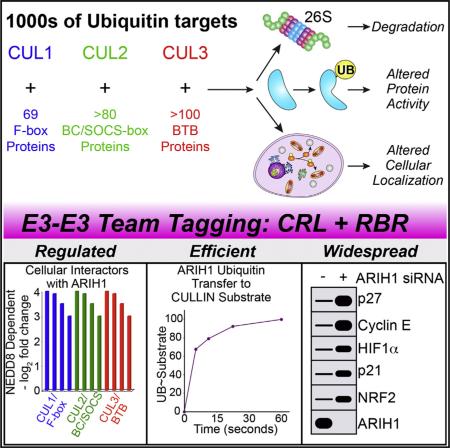
SUMMARY
Hundreds of human cullin-RING E3 ligases (CRLs) modify thousands of proteins with ubiquitin (UB) to achieve vast regulation. Current dogma posits that CRLs first catalyze UB transfer from an E2 to their client substrates and subsequent polyubiquitylation from various linkage-specific E2s. We report an alternative E3-E3 tagging cascade: many cellular NEDD8-modified CRLs associate with a mechanistically distinct thioester-forming RBR-type E3, ARIH1, and rely on ARIH1 to directly add the first UB and, in some cases, multiple additional individual monoubiquitin modifications onto CRL client substrates. Our data define ARIH1 as a component of the human CRL system, demonstrate that ARIH1 can efficiently and specifically mediate monoubiquitylation of several CRL substrates, and establish principles for how two distinctive E3s can reciprocally control each other for simultaneous and joint regulation of substrate ubiquitylation. These studies have broad implications for CRL-dependent proteostasis and mechanisms of E3-mediated UB ligation.
Graphical abstract
INTRODUCTION
A primary mechanism regulating functions of most eukaryotic proteins involves ubiquitin (UB) ligation via E1-E2-E3 enzyme cascades, where E1 catalyzes formation of a thioester-bonded E2~UB intermediate, which subsequently collaborates with an E3 that recruits the specific client substrate for ubiquitylation. In general, E3s are thought to catalyze ubiquitylation in one of two ways. For thioester-forming E3s (e.g., HECTs and RBRs), UB is first transferred from E2~UB to the E3′s catalytic Cys and then from the E3's Cys to the remotely bound substrate. Alternatively, most E3s lack a catalytic Cys but instead display a hallmark RING domain thought to recruit, position, and activate an E2~UB intermediate from which UB is transferred to the substrate (reviewed in Berndsen and Wolberger, 2014; Deshaies and Joazeiro, 2009; Metzger et al., 2014; Streich and Lima, 2014; Vittal et al., 2015).
The largest family of E3s consists of modular, multisubunit cullin-RING ligases (CRLs) nucleated by RBX1 or RBX2 RING proteins bound to specific cullins. A cullin indirectly recruits substrates by assembling with one of numerous interchangeable substrate receptors (SRs) (e.g., ≈70 F-box proteins in CUL1-RBX1-containing CRL1s or >100 BTB proteins in CUL3-RBX1-containing CRL3s) (Lydeard et al., 2013; Zimmerman et al., 2010). Together, the large family of >200 CRLs in humans modifies thousands of proteins to regulate diverse physiology (Emanuele et al., 2011; Harper and Tan, 2012; Kim et al., 2011; Yen and Elledge, 2008).
CRLs are activated by self-modification with the UB-like protein NEDD8 via a conventional mechanism, with the RBX RING activating a NEDD8-specific E2. Cullin neddylation controls 10%–20% of all cellular ubiquitylation by regulating CRL conformations and interactions in a multifaceted mechanism: substrates awaiting ubiquitylation interrupt cycles of neddylation catalyzed by the RBX RING, deneddylation catalyzed by the enzyme CSN, and CUL-RBX-SR assembly catalyzed by CAND1 (reviewed in Lydeard et al., 2013). Ultimately, NEDD8 and SRs are markers of active, substrate-bound CRLs in human cells, whereas entirely blocking neddylation, for example, by treating cells with the MLN4924 inhibitor that is in anti-cancer clinical trials, massively decreases CRL substrate ubiquitylation (Soucy et al., 2009). Ubiquitylation of CRL substrates is also inhibited by NEDD8 Q40E conversion, produced by effector deamidating enzymes during some pathogenic bacterial infections to thwart NEDD8-induced CUL-RBX conformational changes (Yu et al., 2015). Furthermore, CRISPR knockout screens identified NEDD8, RBX1, CULs 1–3, and their neddylating E1 and E2 enzymes as essential for cell proliferation (Hart et al., 2015; Wang et al., 2015). Although the pathways mediating NEDD8-dependent CRL substrate ubiquitylation remain incompletely characterized, neddylated CRLs can activate E2s in the UBCH5 or CDC34 families in vitro (Duda et al., 2008; Saha and Deshaies, 2008; Yamoah et al., 2008), and some CRL substrates do accumulate upon knocking down these E2s (Ceccarelli et al., 2011; Wu et al., 2010). However, UBCH5 and CDC34-family E2s were not essential in the CRISPR screens potentially due to redundancy, or possibly reflecting unknown enzymes contributing to CRL-catalyzed substrate ubiquitylation under some circumstances (Hart et al., 2015; Wang et al., 2015).
We sought to investigate an enigmatic function of CRL neddylation: activation of intrinsic catalytic activity of ARIH1, an Ariadne family RBR E3 UB ligase (Kelsall et al., 2013). ARIH1 displays a typical RBR RING1-IBR-RING2 tri-domain module, where the RING1 domain binds a cognate E2, UBCH7 (aka UBE2L3), in a manner resembling conventional RING-E2 interactions but UB is transferred from the bound UBCH7 to a distinctive RING2 domain catalytic Cys (Wenzel et al., 2011). RBR E3 RING2~UB intermediates in principle are thought to catalyze ubiquitylation of substrate lysines. However, ARIH1 is autoinhibited by intramolecular RING2-Ariadne domain interactions sequestering the catalytic Cys, which in vitro became exposed in the presence of neddylated CUL1-RBX1 but not CUL5-RBX2 (Duda et al., 2013; Kelsall et al., 2013). GFPARIH1 expressed in cultured cells associated with neddylated CULs 1, 2, 3, and 4A but not CUL5, and with RBX1, and this was abolished by MLN4924 treatment, although the functions of these interactions remained elusive (Kelsall et al., 2013).
In an effort to identify cellular targets of ARIH1, we discovered its association with NEDD8 and several CRL SRs, which are hallmarks of assembled, active CRL E3s. We report an efficient, multimodal, and specific mechanism by which neddylated CRLs integrate the relief of ARIH1 autoinhibition with direct ARIH1-mediated monoubiquitylation of CRL client substrates, which can then be rapidly polyubiquitinated by the traditional CRL mechanism involving the E2 CDC34. Importantly, factors regulating CRL substrate ubiquitylation in vivo are required for ARIH1-dependent targeting of CRL substrates in vitro. Furthermore, blocking ARIH1/CRL pathways via dominant-negative ARIH1 or knockdown leads to accumulation of CRL substrates and receptors in vivo and disrupts cell division, implying important roles in CRL biology.
RESULTS
Quantitative Proteomics Reveals ARIH1 Predominantly and Extensively Interacts with Activated Cullin-RING E3s
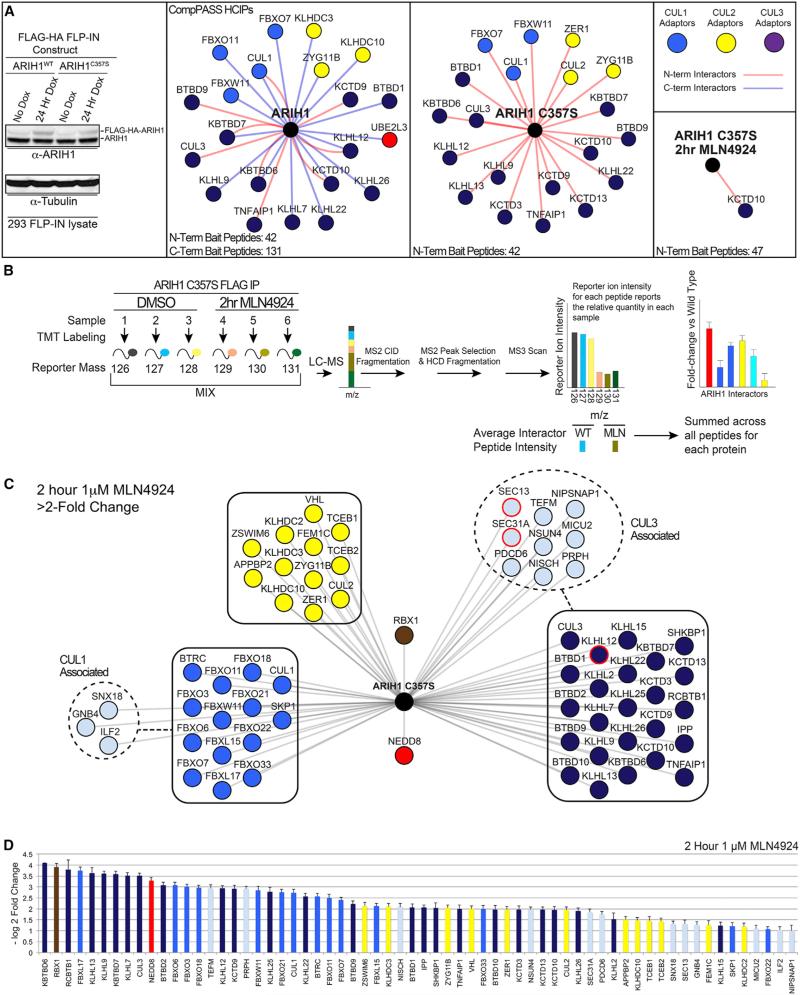
We used proteomics to examine ARIH1's global interactome, using 293T cells expressing HA-FLAG-tagged wild-type (WT), the equivalent C-terminally tagged WT protein, or inactive (catalytic C357S mutant) HA-FLAG-ARIH1 (Figure 1A). Subjecting anti-FLAG immune complexes to mass spectrometry identified high-confidence candidate interacting proteins (HCIPs) based on semiquantitative spectral counting using the CompPASS platform (Figures 1A and S1A; Table S1) (Sowa et al., 2009). Parallel experiments from cells treated with MLN4924 interrogated potential roles for neddylation (Figures 1A and S1A).
Figure 1. ARIH1 Broadly Interacts with Components of Assembled, Neddylated CRLs.
(A) Left to right: immunoblot of low-level induced expression of FLAG-HA-ARIH1 or catalytic C375S mutant in 293 FLP-IN cells; high-confidence interacting proteins (HCIPs) for WT ARIH1 and ARIH1C357S, ± neddylation inhibitor MLN4924, identified by CompPASS (Sowa et al., 2009). ARIH1 predominantly interacts with CRL subunits colored by CRL1, blue; CRL2, yellow; CRL3, purple.
(B) TMT method to identify MLN4924-sensitive/neddylation-dependent ARIH1 HCIPs.
(C) ARIH1C357S HCIPs decreased ≥2-fold by neddylation inhibitor MLN4924 colored by CRL: CRL1, blue; CRL2, yellow; CRL3, violet. CRL3KLHL12 and substrate SEC13-SEC31A are highlighted. Error, ±SEM.
(D) Log2 fold change of ARIH1C357S interactors from the schematic in (C). Error bars, ±SE.
The largest single category of HCIPs for both WT and inactive ARIH1 were components of CRLs, including numerous SRs (F-box, BC/SOCS-box, and BTB proteins) and their respective cullins (Figure 1A). More than a dozen SRs associated with both WT and inactive ARIH1, including 13 found for both. Almost all interactions between ARIH1C357S and CRL components were lost upon treatment with MLN4924 as determined by peptide spectral counting, indicating NEDD8-dependent association (Figures 1A and S1A).
