered at three different places: at a base damage, at a site of base loss, and at a single strand break, all produced by ionizing radiation (Ward, 1985, 1988). Faulty BER has also been implicated in the production of double strand breaks at radiation-induced clustered lesion sites (Blaisdell and Wallace, 2001; Harrison et al., 1999; Sage and Harrison, 2011; Sutherland et al., 2000; Ward, 1981; Yang et al., 2006; Yang et al., 2004). Although many aspects of BER are well-defined, very little is known about the how the first enzymes in this pathway, the DNA glycosylases, locate radiation-damaged purines and pyrimidines in a sea of undamaged bases using only thermal energy, particularly when the damages are often nearly indistinguishable from normal bases.
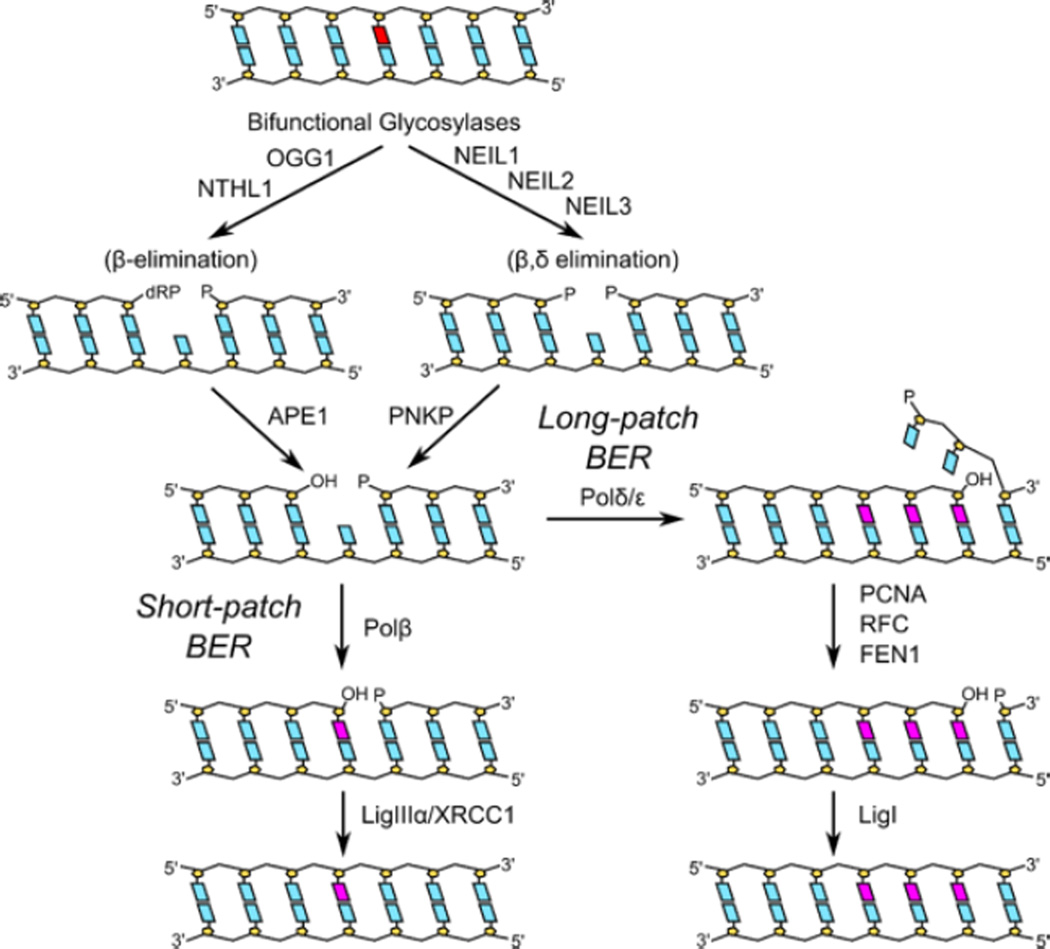
Figure 1. Mammalian short and long patch base excision repair pathways.
Once the DNA glycosylase locates a lesion, the initial action on a damaged base is eversion of the lesion into the enzyme’s active site pocket, then cleavage of the N-glycosyl bond leaving an abasic site, which is the final product when the DNA glycosylase is monofunctional. A bifunctional glycosylase also cleaves the DNA backbone, leaving a polymerase-blocking group attached to the 3' side of the single strand break. The DNA glycosylases that recognize radiation-damaged bases are bifunctional enzymes that belong to either of two structural families, the Fpg/Nei family or the HhH-GPD superfamily, which includes Nth. In bacteria, removal of radiation-damaged pyrimidine bases is initiated by Nth (endonuclease three) or Nei (endonuclease eight) while damaged purines are removed by Fpg (formamidopyrimidine DNA glycosylase). All three of these DNA glycosylases are bifunctional and have a tightly coupled lyase activity. The lyase products, an α,β-unsaturated aldehyde (Nth) or a phosphate (Fpg and Nei), are removed by an apurinic (AP) endonuclease. The gap is filled in by a DNA polymerase and sealed by DNA ligase (Duclos et al., 2012).
In mammalian cells (Hazra et al., 2007; Hegde et al., 2008a; Izumi et al., 2003) (Figure 1), once the glycosylase removes the initial base damage, APE1, an AP endonuclease, cleaves the abasic site left by a monofunctional enzyme or removes the α,β unsaturated aldehyde left by the Nth superfamily members, NTHL1 and OGG1. The phosphate group left by the Fpg/Nei family members (NEIL1, NEIL2 and NEIL3) is removed by polynucleotide kinase phosphatase (PNKP) (Wiederhold et al., 2004). APE1 also initiates repair on free radical-generated sites of base loss as well as on single strand breaks by helping to clean the 3’ end for polymerase action. In short patch repair, DNA polymerase β fills in the resulting gap, usually one or two nucleotides, which is then sealed by ligase IIIα-XRCC1 or Ligase I. Alternatively, repair can be shunted into a longer patch process that utilizes the replicative polymerases, together with PCNA, FEN1 and Ligase I. The repair of free radical-induced base damage occurs primarily by single nucleotide short patch repair, analogous to BER in bacteria (Dogliotti et al., 2001).