ARIH1 association with SRs suggested potentially broader roles in CRL biology than previously recognized. We probed this further by a more sensitive and quantitative Tandem Mass Tagging (TMT)-based proteomic workflow (Figure 1B) (Huttlin et al., 2015; McAlister et al., 2014), using near endogenous HA-FLAG-ARIH1C357S expression levels. Summed intensities for TMT-reporter ions quantified ARIH1C357S interaction with 13 F-box proteins, 9 BC-box proteins, and 23 BTB proteins, all of whose association was reduced with MLN4924 (Figures 1C and 1D). Although relatively fewer CRL SRs were detected with WT ARIH1 by a six-plex TMT approach, the vast majority were also neddylation dependent (Figures S1B and S1C). Retrospective analyses of previous studies of CRL interactomes indicates that inducibly expressed ARIH1 can access a large fraction of endogenous CRL SRs in 293T cells and that endogenous ARIH1 associates with CUL1, NEDD8, and several CRL SRs (Figure S1D; Table S2) (Bennett et al., 2010; Huttlin et al., 2015; Lee et al., 2011). Notably, unlike bulk CRL immune complexes, we did not detect CSN subunits or CAND1 as HCIPs, suggesting that ARIH1-associated CRLs are not actively within the CSN/CAND1 inactivation/disassembly cycle.
We also detected evidence of ARIH1C357S associating with a fully assembled CRL-substrate complex, as SEC31A bound to SEC13 is an established substrate of neddylated CRL3KLHL12 (Jin et al., 2012) (Figure 1C). The persistence of this ligase-substrate interaction may reflect that neddylated CRL3KLHL12 mediates multimonoubiquitylation of SEC31A to regulate macro-molecular architecture rather than proteasomal degradation (Jin et al., 2012).
Overall, the large fraction of HCIPs that are CRL1–3 SRs, elimination of interactions by MLN4924, relative lack of CAND1 and CSN, and observation of a non-degradative CRL substrate collectively suggested that ARIH1 engages some assembled, active CRLs in cells.
ARIH1 Acts as a Monoubiquitin Conjugating Enzyme for Neddylated CUL3KLHL12-RBX1 Substrate SEC13-SEC31A
To gain functional insights, we reconstituted the complex identified by proteomics (Figure 1C) with recombinant neddylated or unneddylated CUL3-RBX1, the SR KLHL12 and its substrate SEC13-SEC31A. ARIH1-dependent UB ligation was selectively monitored by using UBCH7 (UBE2L3) as E2, and following fluorescent *UB's path in pulse-chase assays (Figures 2A, 2B, and S2A). After “pulse” generation of thioester-linked *UB~UBCH7, ARIH1 was added with various components of the RBX1-CUL3KLHL12-SEC13-SEC31A complex. The *UB~UBCH7 intermediate is inert with either a neddylated CRL that cannot itself directly activate substrate ubiquitylation UBCH7, or when mixed with autoinhibited ARIH1 (Duda et al., 2013; Moynihan et al., 1999; Wenzel et al., 2011). However, if ARIH1 is relieved from autoinhibition such as described previously for neddylated CUL1-RBX1 (Kelsall et al., 2013), then *UB is presumably “chased” first onto ARIH1's catalytic Cys, and then from the ARIH1~*UB intermediate onto a suitably placed nucleophilic acceptor (~ refers to covalent bond, either reactive thioester or stable isope-peptide) (Wenzel et al., 2011). Neddylated CRL3KLHL12 relieves ARIH1 autoinhibition and stimulates UB transfer from UBCH7 to ARIH1, with ensuing autoubiquitylation rapidly producing stably modified ARIH1 (Figures 2B, S2A, and S2B). Thus, although it is not possible to isolate the ARIH1~UB thioester-bonded intermediate, initiating reactions with UBCH7~*UB solely probes an activated ARIH1 pathway.
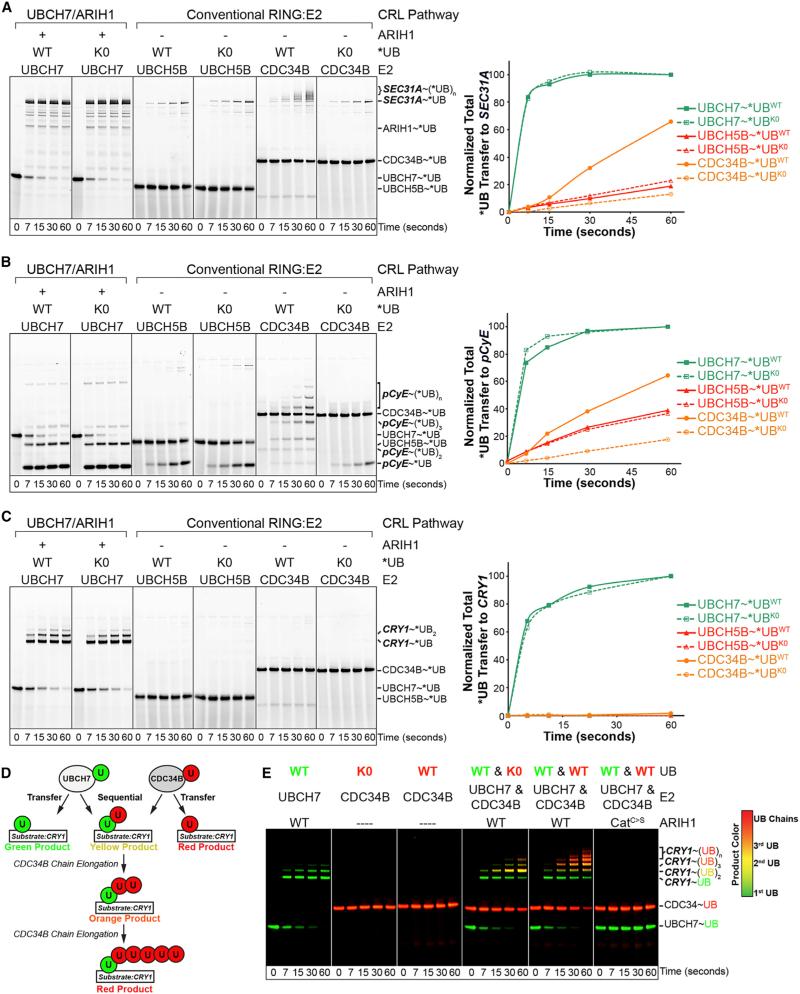
Figure 2. ARIH1 Rapidly, Efficiently, and Specifically Mediates UB Ligation to CRL Substrates.
(A) Pulse-chase reaction scheme. Pulse: thioester-linked UBCH7~*UB is generated and E1 reaction is quenched. Chase: ARIH1 is added; path of fluorescent UB* via ARIH1 is followed in presence or absence of neddylated or non-neddylated CUL-RBX1, SR, and CRL substrate indicated in bold.
(B) Fluorescent scans of gels of assay in (A), showing neddylated CRL3KLHL12 relieves ARIH1 autoinhibition, promotes autoubiquitylation in absence of CRL substrate, and UB ligation to CRL substrate SEC31A depending on neddylation, the SR KLHL12, and ARIH1 catalytic Cys357.
(C) Inferred pathway from (B) and generalization: ARIH1 mediates UB ligation to CRL client substrates depending on CUL neddylation, the substrate receptor, and ARIH1's catalytic Cys.
(D) Fluorescent scans of gels of assay in (A), showing neddylated CRL1FBW7ΔD relieves ARIH1 autoinhibition, promotes autoubiquitylation in absence of CRL substrate, and UB ligation to CRL peptide substrate pCyE depending on neddylation (N8~CUL1), SR, and ARIH1 catalytic Cys.
(E) Assays showing UBCH7-ARIH1-mediated UB ligation to pCyE as a function CRL1FBW7ΔD concentration for kinetic analyses.
(F) Michaelis-Menten analysis of data from (E) to calculate kinetic parameters for ARIH1-mediated UB ligation to pCyE via neddylated CRL1FBW7ΔD.
Remarkably, when neddylated CRL3KLHL12 and SEC13-SEC31A were both added, *UB was preferentially transferred to the KLHL12 substrate SEC31A. This depended on ARIH1's catalytic Cys, and on CUL3 neddylation and KLHL12 in accordance with cellular requirements for an active CRL (Figure 2B). Overall, the requirements of the reaction suggested that UB was transferred from UBCH7 to ARIH1's catalytic Cys to the CRL substrate SEC31A (Figure 2C). Similar products were observed with *UB and *UB-K0 that cannot form Lys-linked polyUB chains in a series of reactions where ARIH1 concentrations were systematically decreased (Figure S2C). Reducing the levels of ARIH1 ensures that it undergoes multiple catalytic cycles to deplete the starting *UB~UBCH7, thus increasing the probability of each molecule of ARIHI1 confronting *UB-modified SEC31A as an alternative substrate if UB chain elongation was been the preferred reaction (Figure S2C). The simplest explanation for the results is that a neddylated RBX1-CUL3KLHL12-UBCH7-ARIH1 pathway preferentially produces multiply monoubiquitylated SEC31A (Figure 2C), mirroring cellular regulation by CRL3KLHL12 (Jin et al., 2012).
ARIH1 Mediates Efficient Monoubiquitin Conjugation to Some Well-Characterized CRL1 Substrates
Remarkably, the UBCH7-ARIH1 mechanism also applied to structurally defined CUL1-based F-box protein-substrate systems. In the presence of the simplified monomeric CRL1FBW7ΔD (Hao et al., 2007), *UB was transferred from UBCH7 via ARIH1 to a peptide derived from phosphorylated human Cyclin E/CCNE1 (CyE), referred to as pCyE (Figures 2C and 2D). This depended on essential features of both the active CRL (neddylation and the SR) and activated ARIH1 (the catalytic Cys), with a Kmapp of 0.58 μM and kcatapp of 0.44 s−1 upon titrating CRL1FBW7ΔD into the reactions (Figures 2D–2F). Although it is difficult to directly compare the efficiency of UBCH7-ARIH1 and the conventional RBX1-E2 reaction mechanisms, the Km of ARIH1 for neddylated CRL1FBW7ΔD is slightly lower than Km values previously reported for the E2s UBCH5 or CDC34 with other neddylated SCF/CRL1 E3s, while ARIH1 exhibited a 10- to 100-fold greater catalytic efficiency for substrate ubiquitylation (Saha and Deshaies, 2008). In order to examine the ARIH1 and conventional pathways side by side, we carried out reactions with protein concentrations that should yield reaction rates at approximately similar, and thus comparable, magnitude below maximal velocity for each system. In these reactions *UB transfer to SEC31 or pCyE was more rapid by the UBCH7-ARIH1-neddylated CRL pathway than by conventional RBX1 RING catalysis with the E2s UBCH5 (aka UBE2D2) or CDC34 (aka UBE2R2), although CDC34 more efficiently generated poly*UB chains after a lag phase (Figures 3A, 3B, and S2D).
Figure 3. ARIH1-CRL Mechanism Efficiently Mediates monoUB Ligation to CUL3 and CUL1 Substrates.
(A) Pulse-chase assays starting with indicated E2~*UB or~*UB K0 (cannot form Lys-linked polyUB chains) transfer to CRL3KLHL12 substrate SEC31A via UBCH7 by ARIH1-CRL pathway or with E2s UBCH5B or CDC34B by canonical CRL RING mechanism. Right, quantification of total *UB or *UB K0 transfer to SEC31A.
(B) As (A), but for CRL1FBW7ΔD and client substrate pCyE.
(C) As (A), but for CRL1FBXL3 and client substrate CRY1.
(D) Scheme of two-color pulse-chase assay tracking UB (WT or K0) transferred from either UBCH7 or CDC34B in the same tube to NEDD8~CRL1FBXL3 client substrate CRY1. In pulse, UBCH7 was loaded with “green” *UB and CDC34 with “red” *UB or *UB K0, and E1 reaction was quenched. Distinguishing green-, yellow-, orange-, and red-colored products in chase reaction allows distinguishing individual E2 reactions and sequential *UB priming by ARIH1/UBCH7 and chain extension by CDC34B.
(E) Reactions from (D) simultaneously monitoring UBCH7 and CDC34-mediated *UB transfer to CRL1FBXL3 client CRY1 indicates priming by UBCH7-ARIH1 and extending by CDC34.
The apparent efficiency of the UBCH7-ARIH1 mechanism was even more striking for the CRL1 substrate cryptochome1 (CRY1) purified in 1:1:1 stoichiometry with its cognate SR complex FBXL3-SKP1 (Busino et al., 2007; Xing et al., 2013). During the time in which all the *UB~UBCH7 was converted into monoubiquitylated CRY1 via the ARIH1-neddylated CRL pathway, little UB transfer occurred by conventional RBX1 activation of the E2s UBCH5 or CDC34 even at 20-fold higher *UB~E2 concentrations that presumably saturate the neddylated CRL1FBXL3 (Figures 3C, S2E, and S2F). This experiment also demonstrated remarkable relative substrate specificity, as 125 lysines in neddylated CUL1-RBX1-SKP1-FBXL3 stoichiometrically present with CRY1 in the reactions were not substantially modified by the ARIH1-UBCH7 path (Figure 3C).