The Target Search on Undamaged DNA
With the advent of structural biology and sophisticated enzymological techniques, much has been learned about BER over the past several decades. However, much less is known about how these DNA repair proteins find their targets amongst the billions of non-target DNA bases. Random three-dimensional diffusion among various locations in the DNA would be highly inefficient for finding low-occurrence damage sites. Instead, it has been suggested that glycosylases maintain contact with the DNA through various specialized interactions. These interactions include sliding processively along the DNA chain, transferring between DNA chains using interstrand or intrastrand crossover, and/or hopping between close sites on the same chain (reviewed in (Lee et al., 2014; Zharkov and Grollman, 2005)). The processive sliding mechanism has been tested using correlated cleavage experiments, and it does appear that several glycosylases are able to sequentially process closely spaced lesions. However, these assays are unable to directly observe long range interactions between the glycosylase and DNA, and they also are unable to characterize glycosylase interactions with undamaged DNA (Bennett et al., 1995; Dowd and Lloyd, 1990; Gruskin and Lloyd, 1988; Higley and Lloyd, 1993; Mechetin and Zharkov, 2011; Porecha and Stivers, 2008; Purmal et al., 1994; Sidorenko et al., 2008).
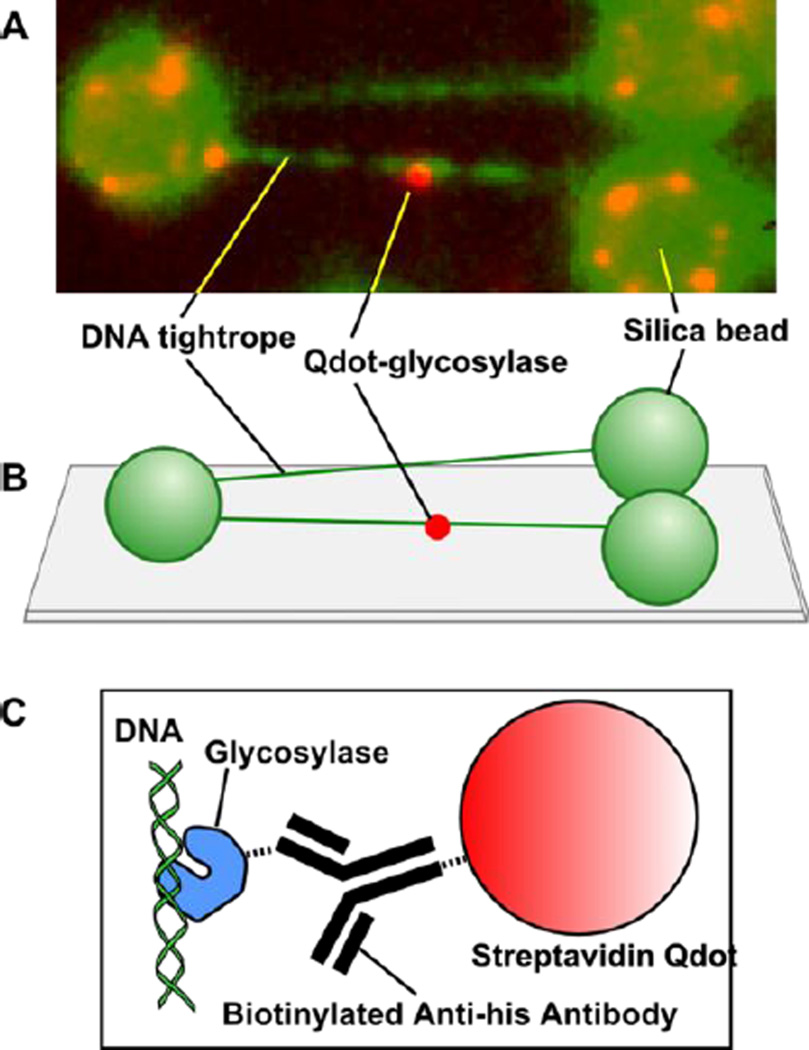
To address this issue, we employed single molecule techniques to investigate the target search in more detail than was formerly possible using classical ensemble-based kinetics. Our single molecule approach utilizes DNA tightropes, DNA glycosylases labeled with quantum dots (Qdots, highly fluorescent semiconductor nanocrystals), and Total Internal Reflectance Fluorescence (TIRF) microscopy which allows us to visualize the initial damage search mechanisms of individual BER glycosylases in real time, with high spatial (10nm) and temporal (30ms) resolution (Figure 2). We have characterized the motion of single molecules of the three bacterial glycosylases that recognize free radical-damaged DNA bases, Fpg, Nei and Nth, as they scan along λ DNA. We observed that the motion of these glycosylases is bidirectional, random, and tracks rotationally along the DNA molecule, which is consistent with the motion of other glycosylases along elongated DNA (Blainey et al., 2009; Blainey et al., 2006).
Figure 2. Single molecule sample configuration.
(A) Image of a single Qdot-labeled glycosylase scanning along elongated λ-DNA. (B) λ-DNA tightropes are elongated between polylysine-coated silica spheres on a PEG-coated microscope coverslip. (C) The 6-his tagged glycosylase is conjugated to a streptavidin Qdot through a biotinylated anti-his antibody.