Efficient Neddylated CRL Substrate Polyubiquitylation Can Be Achieved by UBCH7-ARIH1-Mediated Priming and CDC34-Mediated Chain Elongation
Because pCyE and CRY1 were efficiently monoubiquitylated via UBCH7-ARIH1, but polyubiquitylation was observed only with the traditional CRL-E2 mechanism with CDC34 after a lag phase, we asked whether the two pathways could converge (Figures 3B–3D). We labeled UB with different fluorophores (fluorescein, green; TAMRA, red) with spectral properties allowing simultaneous detection. Individually “colored” thioester-bonded E2~UB intermediates (green *UB~UBCH7 and red WT or K0 *UB~CDC34) were added individually or together to neddylated CUL1-RBX1, ARIH1, and either SKP1-FBXL3 or SKP1-FBW7ΔD and their respective substrates, CRY1 or pCyE. Thus, modification by either UBCH7-ARIH1 or CDC34 is distinguished by loss of a green or red UB~E2 intermediate and concomitant addition of the respective UB to the substrate. Convergence of both paths would yield dual signals, with a gradient from yellow to orange to red with each additional red UB added by CDC34 to a green ARIH1-marked substrate (Figure 3D). As in reactions with CDC34 alone, CDC34 carrying K0 UB was relatively ineffective at substrate ubiquitylation. However, CDC34 efficiently elongated chains initiated by ARIH1, as shown by the yellow di-UB modified product containing one green UB and a red K0 UB, and the progressively more red products of reactions where CDC34 carried a WT UB (Figures 3E and S3A). The relative length and speed of UB chain formation by CDC34 is restricted in these assays by limiting concentrations and sub-saturating starting concentrations of *UB~CDC34, respectively. If CDC34~UB is included in reactions at saturating levels, then the CRY1 marked by *UB from ARIH1 is further rapidly modified by more than ten UBs (Figure S2G). The requirement for ARIH1's active site Cys demonstrated its catalytic role. We also found that this mechanism applies to yet another well-characterized CDC34-dependent pathway, CRL1SKP2-mediated ubiquitylation of p27/CDKN1B (Figures S2H and S3B), although modification of an IκBα peptide by CRL1FBXW11 is slightly more rapid with UBCH5 in agreement with a prior report showing UBCH5 can mediate its priming (Figure S3C) (Wu et al., 2010). Taken together, the experiments revealed that UBCH7-ARIH1 is more efficient than CDC34 for priming several CRL substrates in vitro, and that polyubiquitylation is readily achieved by sequential monoubiquitylation by UBCH7-ARIH1 and subsequent UB chain elongation by CDC34.
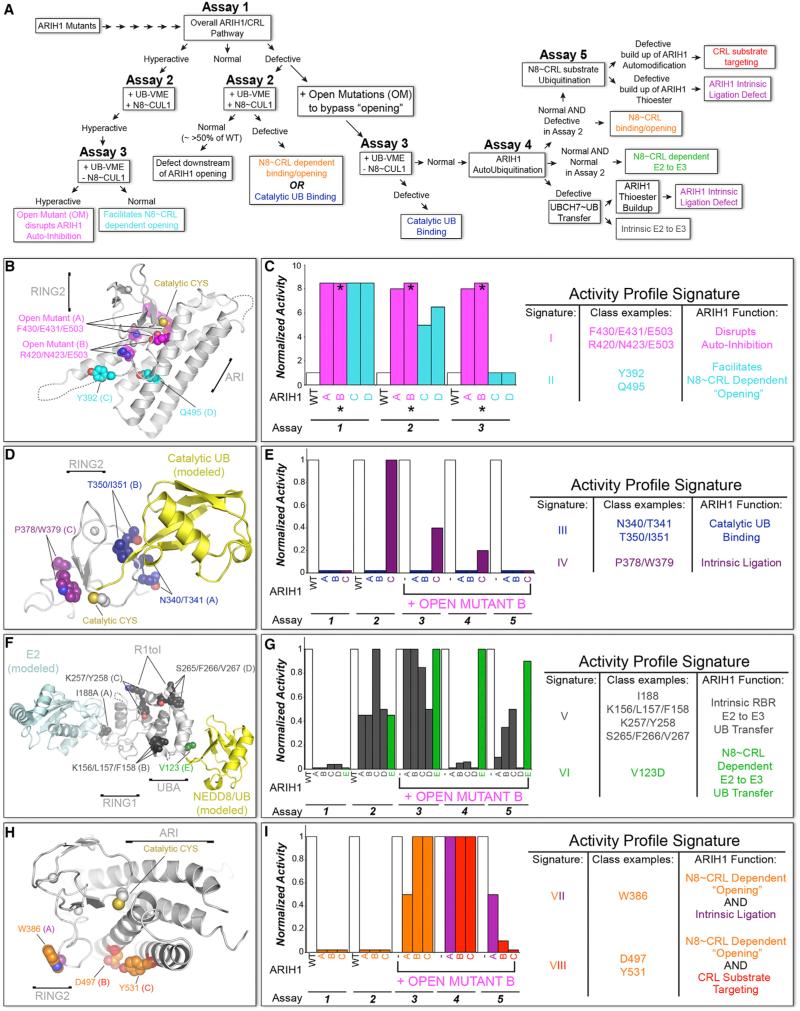
Activity Profiling of Mutants Reveals Mechanisms Establishing ARIH1-Neddylated CRL Ubiquitination Specificity
Legitimate biological pathways typically depend on biochemical specificity. As a first test of specificity, we identified ARIH1/CRL-dependent ubiquitylation sites on p27 by mutagenesis and on CRY1 by mass spectrometry (Figures S3D–S3G). p27 lysines modified by the joint ARIH1/CRL mechanism in vitro match those modified in cell lysates and in cells (Shirane et al., 1999; Yoshida et al., 2015). We found the same sites also modified by the conventional RING-E2 mechanism with UBCH5, albeit more slowly. This suggests that targeting may depend on lysine context due to surrounding sequences or could rely on spatial presentation (Figures S3E–S3G). Interestingly, although numerous lysines and multiple F-box proteins contribute to CRY1 degradation in vivo, in vitro ARIH1 and neddylated CRL1FBXL3 preferentially target sites of CRY1 ubiquitylation detected in cells (K442 and 456; Yoo et al., 2013 and http://www.phosphositeplus.org), which spatially parallel the sole ubiquitylation site in the pCyE peptide and potentially those in p27 as well (Figure S3G).
To understand how ARIH1 specifically targets CRL substrates, we biochemically profiled mutants in a three-tiered strategy. Briefly, after an initial broad survey, selected ARIH1 variants were subjected to a hierarchy of assays dissecting steps in the reaction. Activity profile “signatures” or “fingerprints” assigned roles of mutated residues in (1) mediating ARIH1 autoinhibition; (2) neddylated CRL-dependent relief of ARIH1 autoinhibition; (3) UB transfer from UBCH7 to ARIH1 RING2's catalytic Cys; (4) intrinsic RING2-catalyzed UB ligation to a Lys nucleophile; and/or (5) steering RING2~UB catalytic activity specifically toward the CRL client substrate. Then, deletion and point-mutated versions of neddylated CRLs were tested for corresponding defects. Overall, the data revealed extreme functional amalgamation of ARIH1 and neddylated CRLs, as described below and in Figures 4, 5, and 6 and S4, S5, and S6.
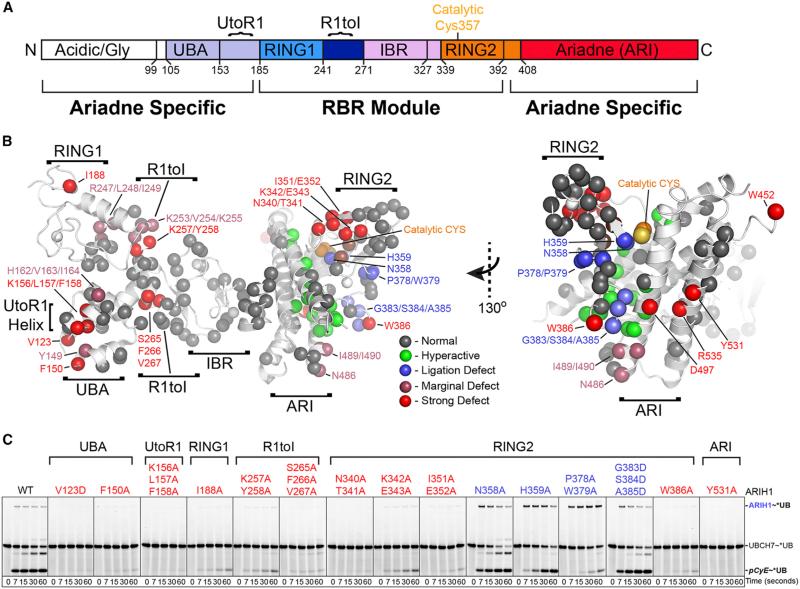
Figure 4. Broad Mutational Survey Reveals Numerous ARIH1 Surfaces Contribute to CRL Substrate Ubiquitylation.
(A) ARIH1 domains.
(B) Sites of Ala mutations as spheres on structure of autoinhibited ARIH1 (Duda et al., 2013), colored by effect on *UB transfer from UBCH7 via ARIH1/neddylated CRL1FBW7ΔD to pCyE (in Figure S4): gray, normal; green, hyperactive; blue, ligation defect/accumulation of normally transient thioester-bonded ARIH1~UB intermediate; raspberry, marginal defect; red, strong defect.
(C) Most defective (red) and ligation-impaired (blue) ARIH1 Ala scan mutants in pulse-chase assays monitoring entire pathway: *UB transfer from UBCH7 via the indicated ARIH1 mutants and neddylated CRL1FBW7ΔD to pCyE. Ligation-impaired mutants are scored by appearance and dissipation of ARIH1~UB thioester intermediate.
Figure 5. Activity Profiling of ARIH1 Mutants: “Signatures” Identifying Functions in UB Ligation to Neddylated CRL Substrates.
(A) Hierarchical assay scheme to profile selected ARIH1 mutants in neddylated CRL pathway. The sum of results from all assays form a signature, or “fingerprint” identifying normal function(s) of a mutated surface. Color coding of functions is maintained in structures and signatures in (B)–(I), except signatures VII and VIII that signify dual functions (orange/purple and orange/red). Details are in Figure S5.
(B) Close up showing ARIH1 catalytic Cys that is normally sequestered by autoinhibitory intramolecular RING2-Ariadne (ARI) domain interface (4KBL.PDB). Hyperactive mutant sites are spheres: magenta, relieve autoinhibition even without neddylated CRL, thus termed “OPEN” due to mutationally opening the RING2-ARI interface; cyan, facilitate “opening” by neddylated CRL, monitored by reactivity with *UB-VME. OPEN mutant B was combined with other mutations for assays 3, 4, and 5 in subsequent profiles.
(C) Activity profile signatures of hyperactive mutants, normalized to WT ARIH1, defined by assays 1–3.
(D) Model of ARIH1 RING2~UB intermediate based on 4KBL.PDB and 4LJO.PDB. Sites of activity profile signatures indicating roles in catalytic UB binding or intrinsic ligation activity are in blue and purple spheres, respectively.
(E) Activity profile signatures for mutants in (D).
(F) ARIH1's RING1 and UBA domains modeled with docked E2 or UBL (NEDD8/UB), respectively (4KBL.PDB, 5EDV.PDB, 4UN2.PDB). Sites of mutants with activity profile signatures indicating roles in UB transfer from E2 (UBCH7) to ARIH1 and neddylated CRL-dependent UB transfer from UBCH7 to ARIH1 are in gray and green spheres, respectively.
(G) Activity profile signatures for mutants in (F).
(H) ARIH1 structure as in (B), rotated ≈60° about x, highlighting residues in RING2 and ARI domains with dual roles in two colors: neddylated CUL-dependent relief of ARIH1 autoinhibition, orange; role in intrinsic RING2-mediated UB ligation to a Lys nucleophile, purple; role in specifically targeting CRL substrate, red.
(I) Activity profile signatures for mutants in (H).