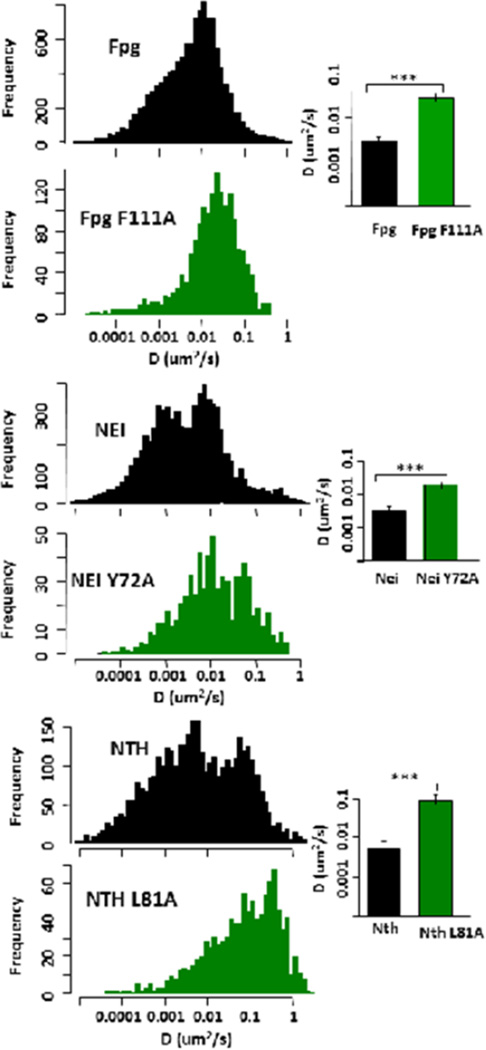
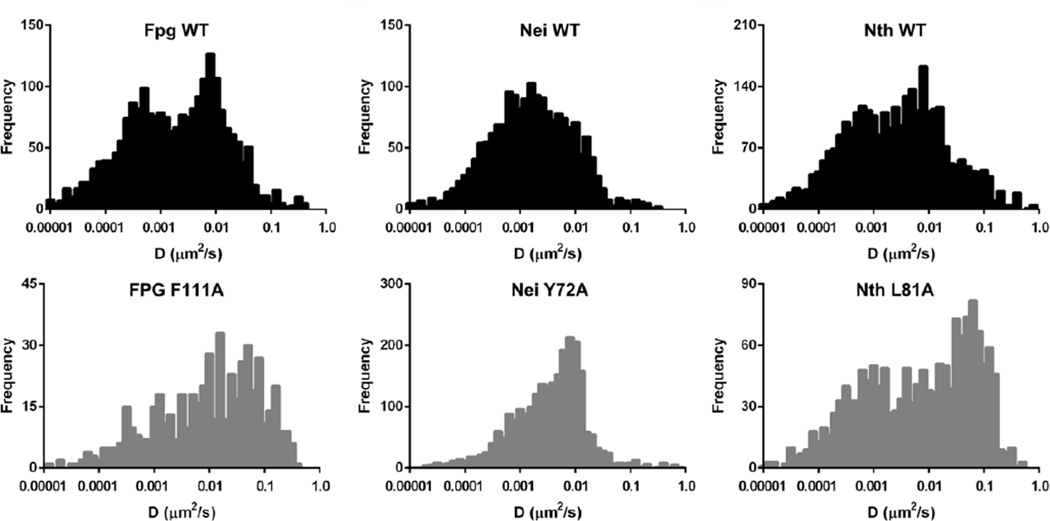
Upon examination of the trajectories of WT Fpg, Nei and Nth glycosylases on undamaged DNA, it was clear that during an individual trajectory, the enzymes transitioned from fast to slow diffusive behavior and vice versa giving a broad spectrum of diffusion constants (Dunn et al., 2011; Nelson et al., 2014). This led us to characterize glycosylase behavior not only by a single diffusion constant (trajectory-weighted) but also by a sliding window approach that allowed us to visualize a time-weighted distribution of diffusion constants (see Figure 3). The latter method clearly showed that the wild type enzymes exhibited both slow and fast diffusive behavior, which may correspond to different search and transport modes along the DNA, begging the question of whether the slow form is related to the damage search.
Figure 3. Time weighted histograms of the diffusive behavior of wild-type and wedge variant glycosylases scanning along undamaged λ DNA (left column).
The bar graphs to the right show the average trajectory-weighted diffusion constant for comparison. See (Nelson et al., 2014) for further details of experimental conditions.
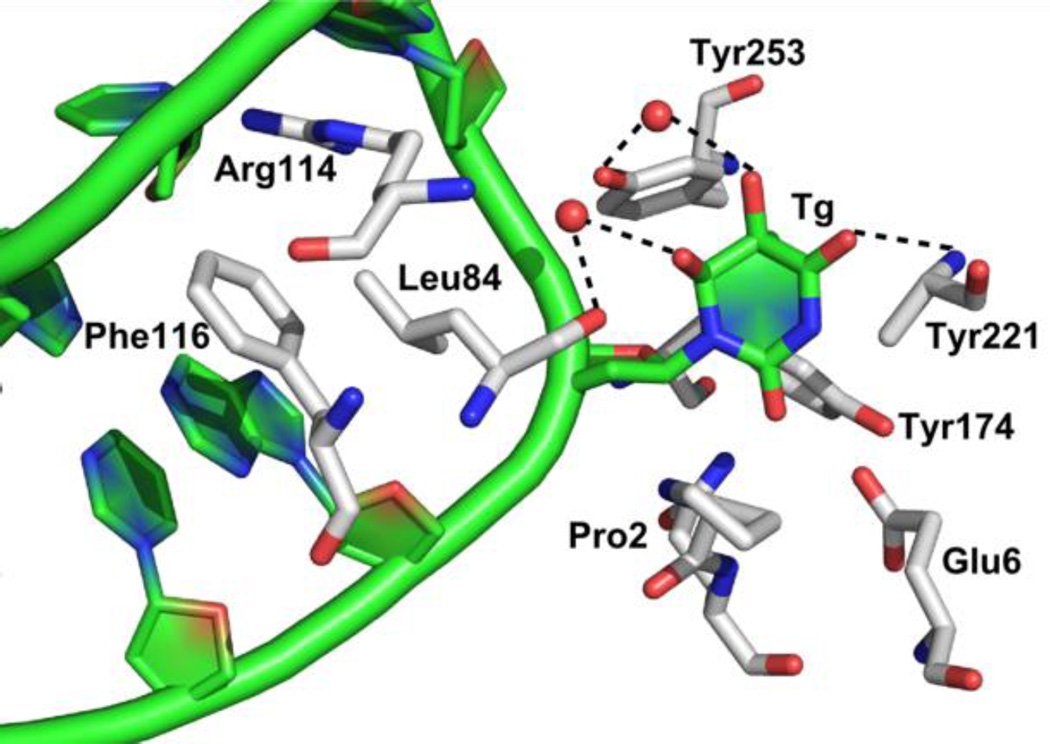
To better understand how glycosylases distinguish between undamaged DNA bases and a lesion of interest, we sought to investigate structural features that would play a role in the glycosylase search. Interestingly, when our colleague, Dr. Doublié, solved the only structure thus far obtained of a free radical-damaged pyrimidine-specific DNA glycosylase with the lesion in the active site, there were relatively few specific interactions between the lesion and the enzyme (Figure 4) (Imamura et al., 2012). Tyr253 interacts via a water molecule with Tg (and anti-5-hydroxyuracil (5-OHU) not shown), Glu6 interacts with syn-5OHU (not shown), the main chain carbonyl of Leu84 via a water molecule interacts with both lesions and Tyr221 forms a direct hydrogen bond with both lesions (Imamura et al., 2012). However, mutants