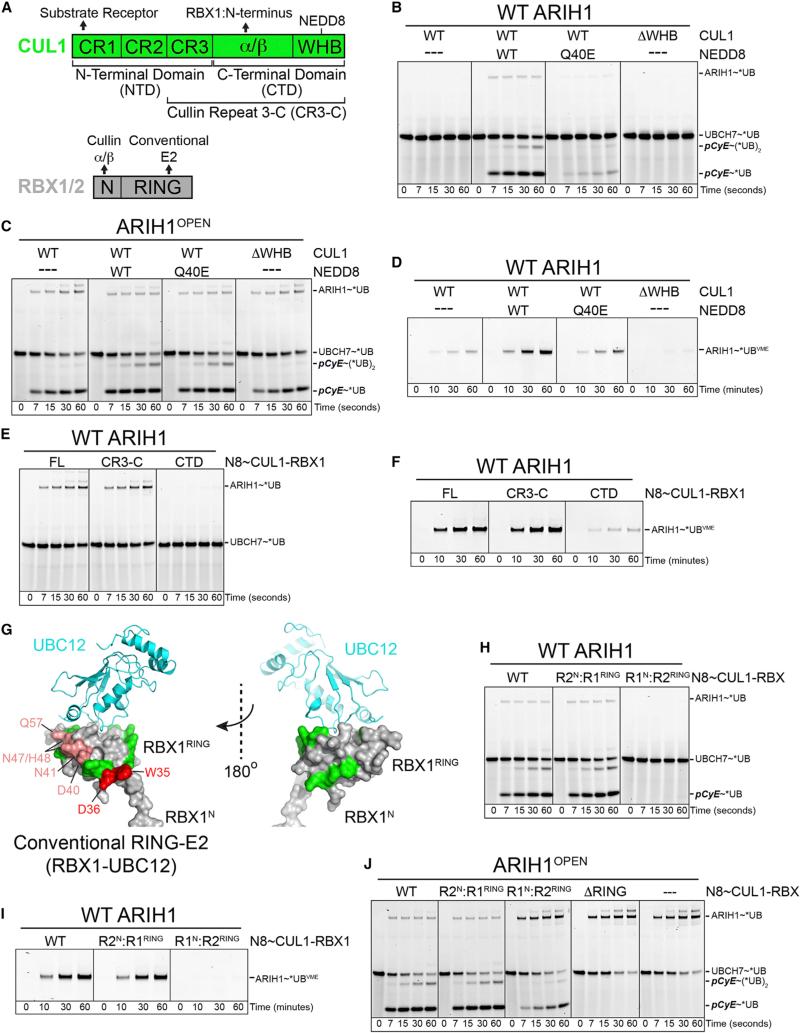
Figure 6. Distinctive and Specialized Roles of Neddylation, CUL1, and RBX1 in Employing ARIH1 to Target a CRL Substrate.
(A) CUL1 and RBX domains (Zheng et al., 2002), denoting previously known functional interactions.
(B) Pulse-chase assay as in Figure 2A testing role of neddylation in UBCH7-ARIH1-mediated *UB transfer to pCyE with indicated versions of CRL1FBW7ΔD (± neddylation, with NEDD8 Q40E mutant that does not support CUL1-RBX1 conformational change, or with CUL1 ΔWHB lacking domain with neddylation site).
(C) Assay as in (B) except with ARIH1OPEN mutant that is not autoinhibited. ARIH1OPEN overcomes defects from lack of neddylation, indicating role of CRL neddylation in relieving ARIH1 autoinhibition.
(D) Roles of neddylation in promoting ARIH1 “opening” as probed reactivity with *UB-VME, showing defect caused by Q40E and that neddylation is not mimicked by deleting CUL1's WHB domain.
(E) Role of CUL1 N-terminal domain, specifically the CR3 subdomain, in relieving ARIH1 autoinhibition, tested by ARIH1 autoubiquitylation in presence of indicated versions of neddylated CUL1-RBX1 (no SR, no substrate).
(F) Role of CUL1 N-terminal domain, specifically the CR3 subdomain, in stimulating ARIH1 “opening,” probed by ability of indicated versions of neddylated CUL1-RBX1 to stimulate ARIH1 reactivity with *UB-VME.
(G) Effects of RBX1 mutants on ARIH1-mediated ubiquitylation of neddylated CRL substrate, mapped on conventional RING E3-E2 (RBX1-UBC12) structure (Scott et al., 2014). Green, normal; raspberry, marginal defect; red, strong defect (data in Figure S6A).
(H) Role of RBX1 RING on ARIH1-neddylated CRL pathway tested by RBX1/RBX2 domain swaps, assayed as in (B).
(I) Role of RBX1 RING in neddylated CUL1-RBX1-dependent “opening” of ARIH1, assayed as in (D).
(J) Second, the distinctive role of RBX1 RING (directing UB transfer from ARIH1 to neddylated CRL substrate) uncovered by use of ARIH1OPEN mutant. In the absence of autoinhibition, this assay directly tests CRL substrate (pCyE) targeting by the indicated RING mutations in neddylated CRL1FBW7ΔD. Right-most lanes, control confirming ARIH1OPEN autoubiquitylation in absence of neddylated CRL.
110 purified ARIH1 variants with mutations encompassing 287 of ARIH1's 556 residues were surveyed with the pulse-chase assay monitoring ARIH1-UBCH7/neddylated CRL1FBW7ΔD-dependent ubiquitylation of pCyE (Figures 4 and S4). In addition to mutations decreasing ubiquitylation, some showed more rapid conversion of *UB~UBCH7 into *UB-modified pCyE (Figures 4B, S4G, and S4I). Enhanced activity of the hyperactive mutants, including two triple mutants previously designed to obliterate autoinhibitory interactions that we term “OPEN,” is explained by their location at the Ariadne domain-RING2 interface that sequesters ARIH1's catalytic Cys357 (Figures 4B, 5B, and 5C) (Duda et al., 2013). The “OPEN” triple mutants completely bypass the need for neddylated CUL-RBX1 to expose the catalytic Cys as monitored by reactivity with the synthetic electrophilic probe *UB-vinyl methyl ester (*UB-vinyl methyl ester [VME]), but they still mediate CRL substrate ubiquitylation (Figures S5A–S5C) (Duda et al., 2013; Kelsall et al., 2013).
We subsequently combined “OPEN” mutations (set B, Figures S5A–S5C) with defective mutants to separate effects on ARIH1 autoinhibition from other steps in ARIH1-UBCH7/neddylated CRL-mediated ubiquitylation. Mutants were tested for reactivity with *UB-VME to screen for (1) “opening” the autoinhibited structure to reveal ARIH's catalytic Cys, (2) catalytic alignment of *UB-VME's C-terminal tail, and (3) integrity of ARIH1's RING2 active site (Figures 5, 6, S2B, S5, and S6B) (Kelsall et al., 2013). Comparing a mutant's reactivity with *UB-VME in the presence of a neddylated CRL, or in the absence of a neddylated CRL after adding OPEN mutations, distinguished between mutational defects in neddylated CRL-dependent “opening” versus inherent RBR functions (e.g., binding the catalytic UB or an intact active site) (Figures S2B, and 5A, assay 2 versus 3). Likewise, after adding OPEN mutations, testing ubiquitylation in the absence and presence of neddylated CUL1-RBX1, the SR FBW7ΔD, and/or pCyE substrate allowed us to distinguish between mutational effects on intrinsic RBR activities (UB transfer from UBCH7 or ligation as monitored by autoubiquitylation) versus neddylated CRL-specific activities (neddylated CRL-dependent UB transfer from UBCH7 to ARIH1, or CRL substrate targeting) (Figure 5A, assay 4 versus 5). Taken together, our classification scheme assigned specific ARIH1 surfaces to eight activity profile signatures including the hyperactive mutants that map to the autoinhibitory interface (signatures I and II, Figures 5B, 5C, and S5A–S5C).
Mutants in ARIH1's UBA-to-RING1 (UtoR1) helix, RING1, RING1-to-IBR (R1toI) helix, and RING2 domains displayed activity profile signatures corresponding to intrinsic RBR E3 activities of UB transfer from UBCH7 to ARIH1 (signature V), alignment of the RING2 active site (signature III), and intrinsic ability of RING2 to mediate ligation (signature IV) (Figures 4, 5D–5G, and S5D–S5G, I–V). While future studies will be required to visualize the extreme structural transformations that accompany ARIH1 remodeling from the autoinhibited structure to an active state, the data are largely consistent with current models for RING1 and RING2 (Dove et al., 2016; Duda et al., 2013; Kelsall et al., 2013; Lechtenberg et al., 2016; Stieglitz et al., 2013), but with the transthiolation architecture also bolstered by ARIH1-specific elements such as the UtoR1-helix.
Specific ARIH1 Surfaces Mediate Distinct Neddylated CRL-Dependent Functions
Three activity profile signatures revealed how neddylated CRLs manipulate ARIH1 and how CRL substrate targeting is achieved. First, mutating the UBA domain's predicted UB-like protein (UBL)-binding site primarily decreased NEDD8-dependent UB transfer from UBCH7 to ARIH1, with minimal effects in other as-says (signature VI, Figures 5A, 5F, 5G, and S5I–S5L). Because the UBA domain, E2-binding RING1 and other elements required for this reaction form a single assembly in the autoinhibited structure of ARIH1, we speculate that neddylated CRL interactions with the UBA domain could promote structural remodeling or stabilize the catalytic architecture for UB transfer from UBCH7 to ARIH1 (Duda et al., 2013) (Figure 5F).
The remaining two activity profile signatures identify surfaces from RING2 represented by W386A and the Ariadne domain represented by Y531A each having dual but distinct functions. Both surfaces are essential for initial neddylated-CRL-dependent ARIH1 “opening,” shown by lack of labeling by *UB-VME (Figure S5Q). However, unique secondary roles for both surfaces were unveiled upon incorporating OPEN mutations. When auto-inhibition was bypassed, the RING2 W386A mutant accumulated as the ARIH1~UB thioester intermediate. A defect in intrinsic reactivity was reflected by the lack of autoubiquitylation in the absence of a neddylated CRL, and relatively slower modification of pCyE with neddylated CRL1FBW7ΔD (signature VII, Figures 5A, 5H, 5I, and S5Q–S5V).
Adding the OPEN mutations to the Ariadne domain Y531A mutant rescued all activities (e.g., reactivity with *UB-VME and autoubiquitylation) except the crucial, specific step in the reaction: targeting the CRL substrate. Thus, this final signature VIII indicates that the Ariadne domain mediates interactions specifying the CRL client as the substrate (signature VIII, Figures 5A, 5H, 5I, and S5Q–S5V).
Specialized Roles of NEDD8, Cullin, and RBX1 in Multimodal Activation and Substrate Targeting via the UBCH7-ARIH1 Pathway
We identified neddylated CRL domains relieving ARIH1 autoinhibition and directing ARIH1-mediated ubiquitylation to CRL clients (Figure 6A). Ubiquitylation with WT ARIH1 was severely impaired by using an unmodified CRL, or a mutant lacking CUL1's winged helix DNA-binding (WHB) subdomain harboring the neddylation site, or a neddylated CRL with the NEDD8 Q40E mutation that blocks cullin-RING conformational changes and inhibits ubiquitylation (Yu et al., 2015). However, these NEDD8-deficient CRLs achieved nearly WT levels of substrate ubiquitylation with ARIH1OPEN that is not autoinhibited (Figures 6B and 6C). Thus, the specific role of neddylation is limited to initially reversing the ARIH1 autoinhibition that prevents UB transfer from UBCH7 to ARIH1 (Figures 6B–6D). Probing effects of CUL1 deletions identified the cullin repeat 3 (CR3) as also required to stimulate “opening” of autoinhibited ARIH1 (Figures 6E and 6F).
Dual roles for RBX1 were discovered from assays with point mutants and a homolog swap mutant replacing the RING sequence with that of RBX2 (Figures 6G–6J). The RBX2 RING swap mutant, and several point mutants mapping to a common RBX1 RING surface, failed to activate ARIH1 reactivity with *UB-VME and obliterated ubiquitylation via the UBHC7-ARIH1 pathway, in spite of structural integrity (Figures 6G–6I and S6A–S6D). Interestingly, use of the ARIH1OPEN mutant revealed a second role of RBX1's RING in directing ARIH1 activity toward the CRL substrate: UBCH7-ARIH1OPEN-mediated *UB ligation to pCyE was substantially impaired for neddylated CRL1FBW7ΔD harboring the RBX2 RING domain. This was completely eliminated by deleting the RING domain from the non-neddylated counterpart, while ARIH1 automodification was increased concomitantly (Figure 6J). Thus, RBX1's RING is both crucial for initially activating ARIH1 and also for steering UB transfer from ARIH1 to the CRL substrate.