of Tyr253 and Glu6 showed no effect on the glycosylase or lyase activities on substrates containing 5-OHU and small effects with Tg-containing substrates. Also, when an undamaged thymine was modeled into the active site of MvNei1, it was found that the bonding contacts would be similar to those of Tg, for example a hydrogen bond was observed with Tyr221 (Imamura et al., 2012). These data suggest that if an undamaged thymine base were in the active site, nothing would prevent its cleavage, and thymine cleavage is never observed. Taken together these findings provide strong evidence that the glycosylase identifies its substrate damage before it is everted into the active site pocket.
Figure 4. Structure of the catalytically inactive Mimivirus Nei (MvNei E3Q) in the presence of a Tg ligand.
Tg is flipped out of the DNA duplex and makes only one direct hydrogen bond to Tyr221. Other bonds are bridged through water molecules to Tyr253 and Leu84. The three intercalation residues, Arg114, Phe116, and Leu84 are shown (Phe116 is the wedge residue).
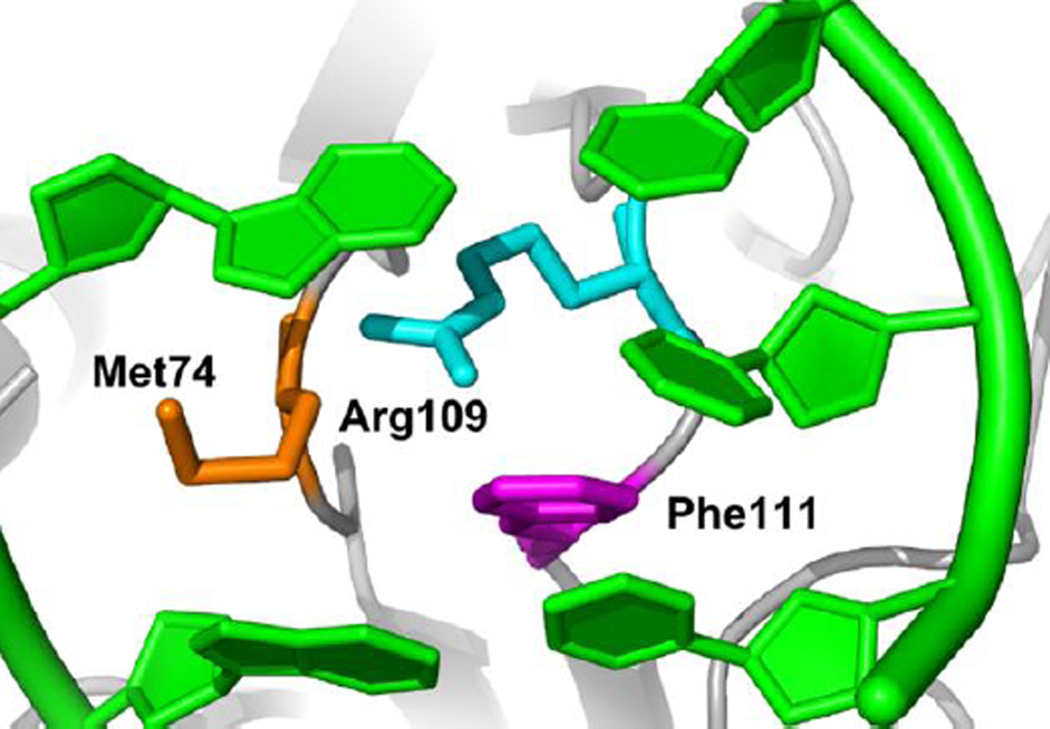
Furthermore, the crystal structures of Nth (Fromme and Verdine, 2003), Fpg (Gilboa et al., 2002), and Nei (Zharkov et al., 2002) glycosylases in a Schiff base complex with DNA lesions show how each of the glycosylases insert three amino acids into the void left in the DNA duplex (Figures 4 and 5). One of these residues is able to “wedge” its way into the base stack opposite the former site of the damage and possibly act as a sensor for physical changes to the DNA that occur when damage is present. Moreover, crystal structures of Geobacter stearothermophilus Fpg glycosylase (Banerjee et al., 2006; Qi et al., 2012) covalently bound to undamaged DNA showed that this “wedge” residue is able to insert into the DNA base stack in the absence of base eversion possibly to probe for damaged bases.
Figure 5. Intercalation of three Fpg WT amino acid residues into the DNA duplex after eversion of the damaged base.
Phe111 is shown inserted between the two undamaged bases opposing the damage.
Using our single molecule approach with undamaged λ DNA, we asked if the three glycosylases rely upon the “wedge residue” for the search process at a step before the damage has been found. We substituted alanine for the wedge residue in each of these glycosylases, and in each case, the variant enzyme scanned along undamaged DNA at a significantly faster rate than the wild-type enzyme (Figure 3). In further support of the amino acid wedge being responsible for the lesion search, (Kuznetsov et al., 2015b) recently showed, using stopped flow kinetics, that the first phase of nonspecific binding of Nth to a lesion may be insertion of Leu81, its wedge residue.
The Fpg Phe111Ala wedge variant loses enzymatic activity, which is consistent with its inability to locate damages as well as its inability to aid in base eversion that is necessary for catalysis. Kuznetsov and coworkers (Kuznetsov et al., 2015a) have recently shown, using a combination of molecular dynamics and stopped flow kinetics, that the Fpg wedge residue buckles the base pairs surrounding the damage, thus destabilizing the intrahelical base. Like Fpg, the analogous wedge variant of Nth, Leu81Ala, loses enzymatic activity. However, the wedge variant of Nei, Tyr72Ala, is enzymatically active. Nei differs from Fpg and Nth in that the other two members of the intercalation triad are in a structural position on the same loop. Since enzyme activities are measured on short damage-containing oligonucleotides where locating the lesion may not be rate-limiting, once the damage is found, the triad-containing loop may still function to evert the lesion and enable catalysis. These findings led to further questions about how wild-type and wedge variant glycosylases behave in the presence of damages on DNA.
Glycosylases Locate DNA Damages
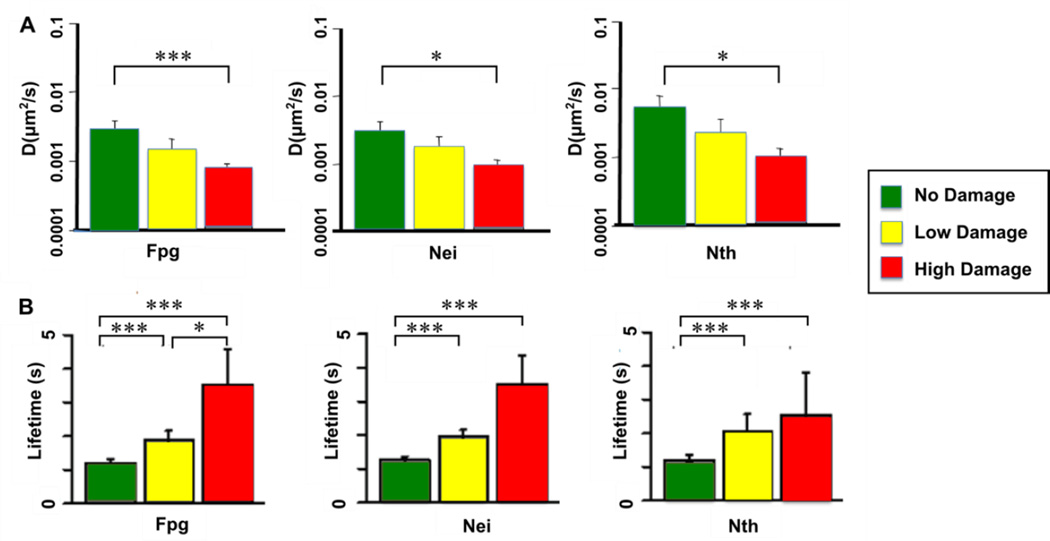
For Nth and Nei, thymine glycols were produced by osmium tetroxide-treatment of the DNA, while for Fpg, methylene blue plus visible light were used to generate damaged purines. The single molecule substrates were tailored to give two damage conditions for comparison with undamaged λ, low dose of approximately 100 lesions per λ molecule and high dose of approximately 280 lesions per λ. What we found was that all of the wild type enzymes diffuse more slowly (Figure 6A) and bind longer (Figure 6B) on DNA that contains the damages that they recognize and remove. In contrast, the mean diffusion constant for Fpg glycosylase on thymine glycol damages present in OsO4-treated DNA was the same as that on undamaged DNA, confirming that glycosylases are able to diffuse past lesions that they do not specifically remove. The decrease in diffusion and increase in binding lifetime in the presence of specific lesions was proportional to the amount of damage present in the substrates. Using our assay, we were also able to observe that after stopping at a lesion, presumably catalyzing its removal, about a third of the Fpg glycosylases resume the damage search without dissociating from the DNA.
Figure 6. Trajectory weighted diffusion constants (A) and binding lifetimes (B) for wild-type glycosylases scanning along damaged λ-DNA.
Low dose damage is approximately 100 damages per λ molecule, while high-dose damage is approximately 300 lesions per λ. See (Nelson et al., 2014) for further details of experimental conditions.
The behavior of the wedge variants on damaged DNA is a little more complex. On one hand, the variants show faster diffusion than the wild-type enzymes in the presence of specific damage (Figure 7), indicating that the wedge plays a critical role in lesion identification and processing. On the other hand, the wedge variants diffuse more slowly on damaged DNA than on undamaged DNA, suggesting that the enzymes still retain some level of damage recognition even without the wedge residue. Since we know that the Fpg and Nth wedge variants are catalytically inactive, we can postulate that the recognition step is independent of the downstream events. This conclusion is supported by the fact that the binding lifetimes of the Fpg and Nth wedge variants in the presence of damage remain short (not shown), which indicates that these variants skip the relatively long catalytic step during their scans. Accordingly, the ability of Nei to undergo catalysis in the absence of the wedge residue is reflected in its longer binding lifetimes on damaged substrates (not shown). Overall, however, although Fpg, Nei and Nth are members of two different structural families, they behave similarly in their diffusive search for damage.
Figure 7. Time-weighted diffusive behavior of wild-type and wedge variant glycosylases scanning along damaged λ-DNA.
Fpg WT and F111A and Nth WT and L81A data are shown at low doses of damage (approximately 100 damages per λ molecule). Nei WT and Y72A are shown at a high dose of damage (approximately 280 lesions per λ). See (Nelson et al., 2014) for further details of experimental conditions.