Accumulation of CRL Substrates upon Knocking Down ARIH1 and Altered Proteome and Cell-Cycle Properties in Cells Overexpressing Dominant-Negative ARIH1
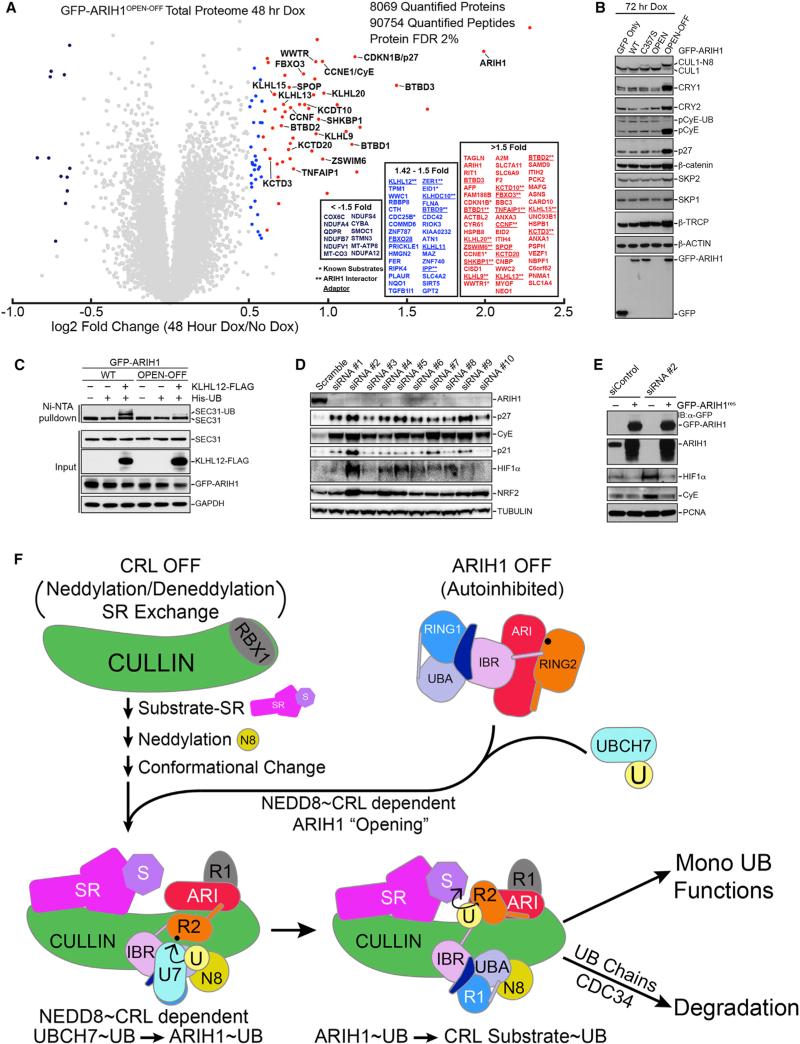
Given the potentially broad impact of ARIH1-mediated ubiquity-lation, we wished to examine its cellular roles. However, we, like others (Hart et al., 2015; Wang et al., 2015), were unable to generate knockout cell lines. Thus, we initially considered that use of dominant-negative mutants has greatly aided understanding of other E3 ligase pathways (Emanuele et al., 2011) and that the mutant combining open and catalytically inactivating (C357S) mutations would provide a suitably potent dominant-negative version of ARIH1 (ARIH1OPEN-OFF). Indeed, ARIH1OPEN modified CRL client substrates yet inhibited CDC34-dependent polyubiquitylation in a manner suggestive of persistent association with neddylated CRLs (Figures S5C and S6E). Furthermore, in a prior study, GFP-ARIH1OPEN was not stable in cells, in agreement with robust self-modification in vitro. However, high-level expression of GFP-ARIH1OPEN-OFF for 72 hr resulted in accumulation of neddylated CUL1 and stabilization of CyE and p27 (Kelsall et al., 2013). Retrospectively, these results are consistent with high-level expression of GFP-ARIH1OPEN-OFF indeed acting as a dominant-negative inhibitor through multiple mechanisms, including altered ARIH1-neddyated CRL on/off rates, out-competing WT ARIH1 for neddylated CRL binding, perturbation of the neddylation/deneddylation cycle, and inhibiting E2s such as CDC34 (Kleiger et al., 2009; Pierce et al., 2013; Wu et al., 2013; Zemla et al., 2013). Inducible low-level overexpression of ARIH1OPEN-OFF partially blocked cullin deneddylation observed after adding MLN4924 (2 hr), in agreement with prior results (Figure S7A) (Kelsall et al., 2013). As with WT ARIH1, ARIH1OPEN-OFF associated with many CRL SRs and SEC13-SEC31A yet exhibited a smaller fold change in interactions compared to WT and C357S ARIH1 following 2-hr treatment with MLN4924 (Figures S7B and S7C).
The ability of ARIH1OPEN-OFF to bind a cohort of CRLs in cells and inhibit CRL activity in vitro led us to examine the effects of GFP-ARIH1OPEN-OFF expressed at ~10-fold higher levels than endogenous ARIH1 on global proteome homeostasis (Figures 7A and S7A). We employed 6-plex TMT in 293T-GFP-ARIH1OPEN-OFF cells with or without Dox treatment (48 hr) in biological triplicate. In total, 9,112 proteins were identified, of which 8,069 were quantified at a false discovery rate (FDR) of 2% (Table S3). The abundance of ≈50 proteins increased between 1.5- and 5-fold, and the abundance of 12 primarily mitochondrial proteins decreased more than 50%. Among proteins whose abundance increased were several CRL SRs and known CRL substrates (CyE, CRL1FBXW7 target; Figures 2, 3, 4, 5, and 6; p27, CRL1SKP2 target; Figures S2H, S3B, and S3D; and WWTR1/TAZ, CRL1β-TRCP target) (Figure 7A). Eight of the 32 other proteins that increased 1.42- to 1.5-fold are CRL SRs or candidate substrates (Figure 7A; Table S3). Immunoblots from cells expressing GFP-ARIH1OPEN-OFF for 72 hr also showed increased levels of targets of reconstituted ARIH1-neddylated CRL-dependent ubiquitination (pCyE, p27, and CRY1; [Figures 3B, 3C, and S2H], which increased 1.35-fold at 48 hr post-induction by proteomics, Table S3), as well as CRY2, a substrate of CRL1FBXL3, HIF1α, a substrate of CRL2VHL, and IκBα, a substrate of CRL1β-TRCP in response to TNFα stimulation (Figures 7B, S7D, and S7E). Consistent with the dominant-negative activity of ARIH1OPEN-OFF, we found that the ubiquitylation of β-catenin (a substrate of SCFβ-TRCP), HIF1a, and p27, which was normally detectable upon adding MG132 or by enrichment on immobilized UBA domains was inhibited in cells overexpressing GFPARIH1OPEN-OFF (Figures S7D and S7F). Furthermore, the level of monoubiquitylated SEC31A detected by immunoblotting dramatically decreased post-induction of GFP-ARIH1OPEN-OFF for 72 hr, in agreement with our in vitro reconstitution of multi monoubiquitylation via the ARIH1-neddylated CRL3KLHL12 (Figures 2A and 7C).
Figure 7. Widespread Effects of Dominant-Negative ARIH1OPEN-OFF or ARIH1 Knockdown on CRL Pathways and Model for Extreme Amalgamation of ARIH1 and Neddylated CRLs in Mediating Regulation.
(A) TMT-based quantification of the total proteome following expression of GFP-ARIH1OPEN-OFF for 48 hr. Whole-cell lysates were prepared in triplicate for each condition and pooled following TMT labeling. ≥1.5-fold enrichment in GFP-ARIH1OPEN-OFF expressing cells, red, 1.42- to 1.5-fold increase, blue. *Previously reported CRL substrates. **ARIH1 interactors.
(B) Immunoblots of indicated CRL1 substrates following expression of GFP-ARIH1OPEN-OFF for 72 hr.
(C) Overexpression of GFP-ARIHOPEN-OFF inhibits cellular ubiquitylation of SEC31A.
(D) Depletion of ARIH1 in 293T cells with ten different siRNAs for 72 hr increases levels of CRL substrates detected by indicated immunoblots.
(E) Expressing siRNA-resistant GFP-ARIH1 reverses increase in indicated CRL substrate levels after ARIH1 knockdown in 293T cells.
(F) Model for ARIH1 and neddylated CRL acting in unison for joint E3-E3-mediated substrate ubiquitylation. In the absence of a substrate, CRL is OFF (unneddylated, unassembled, and inhibited) and ARIH1 is OFF (autoinhibited). Substrate binding to SR stimulates the pathway, promoting CRL assembly, neddylation, and conformational change, which, in turn, enables NEDD8, the cullin, and RBX1 to activate ARIH1 via ARIH1's UBA, RING2, and Ariadne (ARI) domains. This “opens” the ARIH1 structure and allows UB transfer from UBCH7 to ARIH1's RING2 catalytic Cys. Subsequently, specific surfaces from the CRL's RBX1 RING domain and from ARIH1's Ariadne domain promote monoUB transfer from ARIH1's RING2 catalytic Cys to the CRL's substrate receptor (SR)-bound substrate. The monoubiquitylated substrate is primed for altered function, further multimonoubiquitylation via repeated cycles through the ARIH1-CRL path, polyubiquitylation via a canonical RING-E2 mechanism directing proteasomal degradation, or other fates.
Importantly, depletion of ARIH1 by multiple siRNAs in 293T cells resulted in accumulation of p27, p21/CDKN1A, CyE, NRF2/NFE2L2, and HIF1a (Figure 7D, and p27, phosphor-CyE, and HIF1a in U2OS cells, Figure S7G), and turnover of phospho-CyE and HIF1a in 293T cells was rescued upon expression of an RNAi-resistant ARIH1 cDNA (Figure 7E), indicating that the effects are on target.
Associated with the disturbances seen in the proteome upon GFP-ARIH1OPEN-OFF overexpression were changes in cell size, cell-cycle progression, nuclear size, and DNA content (Figures S7H and S7I). Cell cycle defects seen upon overexpressing ARIH1OPEN-OFF are consistent with recent genome-wide CRISPR/Cas9 studies suggesting that ARIH1, like neddylated CRL subunits, is essential for cell proliferation in many cell lines (Hart et al., 2015; Marcotte et al., 2016; Wang et al., 2015). Taken together, the results indicate that inhibiting the ARIH1 pathway in cells lead to numerous defects possibly reflecting roles of ARIH1 in regulating CRLs linked with cell division and cell signaling, as summarized in Table S2.
DISCUSSION
Our results suggest that ARIH1 is a component of several human CRLs. Requirements for CRL substrate ubiquitylation in vivo (cullin-SR assembly, neddylation, integrity of NEDD8 Gln40, RBX1) are integral to the multifaceted mechanism by which neddylated CRL and ARIH1 domains together direct efficient and specific monoubiquitylation of CRL client substrates in vitro (Figures 4, 5, and 6). Furthermore, like core CRL subunits, UBCH7 and ARIH1 are essential for cell proliferation (Hart et al., 2015; Wang et al., 2015).
We speculate that the ARIH1-CRL mechanism could be important for pathways requiring tight control by neddylation. Both E3s are suppressed until cullin neddylation directly alters the conformation of CUL-RBX1 (Duda et al., 2008; Saha and Deshaies, 2008; Yamoah et al., 2008; Yu et al., 2015), and via E3-E3 interactions neddylation also promotes an activating conformational change in ARIH1 (Kelsall et al., 2013) (Figures S2 and S5). Dual NEDD8-dependent conformational switches would greatly increase the regulatory capacitance of the single NEDD8 modification. We postulate that the ARIH1-CRL pathway could proceed in cells as in Figure 7F: (1) substrate binding would stabilize a neddylated CUL-RBX1-SR assembly (Pierce et al., 2013; Wu et al., 2013; Zemla et al., 2013); (2) a cullin's linked NEDD8 and CR3 domain, RBX1's RING, and perhaps other CRL regions would activate ARIH1 via its UBA, RING2, and Ariadne domains. Given their proximities in the respective inactive structures (Duda et al., 2008, 2013; Zheng et al., 2002), the CRL's cullin and RING regions may co-encounter ARIH1's RING2 and Ariadne domains for conformational activation, while interactions between NEDD8 and ARIH1's UBA domain may buttress the catalytic intermediate mediating UB transfer from UBCH7 to ARIH1's RING2 Cys357; (3) after the thioester-linked ARIH1~UB intermediate is formed, neither NEDD8 nor the N-terminal half of ARIH1 substantially impacts the ligation reaction (Figures 5F, 5G, and 6C). Instead, the RING2~UB intermediate would be co-steered by ARIH1's Ariadne domain and RBX1's RING to modify the SR-bound substrate (Figures 5H, 5I, 6G, 6H, and 6J).