Finally, using a simple chemomechanical simulation based our data and kinetic data from the Federova lab (Koval et al., 2010; Koval et al., 2004; Kuznetsov et al., 2007; Kuznetsov et al., 2012; Kuznetsov et al., 2009), we suggest that the wild-type glycosylases scan rotationally along the DNA backbone at a rate of 0.05 µm2/s, periodically stopping to insert the wedge at a rate of 2000 s−1 to interrogate for damage. The wedge is removed at a rate of 500 s−1, unless the enzyme detects a damaged base in which case it remains bound to the lesion. Spot checking of undamaged DNA is highly redundant, which would allow for discovery of the few damage sites within the large number of undamaged bases. Based on our measured binding lifetimes and diffusion constants, we predict that 450–600 base pairs are scanned during each encounter, which agrees well with values measured in correlated cleavage experiments. Overall, our model predicts that at physiological concentrations and scanning rates, each of these glycosylases would be able to scan both strands of the entire Escherichia coli genome every 10 minutes.
Where We Are Going: Human DNA Glycosylases
In human cells, NTHL1 and OGG1 appear to be housekeeping glycosylases scanning the genome for free radical-damaged pyrimidines and purines, respectively, and then removing them. It had previously been established (Blainey et al., 2009; Blainey et al., 2006) that OGG1 scans DNA in a manner consistent with rotation around the helix, and our preliminary data (unpublished observations) on undamaged DNA suggest that like its functional bacterial homolog Fpg, and its structural relative Nth, OGG1 engages in very similar diffusive behavior with both fast and slow modes and employing a wedge residue to scan DNA for damage. Also, like the other glycosylases, OGG1 stops upon encountering a lesion (unpublished observations). NTHL1 appears to be more complicated since it dimerizes at high concentrations (Liu et al., 2003; Liu and Roy, 2002) similar to the concentrations found in vivo but it is not clear if this dimer is the structure that searches the DNA for damage.
The human Nei homologs, NEIL1, NEIL2 and NEIL3, remove free radical-damaged pyrimidines, formamidopyrimidines and the further oxidation products of 8-oxoguanine, spiroiminodihydantoin and guanidinohydantoin (Bandaru et al., 2002; Hailer et al., 2005; Hazra et al., 2002; Krishnamurthy et al., 2008; Liu et al., 2010; Wallace et al., 2003; Zhao et al., 2010). Unlike NTHL1 and OGG1, the NEIL glycosylases remove lesions from single-stranded DNA as well as duplex DNA; in fact, both NEIL2 and NEIL3 prefer lesions in single-stranded DNA (Dou et al., 2008; Liu et al., 2010). Importantly, the NEIL glycosylases appear to have evolved specialized roles in DNA repair. For example, NEIL1 appears to act as a cow-catcher removing lesions in front of the replication fork (Dou et al., 2008; Hegde et al., 2013; Hegde et al., 2008b; Theriot et al., 2010), NEIL2 has been linked to repair during transcription (Banerjee et al., 2011) and NEIL3 to repair of telomere DNA (Fleming et al., 2015; Zhou et al., 2015; Zhou et al., 2013). Interestingly, NEIL3 is missing two out of three of the intercalation residues including the wedge residue (Liu et al., 2013). Taken together, the human Nei homologs portend to be very interesting targets for further single molecule studies.
We are planning to examine not only the human glycosylase search but also the interactions among the glycosylases and the downstream enzymes in the pathway (Figure 1). To accomplish this, we have constructed a plasmid engineered to introduce our lesion of choice and will use a strategy similar to the one described by Gorman et al. for mismatch repair (Gorman et al., 2010) and also used for NER (Ghodke et al., 2014).
Summary
Using single molecule biophysical approaches coupled to TIRF microscopy, we have made major inroads into our understanding of how DNA glycosylases search for damage, by inserting a wedge amino acid into the DNA helix to probe for alterations in base pairing, base stacking, and sugar puckers produced by the lesion. Sophisticated chemo-mechanical modeling has also allowed for novel ways to interpret the data. Although our initial studies focused on three bacterial glycosylases that recognize free radical-damaged DNA bases, the totality of the human BER enzymes engaged in short-patch repair of radiation-damaged DNA is our next goal.
Highlights.
TIRF microscopy provides insight into the DNA damage search by DNA glycosylases
Glycosylases rotationally slide along the DNA backbone with a broad spectrum of diffusion constants
Glycosylases diffuse randomly along DNA inserting a wedge amino acid into the duplex to locate damage
Glycosylases stop upon encountering their substrate damage
Glycosylases from different structural families exhibit the same search behavior
Acknowledgments
The authors would like to thank the National Institutes of Health Grant P01 CA098993, awarded by the National Cancer Institute, for supporting the work in our laboratory. The authors are also grateful to our collaborator, Dr. David Warshaw, for his expertise, to Dr. Shane Nelson for computational simulation of the data and to Dr. Andrew Dunn for initiating these studies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311:1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Mandal SM, Das A, Hegde ML, Das S, Bhakat KK, Boldogh I, Sarkar PS, Mitra S, Hazra TK. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J Biol Chem. 2011;286:6006–6016. doi: 10.1074/jbc.M110.198796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SE, Sanderson RJ, Mosbaugh DW. Processivity of Escherichia coli and rat liver mitochondrial uracil-DNA glycosylase is affected by NaCl concentration. Biochemistry. 1995;34:6109–6119. doi: 10.1021/bi00018a014. [DOI] [PubMed] [Google Scholar]
- Blainey PC, Luo G, Kou SC, Mangel WF, Verdine GL, Bagchi B, Xie XS. Nonspecifically bound proteins spin while diffusing along DNA. Nat Struct Mol Biol. 2009;16:1224–1229. doi: 10.1038/nsmb.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc Natl Acad Sci U S A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaisdell JO, Wallace SS. Abortive base-excision repair of radiation-induced clustered DNA lesions in Escherichia coli. Proc Natl Acad Sci U S A. 2001;98:7426–7430. doi: 10.1073/pnas.131077798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David SS, O'Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogliotti E, Fortini P, Pascucci B, Parlanti E. The mechanism of switching among multiple BER pathways. Prog Nucleic Acid Res Mol Biol. 2001;68:3–27. doi: 10.1016/s0079-6603(01)68086-3. [DOI] [PubMed] [Google Scholar]
- Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem. 2008;283:3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- Dowd DR, Lloyd RS. Biological significance of facilitated diffusion in protein-DNA interactions. Applications to T4 endonuclease V-initiated DNA repair. J Biol Chem. 1990;265:3424–3431. [PubMed] [Google Scholar]
- Duclos S, Doublié S, Wallace SS. Consequences and Repair of Oxidative DNA Damage. In: Greim H, Albertini RJ, editors. The Cellular Response to the Genotoxic Insult: The Question of Threshold for Genotoxic Carcinogens. Cambridge, United Kingdom: The Royal Society of Chemistry; 2012. pp. 109–153. [Google Scholar]
- Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011;39:7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AM, Zhou J, Wallace SS, Burrows CJ. A Role for the Fifth G-Track in G-Quadruplex Forming Oncogene Promoter Sequences during Oxidative Stress: Do These "Spare Tires" Have an Evolved Function? ACS Cent Sci. 2015;1:226–233. doi: 10.1021/acscentsci.5b00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–3471. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodke H, Wang H, Hsieh CL, Woldemeskel S, Watkins SC, Rapic-Otrin V, Van Houten B. Single-molecule analysis reveals human UV-damaged DNA-binding protein (UV-DDB) dimerizes on DNA via multiple kinetic intermediates. Proc Natl Acad Sci U S A. 2014;111:E1862–E1871. doi: 10.1073/pnas.1323856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002;277:19811–19816. doi: 10.1074/jbc.M202058200. [DOI] [PubMed] [Google Scholar]
- Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17:932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruskin EA, Lloyd RS. Molecular analysis of plasmid DNA repair within ultraviolet-irradiated Escherichia coliI. I. T4 endonuclease V-initiated excision repair. J Biol Chem. 1988;263:12728–12737. [PubMed] [Google Scholar]
- Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 2005;4:41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Harrison L, Hatahet Z, Wallace SS. In vitro repair of synthetic ionizing radiation-induced multiply damaged DNA sites. J Mol Biol. 1999;290:667–684. doi: 10.1006/jmbi.1999.2892. [DOI] [PubMed] [Google Scholar]
- Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst) 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci U S A. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Hazra TK, Mitra S. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res. 2008a;18:27–47. doi: 10.1038/cr.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Hegde PM, Bellot LJ, Mandal SM, Hazra TK, Li GM, Boldogh I, Tomkinson AE, Mitra S. Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc Natl Acad Sci U S A. 2013;110:E3090–E3099. doi: 10.1073/pnas.1304231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Theriot CA, Das A, Hegde PM, Guo Z, Gary RK, Hazra TK, Shen B, Mitra S. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. J Biol Chem. 2008b;283:27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higley M, Lloyd RS. Processivity of uracil DNA glycosylase. Mutat Res. 1993;294:109–116. doi: 10.1016/0921-8777(93)90019-d. [DOI] [PubMed] [Google Scholar]
- Imamura K, Averill A, Wallace SS, Doublié S. Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA. J Biol Chem. 2012;287:4288–4298. doi: 10.1074/jbc.M111.315309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, Mitra S, Hazra TK. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193:43–65. doi: 10.1016/s0300-483x(03)00289-0. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Wilson DM., 3rd Overview of base excision repair biochemistry. Curr Mol Pharmacol. 2012;5:3–13. doi: 10.2174/1874467211205010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koval VV, Kuznetsov NA, Ishchenko AA, Saparbaev MK, Fedorova OS. Real-time studies of conformational dynamics of the repair enzyme E. coli formamidopyrimidine-DNA glycosylase and its DNA complexes during catalytic cycle. Mutat Res. 2010;685:3–10. doi: 10.1016/j.mrfmmm.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Koval VV, Kuznetsov NA, Zharkov DO, Ishchenko AA, Douglas KT, Nevinsky GA, Fedorova OS. Pre-steady-state kinetics shows differences in processing of various DNA lesions by Escherichia coli formamidopyrimidine-DNA glycosylase. Nucleic Acids Res. 2004;32:926–935. doi: 10.1093/nar/gkh237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47:7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov NA, Bergonzo C, Campbell AJ, Li H, Mechetin GV, de los Santos C, Grollman AP, Fedorova OS, Zharkov DO, Simmerling C. Active destabilization of base pairs by a DNA glycosylase wedge initiates damage recognition. Nucleic Acids Res. 2015a;43:272–281. doi: 10.1093/nar/gku1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov NA, Kladova OA, Kuznetsova AA, Ishchenko AA, Saparbaev MK, Zharkov DO, Fedorova OS. Conformational Dynamics of DNA Repair by Escherichia coli Endonuclease III. J Biol Chem. 2015b;290:14338–14349. doi: 10.1074/jbc.M114.621128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov NA, Koval VV, Zharkov DO, Vorobjev YN, Nevinsky GA, Douglas KT, Fedorova OS. Pre-steady-state kinetic study of substrate specificity of Escherichia coli formamidopyrimidine--DNA glycosylase. Biochemistry. 2007;46:424–435. doi: 10.1021/bi060787r. [DOI] [PubMed] [Google Scholar]