Although future studies will be required to determine when, how, and which cellular CRLs employ ARIH1 for substrate priming, or whether the ARIH1 pathway is particularly geared toward substrates that are multi-monoubiquitylated like SEC31A (Jin et al., 2012) (Figures 1C and 2B), our data raise the possibility that the CRL-ARIH1 mechanism could be widespread. ARIH1 preferentially associates with subunits of diverse, active CRLs in human cells, based on neddylation-dependent binding of CUL1 and CUL3 and various SRs, and their abundance relative to CAND1 and CSN (Figures 1 and S1). ARIH1C357S associated with 12 of 26 F-box, seven of ten BC-box, and 23 of 49 BTB found previously to interact with CUL1, CUL2, and CUL3 in 293T cells using the CompPASS approach, consistent with catalytically inactive enzymes often forming relatively stable assemblies in cells (Bennett et al., 2010). Indeed, of the 95 CUL1, CUL2, and CUL3 SRs detectable in the 293T cell proteome (Huttlin et al., 2015), 41 were high-confidence interactors with ARIH1C357S expressed at low levels (Table S2), and retrospective inspection of published proteomic experiments revealed endogenous ARIH1 as an interactor of CUL1, CUL3, NEDD8, and several SRs (Bennett et al., 2010; Huttlin et al., 2015; Lee et al., 2011). Thus, ARIH1 may access a substantial percentage of CRL receptors in cells. Alternatively, the ARIH1 mechanism could apply selectively to particular CRL pathways, as implied by genetic interactions between the C. elegans ARIH1 ortholog ari-1 and the one of 21 SKP1 orthologs that regulates pharyngeal development (Polley et al., 2014). Although at this point there is no evidence that lower eukaryotes utilize a joint RBR-CRL mechanism despite conservation of an Ariadne-family E3, higher eukaryotes appear to have expanded related team-tagging pathways three times during evolution. In addition to ARIH1 functioning with RBX1-based CRLs, ARIH1's closest relative ARIH2 was found to specifically bind CUL5-RBX2 (Kelsall et al., 2013). Our finding of dual roles for RBX1's RING domain in relieving autoinhibition and directing ARIH1 to target the CRL substrate (Figures 6G, 6H, and 6J) may provide a molecular explanation for why CUL5-RBX2, with a distinct RING, would require its own RBR E3 partner. Furthermore, CUL9 encodes both CRL and RBR functionalities in a single polypeptide arising from a gene fusion event between a cullin and an Ariadne-family RBR E3 (Marín, 2009).
Team tagging is emerging as a common theme in UB and UBL pathways.
Although early studies identified sequential E1, E2, and E3 activities, it is now clear that some pathways operate via nonlinear enzyme configurations. For example, multiple E3s can act successively on a common target, with the first enzyme priming a substrate with a form of UB that is further modified by the next (Hwang et al., 2010; Kirisako et al., 2006; Koegl et al., 1999). In other cases, a single E3 enzyme may function sequentially with two E2s that each modify a substrate with different forms of UB, such as mono- or polyUb to increase capacity for regulating the rate at which substrates acquire degradative signals (Rodrigo-Brenni and Morgan, 2007). The tandem ARIH1 RBR-neddylated CRL mechanism reported here presents an exceptional variation on the classic E1 > E2 > E3 view of UB conjugation with an E3 in one class (RING) binding and employing and E3 of another (RBR) as a UB conjugating enzyme (Figures 2 and 3), by the unprecedented function of a RING domain to steer substrate targeting by another E3 (Figure 6), and in the potentially widespread use of this mechanism by a massive number of CRLs in human cells or even more in other organisms (Figure 1). It seems likely that integration of multiple distinctive types of E3 ligases to act simultaneously, efficiently, and specifically will emerge as a common mechanism ensuring exquisite regulation of UB ligation via team tagging.
STAR ★ METHODS
Detailed methods are provided in the online version of this paper and include the following:
● KEY RESOURCES TABLE
● CONTACT FOR REAGENT AND RESOURCE SHARING
- ● EXPERIMENTAL MODEL AND SUBJECT DETAILS
- ○ Cell Lines
- ● METHOD DETAILS
- ○ Constructs
- ○ siRNA Knockdown
- ○ Cell Lysis and Western Blotting
- ○ Rescue Experiment of siARIH1
- ○ Ubiquitylation of Cellular SEC31A
- ○ TUBE IPs
- ○ Protein Expression and Purification
- ○ Flow Cytometry and Imaging
- ○ CompPASS Interaction Proteomics
- ○ Tandem Mass Tagging Interaction Proteomics
- ○ TMT Total Proteome Analysis
- ○ Biochemical Assays
- ● QUANTIFICATION AND STATISTICAL ANALYSIS
- ○ TMT Data Analysis
- ○ Statistical Analysis
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-GFP (clone 7.1 and 13.1) antibody | Roche | Cat#11814460001; RRID: AB_390913 |
| Mouse monoclonal anti-Hif1α (clone #241809) antibody | R&D Systems | Cat#MAP1536; RRID: AB_2116983 |
| Rabbit polyclonal anti-CRY2 | Bethyl Laboratories | Cat#A302-615A; RRID: AB_10554665 |
| Mouse monoclonal anti-β-Catenin | BD Biosciences | Cat#610154; RRID: AB_397555 |
| Rabbit polyclonal anti-Skp1 antibody | Cell Signaling | Cat#2156; RRID: AB_2270271 |
| Rabbit polyclonal anti-Skp2 antibody | Cell Signaling | Cat#4313; RRID: AB_2187641 |
| Rabbit polyclonal anti-Cul1 antibody | Invitrogen | Cat#71-8700; RRID: AB_87955 |
| Rabbit monoclonal anti-β-TrCP (clone D13F10) antibody | Cell Signaling | Cat#4394S; RRID: AB_1054763 |
| Mouse monoclonal anti-Sec31A (clone 32/Sec31A) antibody | BD Biosciences | Cat#612350; RRID: AB_399716 |
| Mouse monoclonal anti-KLHL12 (clone 2G2) antibody | Thermo Fisher | Cat#MA5-15649; RRID: AB_11004907 |
| Rabbit monoclonal anti-phospho-NF-κB p65 (Ser536) (clone 93H1) antibody | Cell Signaling | Cat#3033L; RRID: AB_331285 |
| Mouse monoclonal anti-IκBα (clone L35A5) antibody | Cell Signaling | Cat#4814S; RRID: AB_10693636 |
| Rabbit polyclonal anti-Cul2 antibody | Thermo Fisher | Cat#51-1800;RRID: AB_2533898 |
| Mouse monoclonal anti-PCNA (clone PC10) antibody | Sigma-Aldrich | Cat#P8825; RRID: AB_477413 |
| Goat polyclonal anti-ARIH1 antibody | Abcam | Cat#ab3891; RRID:AB_304129 |
| Mouse monoclonal anti-HIF1α αντιβοδψ | BD Biosciences | Cat#610958; RRID:AB_398271 |
| Rabbit polyclonal anti-NEDD8 antibody | Cell Signaling | Cat#2745S; RRID:AB_10695300 |
| Rabbit monoclonal anti-p27 (Kip1) antibody | Cell Signaling | Cat#3686S; RRID:AB_2077850 |
| Mouse monoclonal anit-Cyclin E1 antibody | Cell Signaling | Cat#4129S; RRID:AB_2071200 |
| Anti-Flag M2 Magnetic Beads | Sigma | Cat#M8823 |
| Mouse monoclonal anti-Tubulin antibody | Sigma | Cat#T9026; RRID:AB_477593 |
| Rabbit polyclonal anti-CRY1 antibody | Bethyl | Cat#A302-614; RRID:AB_10555376 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MG132 Proteasome inhibitor | InvivoGen | Cat#tlrl-mg132; CAS: 133407-82-06 |
| TNF (murine recombinant) | Peprotech | Cat#315-01A |
| Tetracycline | Bioline | Cat#BIO-87030; CAS: 64-75-5 |
| 1,10-Phenanthroline | Sigma-Aldrich | Cat#131377-5G; CAS: 66-71-7 |
| HALT protease and phosphatase inhibitor cocktail | Thermo Fisher | Cat#78441 |
| Fluorescein-5-Maleimide | Thermo Fisher | Cat#F150 |
| Tetramethylrhodamine-5-Maleimide (single isomer) | Thermo Fisher | Cat#T6027 |
| Hoechst Solution | Thermo Fisher | Cat#62249 |
| Blasticidin S HCl | Thermo Fisher | Cat#A1113903 |
| Zeocin Selection Reagent | Thermo Fisher | Cat#R25001 |
| Hygromycin B | Thermo Fisher | Cat#10687010 |
| Puromycin Dihydrochloride | Thermo Fisher | Cat#A1113803 |
| Roche mini complete EDTA free | Roche | Cat#11836170001 |
| Roche complete | Roche | Cat#4693116001 |
| phosSTOP inhibitor | Roche | Cat#04-906-837-001 |
| 1,10-Phenanthroline | Sigma | Cat#131377 |
| MLN4924 | EMD Millipore | Cat#5054770001 |
| 3x FLAG Peptide | LabPe | Cat#L1033 |
| TMT10plex Label Reagent Set | Thermo Fisher | Cat#90406 |
| TMT6plex Label Reagent Set | Thermo Fisher | Cat#90068 |
| Trypsin Gold, MS grade | Promega | Cat#V5280 |
| Lysyl Endopeptidase | Wako Chemicals | Cat#129-02541 |
| Gateway LR Clonase Enzyme mix | Thermo Fisher | Cat#11791019 |
| Experimental Models: Cell Lines | ||
| U2-OS | ATCC | ATCC#HTB-96; RRID:CVCL_0042 |
| Flp-In T-Rex 293 Cell Line | Thermo Fisher | Cat#R78007; RRID:CVCL_U427 |
| HEK293T | ATCC | ATTC#CRL-3216; RRID:CVCL_0063 |
| Experimental Models: Organisms/Strains | ||
| E. coli BL21 Gold (DE3) | Thermo Fisher | Cat#50-125-348 |
| E. coli Rosetta 2 (DE3) | EMD Millipore | Cat#71400 |
| Sf9 Insect cells | Thermo Fisher | Cat#11496015 |
| High Five Insect cells | Thermo Fisher | Cat#B85502 |
| Recombinant DNA | ||
| pGEX4T1 GST-Thrombin-UBCH5B | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-UBCH7 | Duda et al., 2013 | N/A |
| pGEX4T1 GST-Thrombin-UBC12 | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-UBE2F | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-APPBP1-UBA3 | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-RBX1:His-Thrombin-CUL1CTD | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-RBX1:His-Thrombin-CUL1ΔWHB | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-RBX1ΔRING:His-Thrombin-CUL1CTD | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-RBX1(W35A):His-Thrombin-CUL1CTD | Scott et al., 2014 | N/A |
| pGEX4T1 GST-Thrombin-RBX1(D36P):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1(I37A):His-Thrombin-CUL1CTD | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-RBX1(V38P):His-Thrombin-CUL1CTD | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-RBX1(V39G):His-Thrombin-CUL1CTD | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-RBX1(D40G):His-Thrombin-CUL1CTD | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-RBX1(N41G):His-Thrombin-CUL1CTD | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-RBX1(N47A:H48A):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1(D51A):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1(I54A):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1(Q57A):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1(F81N):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1(T90A:R91A):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1(V93R):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1(N98Q):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1(E102V):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1(F103V):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1(Y106I):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1(H108K):His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX1N-RBX2RING:His-Thrombin-CUL1CTD | This Study | N/A |
| pGEX4T1 GST-Thrombin-RBX2N-RBX1RING:His-Thrombin-CUL1CTD | This Study | N/A |
| pAL His-Thrombin-CUL1NTD | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-CKS1 (5-73) | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-NEDD8 | Duda et al., 2008 | N/A |
| pGEX4T1 GST-Thrombin-NEDD8 (Q40E) | This Study | N/A |
| pGEX2TK GST-Thrombin-UB (S > C) | Scott et al., 2014 | N/A |
| pGEX2TK GST-Thrombin-UB (S > C) (I44A) | This Study | N/A |
| pGEX2TK GST-Thrombin-UB (S > C) (L71A) | This Study | N/A |
| pGEX2TK GST-Thrombin-UB (S > C) (L73A) | This Study | N/A |
| pGEX2TK GST-Thrombin-UB (S > C) (LR74A) | This Study | N/A |
| pGEX2TK GST-Thrombin-UBK0 (S > C) | This Study | N/A |
| pGEX4T1 GST-TEV-CDC34B | Duda et al., 2008 | |
| pGEX4T1 GST-TEV-CUL1CR3-C:His-MPB-TEV-RBX1 (CUL1 residues 332-C) | This Study | N/A |
| pGEX4T1 GST-TEV-CUL1CTD:His-MPB-TEV-RBX1 (CUL1 residues 441-C) | Jubelin et al., 2010 | N/A |
| pGEX4T1 GST-TEV-SKP2101-C:SKP1 | Duda et al., 2008 | N/A |
| pGEX4T1 GST-TEV-FBW7ΔD:SKP1 (FBW7 residues 263-C) | Jubelin et al., 2010 | N/A |
| pGEX4T1 GST-TEV CYCLINA | Duda et al., 2008 | N/A |
| pGEX4T1 GST-TEV-CDK2:CIV1 | Duda et al., 2008 | N/A |
| pRSFDUET His-TEV-p27 (S10A:F62A:F64A) | Duda et al., 2008 | N/A |
| pRSFDUET His-TEV-p27 K > R (All LYS Mutated to ARG; DNA insert synthesized by Open Biosystems) | Open Biosystems | N/A |
| pRSFDUET His-TEV-p27 K > R (S10A:F62A:F64A: K > R All LYS mutated to ARG) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R25K) (S10A:F62A:F64A) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R47K) (S10A:F62A:F64A) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R59K) (S10A:F62A:F64A) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R68K) (S10A:F62A:F64A) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R75K) (S10A:F62A:F64A) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R81K) (S10A:F62A:F64A) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R96K) (S10A:F62A:F64A) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R100K) (S10A:F62A:F64A) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R134K) (S10A:F62A:F64A) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R153K) (S10A:F62A:F64A) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R165K) (S10A:F62A:F64A) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R189K) (S10A:F62A:F64A) | This Study | N/A |
| pRSFDUET His-TEV-p27 K > R(R190K) (S10A:F62A:F64A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 | Duda et al., 2013 | N/A |
| pGEX4T1 GST-TEV-ARIH190-C (ARIH1 residues 90-C) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (R118A:E119A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (V123D) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Q125) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (I130A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (H137A:N139A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (E143A:K144A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (E147A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Y149A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (F150A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N153A:L154A:E155A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K156A:L157A:F158A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (E160A:C161A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (H162A:V163A:I164A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N165A:P166A:S166A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K168A:K169A:S170A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (R171A:T172A:R173A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Q174A:M175A:N176A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (T177A:R178A:S179A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (I188A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (D241A:D242A:N243A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (T244A:V245A:M246A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (R247A:L248A:I249A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (T250A:D251A:S252A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K253A:V254A:K255A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K257A:Y258A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Q259A:H260A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (L261A:I262A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (T263A:N264A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (S265A:F266A:V267A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (E268A:C269A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N270A:R271A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (L272A:L273A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K274A:W275A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (P279A:D280A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (H282A:H283A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Q288A:Y289A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (D291A:K293A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (R296A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K298A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Q302A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (D313A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (W319A:L320A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K321A:K322A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (W323A:I324A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K325A:K326A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (C327A:D328A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (D329A:D330A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (S331A:E332A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (T333A:S334A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N335A:W336A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N340A:T341A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K342A:E343A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (P345A:K346A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (H348A:V349A:T350A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (I351A:E352A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N358A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (H359A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (M360A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (V361D) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (R363A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N364A:Q365A:N366A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K368A:E370A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (W373A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (V374A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (P378A:W379A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (E380A:P381A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (G383D:S384D:A385D) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (W386A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N388A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N390A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (R391A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Y392A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (D395A:D396A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (A397D:K398I:A399V) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (A400N:R401Q:D402S) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (A403Q:E405A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Q412A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (L415A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Y417A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N419A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (R420A:N423A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (H424A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (R429A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (F430A:E431A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (H432A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Y435A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (M455A:Q456A:Q457A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (W452A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K483A:K484A:N485A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Y476A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N486A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (I489A:I490A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (E492A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N493A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Q495A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (D497A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (E499A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N500A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K522A:Q523A:K524A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Q526A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (R530A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Y531A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (R535A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (R537A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (H452A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Y547A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (F430A:E431A:N503A) Open Mutant A | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (R420A:N423A:N503A) Open Mutant B | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (V123D:R420A:N423A:N503A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K156A:L157A:F158A:R420A:N423A:N503A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (I188A:R420A:N423A:N503A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (K257A:Y258A:R420A:N423A:N503A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (S265A:F266A:V267A:R420A:N423A:N503A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (N340A:T341A:R420A:N423A:N503A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (IT350A:I351A:R420A:N423A:N503A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (P378A:W379A:R420A:N423A:N503A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (W386A:R420A:N423A:N503A) | This Study | N/A |
| pGEX4T1 GST-TEV-ARIH1 (Y531A:W379A:R420A:N423A:N503A) | This Study | N/A |
| pMal MBP-TEV-KLHL12-His | Jin et al., 2012 | N/A |
| pFastbac1 CUL1 | This Study | N/A |
| pFastbacHTB His-TEV-CUL3 | This Study | N/A |
| pFastbac1 SKP1 | This Study | N/A |
| pFastbac GST-TEV-FBXL3 | This Study | N/A |
| pFastbac GST-TEV-RBX1 | This Study | N/A |
| pFastbac1 CRY1ΔC (CRY1 residues 1-532) | This Study | N/A |
| pFastbac GST-TEV-FBXW11 | This Study | N/A |
| pFastbac SEC13 | Jin et al., 2012 | N/A |
| pFastbac His-TEV-SEC31A | Jin et al., 2012 | N/A |
| pcDNA5 KLHL12-FLAG | Jin et al., 2012 | N/A |
| pCS2 His-UB | Jin et al., 2012 | N/A |
| pcDNA FRT/TO | Thermo Fisher | V652020 |
| pDONR223 ARIH1 | Orfeome V8.1 | HsCD00378777 |
| pOG44 | Thermo Fisher | V600520 |
| pHAGE-Puro-C-Flag-HA | Huttlin et al., 2015 | N/A |
| pcDNA5-FRT/TO | Invitrogen | Cat#V6520-20 |
| pcDNA5-FRT/TO+GFP | Kelsall et al., 2013 | N/A |
| pcDNA5-FRT/TO+GFP-ARIH1 | Kelsall et al., 2013 | N/A |
| pcDNA5-FRT/TO+GFP-ARIH1 (C357S) | Kelsall et al., 2013 | N/A |
| pcDNA5-FRT/TO+GFP-ARIH1 (siRNA resistant) | This study | N/A |
| Plasmid: pcDNA5-FRT/TO+GFP-ARIH1 (F430A, E431A, E503A) | Kelsall et al., 2013 | N/A |
| Plasmid: pcDNA5-FRT/TO+GFP-ARIH1 (C357S, F430A, E431A, E503A) | Kelsall et al., 2013 | N/A |
| Sequence-Based Reagents | ||
| siGENOME Non-targeting siRNA #1 | Dharmacon | Cat#D-001210-01-05 |
| Set of 4 upgrade: siGENOME ARIH1 siRNA; siRNA #1: GUGCUGGAGUGAAUAUUUA; siRNA #2: CGAGAUAUUUCCCAAGAUU; siRNA #3: ACACUUAUGUCUUCGCUUU; siRNA #4: GGAUAUGCCUUGUCAGAUC | Dharmacon | Cat#MU-019984-00-0002 |
| Silencer Select Pre-Designed siRNA ARIH1: siRNA ID: s223658; siRNA #5: ACACGAAAUAGUGUGACCCat | Thermo Fisher | Cat#4392420 |
| Silencer Select Pre-Designed siRNA ARIH1: siRNA ID: s223660; siRNA #6: UUCCAUCAAAGUACCUUUCca | Thermo Fisher | Cat#4392420 |
| FlexiTube GeneSolution GS25820 for ARIH1; siRNA #7: CTGCTACTTGAACTACCCTAA; siRNA #8: CTGCGCTTTGAGCACAAACTA; siRNA #9: GCGCCTGATCACAGATTCAAA; siRNA #10: TAGAGTGCAATCGACTGTTAA | QIAGEN | Cat#GS25820 |
| Software and Algorithms | ||
| Prism | GraphPad | ver 5; RRID: SCR_002798 |
| R statistical package | http://www.rproject.org/ | ver 3.2.3; RRID: SCR_001905 |
| Bioconductor | https://www.bioconductor.org | ver 3.3; RRID: SCR_006442 |
| Other | ||
| DMEM, high glucose, pyruvate | Thermo Fisher | Cat#11995-073 |
| Lipofectamine 3000 | Life Technologies | Cat#L3000008 |
| Lipofectamine RNAi Max | Life Technologies | Cat#13778075 |
| FxCycle PI/Rnase Staining Solution | Life Technologies | Cat#F10797 |
| C-18 Empore extraction Disks | Thermo Fisher | Cat#14-386-2 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Brenda Schulman (Brenda.Schulman@stjude.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
HEK293T Cells
Cells were maintained in DMEM (High glucose, L-glutamine, Pyruvate) with 10% fetal bovine serum, 50 μL/mL penicillin and 50 μL/mL streptomycin.
293T Flp-In T-Rex Cells
Cells were maintained in DMEM (High glucose, L-glutamine, Pyruvate) with 10% fetal bovine serum, 15 μg/mL Blasticidin and 100 μg/mL Zeocin. To generate cell lines of interest, pcDNA5-FRT/TO-FLAG-HA or pcDNA-FRT/TO-GFP based vectors containing open reading frames of interest were transfected using Lipofectamine 3000 along with pOG44 Flp-Recombinase expression vector followed by selection with 200 μg/mL Hygromycin and 15 μg/mL Blasticidin. To induce protein expression, cells were treated with 0.5 μg/mL doxycycline for the times indicated.
U2OS Cells
Cells were maintained in DMEM supplemented with 10% fetal bovine serum and 2 mmol/L L-glutamine.
METHOD DETAILS
Constructs
Expression constructs generated for this study were prepared by standard molecular biology techniques and coding sequences entirely verified. Mutant versions used in this study were generated by QuickChange (Stratagene). To generate FLAG-HA tagged cell lines for immunoprecipitation experiments, ARIH1 or ARIH1 mutants were transferred to recipient vectors (pcDNA5-FRT-TO-FLAG-HA) using λ-recombinase. To generate ARIH1 resistant to siRNA #2 wild-type target sequence CGAGATATTTCCCAAGATT was changed to AGGGACATCAGTCAGGACT by QuickChange (Stratagene).Constructs for KLHL12 and SEC13–SEC31A expression and purification were kind gifts from Michael Rape's lab (HHMI, UC Berkeley). For expression of SKP1-FBXW11, we cloned full-length FBXW11 into a modified pFastbac vector as a GST-fusion with a TEV protease cleavage site, for baculovirus co-infection with an untagged full-length SKP1 virus. Similarly, FBLX3 and CRY1ΔC (corresponding to residues 1-532 of CRY1) were cloned as a GST-TEV-cleavable fusion in a modified pFastbac vector and untagged in pFastbac1, respectively. The DNA sequence encoding a variant of p27 with all lysine residues mutated to arginines was synthesized by Geneart and cloned into pRSFDuet (Novagen) with an N-terminal His-TEV tag. We introduced by site-directed mutagenesis the S10A plus kinase inhibitory domain mutant (F62A/F64A) that facilitates phosphorylation by CyclinA-CDK2. Individual lysine residues in the p27 sequence were then reintroduced by site-directed mutagenesis to produce the single lysine variants.
siRNA Knockdown
siRNA transfections were carried out using Lipofectamine RNAiMax using reverse transfection.