- Kuznetsov NA, Vorobjev YN, Krasnoperov LN, Fedorova OS. Thermodynamics of the multi-stage DNA lesion recognition and repair by formamidopyrimidine-DNA glycosylase using pyrrolocytosine fluorescence--stopped-flow pre-steady-state kinetics. Nucleic Acids Res. 2012;40:7384–7392. doi: 10.1093/nar/gks423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov NA, Zharkov DO, Koval VV, Buckle M, Fedorova OS. Reversible chemical step and rate-limiting enzyme regeneration in the reaction catalyzed by formamidopyrimidine-DNA glycosylase. Biochemistry. 2009;48:11335–11343. doi: 10.1021/bi901100b. [DOI] [PubMed] [Google Scholar]
- Lee AJ, Warshaw DM, Wallace SS. Insights into the glycosylase search for damage from single-molecule fluorescence microscopy. DNA Repair (Amst) 2014;20:23–31. doi: 10.1016/j.dnarep.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci U S A. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Imamura K, Averill AM, Wallace SS, Doublié S. Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure. 2013;21:247–256. doi: 10.1016/j.str.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Choudhury S, Roy R. In vitro and in vivo dimerization of human endonuclease III stimulates its activity. J Biol Chem. 2003;278:50061–50069. doi: 10.1074/jbc.M309997200. [DOI] [PubMed] [Google Scholar]
- Liu X, Roy R. Truncation of amino-terminal tail stimulates activity of human endonuclease III (hNTH1) J Mol Biol. 2002;321:265–276. doi: 10.1016/s0022-2836(02)00623-x. DOI: S002228360200623X [pii] [DOI] [PubMed] [Google Scholar]
- Mechetin GV, Zharkov DO. Mechanism of translocation of uracil-DNA glycosylase from Escherichia coli between distributed lesions. Biochem Biophys Res Commun. 2011;414:425–430. doi: 10.1016/j.bbrc.2011.09.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SR, Dunn AR, Kathe SD, Warshaw DM, Wallace SS. Two glycosylase families diffusively scan DNA using a wedge residue to probe for and identify oxidatively damaged bases. Proc Natl Acad Sci U S A. 2014;111:E2091–E2099. doi: 10.1073/pnas.1400386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porecha RH, Stivers JT. Uracil DNA glycosylase uses DNA hopping and short-range sliding to trap extrahelical uracils. Proc Natl Acad Sci U S A. 2008;105:10791–10796. doi: 10.1073/pnas.0801612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash A, Doublié S, Wallace SS. The Fpg/Nei family of DNA glycosylases: substrates, structures, and search for damage. Prog Mol Biol Transl Sci. 2012;110:71–91. doi: 10.1016/B978-0-12-387665-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purmal AA, Lampman GW, Pourmal EI, Melamede RJ, Wallace SS, Kow YW. Uracil DNA N-glycosylase distributively interacts with duplex polynucleotides containing repeating units of either TGGCCAAGCU or TGGCCAAGCTTGGCCAAGCU. J Biol Chem. 1994;269:22046–22053. [PubMed] [Google Scholar]
- Qi Y, Nam K, Spong MC, Banerjee A, Sung RJ, Zhang M, Karplus M, Verdine GL. Strandwise translocation of a DNA glycosylase on undamaged DNA. Proc Natl Acad Sci U S A. 2012;109:1086–1091. doi: 10.1073/pnas.1111237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage E, Harrison L. Clustered DNA lesion repair in eukaryotes: relevance to mutagenesis and cell survival. Mutat Res. 2011;711:123–133. doi: 10.1016/j.mrfmmm.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko VS, Mechetin GV, Nevinsky GA, Zharkov DO. Correlated cleavage of single- and double-stranded substrates by uracil-DNA glycosylase. FEBS Lett. 2008;582:410–414. doi: 10.1016/j.febslet.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Sutherland BM, Bennett PV, Sidorkina O, Laval J. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc Natl Acad Sci U S A. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot CA, Hegde ML, Hazra TK, Mitra S. RPA physically interacts with the human DNA glycosylase NEIL1 to regulate excision of oxidative DNA base damage in primer-template structures. DNA Repair (Amst) 2010;9:643–652. doi: 10.1016/j.dnarep.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SS. Enzymatic processing of radiation-induced free radical damage in DNA. Radiat Res. 1998;150:S60–S79. [PubMed] [Google Scholar]
- Wallace SS. DNA glycosylases search for and remove oxidized DNA bases. Environ Mol Mutagen. 2013;54:691–704. doi: 10.1002/em.21820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA Repair (Amst) 2003;2:441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- Ward JF. Some biochemical consequences of the spatial distribution of ionizing radiation-produced free radicals. Radiat Res. 1981;86:185–195. [PubMed] [Google Scholar]
- Ward JF. Biochemistry of DNA lesions. Radiat Res Suppl. 1985;8:S103–S111. [PubMed] [Google Scholar]
- Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Wilson DM, 3rd, Sofinowski TM, McNeill DR. Repair mechanisms for oxidative DNA damage. Front Biosci. 2003;8:d963–d981. doi: 10.2741/1109. [DOI] [PubMed] [Google Scholar]
- Yang N, Chaudhry MA, Wallace SS. Base excision repair by hNTH1 and hOGG1: a two edged sword in the processing of DNA damage in gamma-irradiated human cells. DNA Repair (Amst) 2006;5:43–51. doi: 10.1016/j.dnarep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Yang N, Galick H, Wallace SS. Attempted base excision repair of ionizing radiation damage in human lymphoblastoid cells produces lethal and mutagenic double strand breaks. DNA Repair (Amst) 2004;3:1323–1334. doi: 10.1016/j.dnarep.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Zhao X, Krishnamurthy N, Burrows CJ, David SS. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010;49:1658–1666. doi: 10.1021/bi901852q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002;21:789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zharkov DO, Grollman AP. The DNA trackwalkers: principles of lesion search and recognition by DNA glycosylases. Mutat Res. 2005;577:24–54. doi: 10.1016/j.mrfmmm.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Zhou J, Fleming AM, Averill AM, Burrows CJ, Wallace SS. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015;43:4039–4054. doi: 10.1093/nar/gkv252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu M, Fleming AM, Burrows CJ, Wallace SS. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J Biol Chem. 2013;288:27263–27272. doi: 10.1074/jbc.M113.479055. [DOI] [PMC free article] [PubMed] [Google Scholar]