Cell Lysis and Western Blotting
To assess IκBα stability, 293T Flp-In T-Rex cells expressing GFP-ARIH1 versions for 72 hr were either mock treated or treated with TNF-α (50 ng/ml) for 20 min. Whole-cell extracts were prepared by lysis in 40 mM HEPES pH 7.4, 120 mM NaCl, 1mM EDTA, 1% Triton X-100, “HALT” protease/phosphatase inhibitor cocktail (Thermo Fisher), and 5 mM OPT by mechanical disruption by passing through a 25G needle. Lysates were clarified by centrifugation, and subjected to immunoblot analysis using indicated antibodies.
U2OS cells were lysed in RIPA buffer (1 × PBS, 1% NP40, 0.5% Na deoxycholate, 0.1% SDS, “HALT” protease/phosphatase inhibitor cocktail (Thermo Fisher), 5 mM NEM, and 5 mM OPT.
Rescue Experiment of siARIH1
293T Flp-In T-Rex cells were transfected twice within 24 hr with 20 ng/ml siARIH1#2. After 24 hr cells were mock or transiently transfected with siRNA-resistant GFP-ARIH1R, incubated for 48 hr prior to lysis in RIPA buffer (1 × PBS, 1% NP40, 0.5% Na deoxycholate, 0.1% SDS) supplemented with “HALT” protease inhibitor cocktail (Thermo Fisher), 5 mM NEM, and 5 mM OPT. Lysates were clarified by centrifugation, and subjected to immunoblot analysis using indicated antibodies.
Ubiquitylation of Cellular SEC31A
In order to assay cellular ubiquitylation of SEC31A, 293T Flp-In T-Rex cells expressing GFP-ARIH1WT or GFP-ARIH1OPEN-OFF were grown in 15-cm dishes until approx 60-80% confluent then transfected with 6 μg pcDNA5-KLHL12-FLAG and/or 6 μg pCS2-6His-UB, as indicated, using polyethylenimine as a gene transfer reagent. 24 hr later GFP-ARIH1 expression was induced by addition of 1 μg/ml doxycycline and the cells were incubated for a further 16 hr. Cells were treated with 20 μM MG132 for 3 hr and harvested by gently scraping into PBS followed by centrifugation at 300 × g for 5 min and resuspension in 6 M guanidinium-HCl, 100 mM sodium phosphate buffer pH 8.0, 10 mM imidazole and 5 mM β-mercaptoethanol. A smaller aliquot of the harvested cells (one tenth of the total cell number) was taken prior to resuspension in denaturing buffer and lysed in RIPA buffer to allow analysis of protein INPUT levels. The cells were resuspended in guanidinium hydrochloride and subjected to sonication for 30 s, and the resultant lysate was incubated with 25 μl Ni-NTA agarose for 3 hr at room temperature. The beads were washed once with lysis buffer containing 0.1% Triton X-100, once with pH 8.0 wash buffer (8 M urea, 10 mM Tris, 100 mM sodium phosphate buffer pH 8.0, 0.1% Triton X-100, 5 mM β-mercaptoethanol), four times with pH 6.3 wash buffer (8 M urea, 10 mM Tris, 100 mM sodium phosphate buffer pH 6.3, 0.1% Triton X-100, 5 mM β-mercaptoethanol) and eluted by heating at 95°C for 5 min in 60 μl 1X NuPAGE LDS sample buffer (reducing) supplemented with 300 mM imidazole. Samples were separated by SDS-PAGE and analyzed by immunoblotting.
TUBE IPs
To enrich for ubiquitinated proteins from whole cell lysates, we adapted a TUBE approach. The TUBE, His-Halo-UBA (4xUBQLN1) was expressed in E. coli BL21 cells, affinity purified with Ni-NTA-agarose, and dialyzed into 50 mM HEPES ph 7.5, 10% glycerol, 150 mM NaCl, 1mM DTT. 21 nmol of Halo-UBA was incubated with 200 μl HaloLink resin (Promega) in Halo Binding Buffer (HBB; 50 mM Tris-HCl, pH7.5, 150 mM NaCl, 0.05% NP-40, and 1mM DTT) overnight at 4°C. Beads were washed three times with HBB. 293T Flp-In T-Rex cells expressing GFP-ARIH1WT or GFP-ARIH1OPEN-OFF (GFP-ARIH1 expression was induced by addition of 1 μg/ml doxycycline for 48 hr) were treated with 20 μM MG132 for 3 hr prior to lysis in lysis buffer (50 mM Tris pH7.5, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 0.27 M sucrose, 200 mM 2-Chloro-acetoamide, and “HALT” protease inhibitor cocktail [Thermo Fisher]) by passing 10x through a 19G needle. 1 mg (1 mg/ml) of cleared lysate was incubated with 30 μl of Halo-UBA-coupled beads for 4 hr at 4°C. After incubation, beads were extensively washed with lysis buffer supplemented with 500 mM NaCl, and precipitated proteins eluted by heating at 95°C for 5 min in 1X reducing NuPAGE LDS sample buffer. Samples were separated by SDS-PAGE and analyzed by immunoblotting.
Protein Expression and Purification
UBCH5B, UBCH7, UBC12 (aka UBE2M), UBE2F, the NEDD8 E1 APPBP1-UBA3 (aka NAE1-UBA3), full-length split and co-express (sce) CUL1-RBX1, full-length sce CUL1-RBX2NRBX1RING, and full-length sce CUL1-RBX1NRBX2RING were expressed in E. coli BL21 Gold (DE3) cells as GST-Thrombin fusion proteins. Fusion proteins were purified from cell lysates by glutathione affinity chromatography and liberated from GST by thrombin cleavage overnight at 4°C. Cleavage reactions were further purified by ion-exchange and size exlustion chromatography in 25 mM HEPES, 200 mM NaCl, 1 mM DTT pH = 7.5 (Buffer A). NEDD8, UB, and CKS1 (residues 5-73) were expressed in E. coli BL21 Gold (DE3) cells as GST-tagged fusion proteins with a thrombin cleavage site. Fusion proteins were purified from cell lysates by glutathione affinity chromatography and liberated from GST by thrombin cleavage during extensive dialysis overnight in Buffer A at 4°C. Cleavage reactions were passed back over a glutathione affinity resin to remove free GST and any remaining uncleaved GST-fusion protein. Protein collected in the flow fraction was concentrated with an Amicon Ultra filtration unit and further purified by size exclusion chromatography in Buffer A. CDC34B, ARIH1 (and mutant derivatives), CUL3CR3-C-RBX1 (CUL1-residues 332-776), CUL1CTD–RBX1 (CUL1 residues 411-776), SKP1–SKP2 (residues 101-C), and SKP1–FBW7ΔD (simplified monomeric version containing FBW7 residues 263 X-terminus) were expressed in E. coli BL21 Gold (DE3) cells as GST-TEV fusion proteins. Fusion proteins were purified from cell lysates by glutathione affinity chromatography and liberated from GST by TEV cleavage overnight at 4°C. Cleavage reactions were further purified by ion exchange and size exclusion chromatography in Buffer A. CyclinA and CDK2 (phosphorylated by coexpressed S. cerevisiae Cak1p) were expressed separately in E. coli BL21 Gold (DE3) cells as GSTTEV fusion proteins. Fusion proteins were purified from cell lysates by glutathione affinity chromatography. Stoichiometric amounts of GST-TEV-CyclinA and GST-TEV-CDK2 were mixed and liberated from GST by TEV cleavage overnight during extensive dialysis at 4°C. Cleavage reactions were passed back over a glutathione affinity resin to remove free GST and any remaining uncleaved GST-fusion protein. The CyclinA-CDK2 complex collected in the flow fraction was further purified by ion exchange and size exclusion chromatography in Buffer A. p27 and mutant derivatives (all versions contain additional S10A/k− mutations that can be phosphorylated on T187 by CDK2) were expressed in E. coli BL21 Gold (DE3) cells with a N-terminal 6XHis-TEV tag. Tagged protein was purified from cell lysates by Ni affinity chromatography. Bound proteins were eluted from the Ni resin with 250mM Immidazole and immediately subjected to desalting over a PD-10 column (GE Healthcare) into Buffer A as long term exposure to high imidazole levels caused the protein to precipitate from solution. The 6X Histidine tag was liberated by cleavage with TEV overnight at 4°C. Cleavage products were further purified by size exclusion chromatography in Buffer A. The UB E1 UBA1 was expressed in E. coli Rosetta 2 (DE3) cells with a N-terminal 6XHis-TEV tag. Tagged protein was purified from cell lysates by Ni affinity chromatography. The N-terminal 6XHis tag was liberated with TEV overnight at 4°C. Cleavage reactions were further purified by ion exchange and size exclusion chromatography in Buffer A. GST-RBX1-CUL1, GST-RBX1-CUL3, GST-FBXW11-SKP1, and GST-FBXL3-CRY1ΔC (residues 1-532)-SKP1 were expressed in insect cells. The complexes were purified from cell lysates by glutathione affinity chromatography capturing the GST-tagged subunit, and GST was liberated by TEV cleavage overnight at 4 C. Cleavage reactions were further purified by ion exchange and size exclusion chromatography in Buffer A. SEC13-SEC31A was expressed in insect cells with a N-terminal 6XHis tag on SEC31A. Tagged protein was purified from cell lysates by Ni affinity chromatography. Ni elutions were further purified over a HiTrap Q (GE Healthcare) ion exchange column.
Neddylation and purification of CUL1–RBX1, CUL1-RBX2NRBX1RING, CUL1-RBX1NRBX2RING, and CUL3–RBX1 variants was performed as described previously (Scott et al., 2014). Briefly, the final concentrations of components in the neddylation reactions were as follows: 12 μM CUL1-RBX1, CUL3-RBX1, CUL1-RBX2NRBX1RING, or mutant versions as indicated, 1 μM UBC12, 0.1 μM APPBP1-UBA3, and 20 mM NEDD8 in 25mM HEPES, 200mM NaCl, 10mM MgCl2, 1mM ATP, pH = 7.5. Reactions were initiated at room temperature by the addition of NEDD8 and incubated for five minutes prior to quenching by the addition of DTT to 10mM. Quenched reactions were spun at 13K rpm for 10 min and immediately applied to a Superdex SD200 column to purify NEDD8~CUL-RBX away from reaction components. Neddylation and purification of NEDD8 CUL1-RBX1NRBX2RING was performed as described above except the RBX2RING activated NEDD8 E2 UBE2F was used in place of UBC12.
To introduce a cysteine for fluorescent labeling of UB and UBK0, the Protein Kinase A phosphorylation site in the pGEX2TK backbone was changed, from the sequence (Scott et al., 2014). UB or UBK0 purified from this expression construct were labeled with Fluorescein-5-Maleimide or Tetramethylrhodamine-5-Maleimide (single isomer) as previously described (Scott et al., 2014). Briefly, DTT was added to UB or UBK0 at a final concentration of 10mM and incubated on ice for 20 min to completely reduce cysteines for labeling. DTT was removed by buffer exchange over a NAP-5 column (GE Healthcare) in labeling buffer (25mM HEPES, 200mM NaCl). Labeling reactions consisted of UB or UBK0 at 150 μM final concentration and were initiated by the addition of 600 μM Fluorescein-5-Maleimide or Tetramethylrhodamine-5-Maleimide (4X excess over labeling target and < 5% final DMSO concentration). Reactions were incubated at room temperature for 2 hr and quenched by the addition of DTT to 10mM. Quenched reactions were desalted over a PD-10 column in labeling buffer containing 1mM DTT to remove unreacted probe. Desalted protein was concentrated in an Amicon Ultra filtration unit and further purified over a Superdex SD75 column.
Flow Cytometry and Imaging
For cell cycle analysis, cells were fixed in ethanol followed by a 1x PBS wash. Cells were subsequently stained using propidium iodide and treated with RNase prior to analysis on a BD LSR Fortessa instrument. To determine nuclear size, 293T FLP-In cells with or without induction of GFP ARIH1-OPEN-OFF (48 hr) were stained with Hoechst dye. Cells were then imaged using a Yokogawa CSUX1 spinning disk confocal lens on a Nikon Ti-E inverted microscope in the Nikon Imaging Center at Harvard Medical School.
CompPASS Interaction Proteomics
Interaction proteomics using the CompPASS platform was performed as described (Sowa et al., 2009) with modifications. Briefly, 293T Flp-In or 293T cells containing N or C-terminally FLAG-HA tagged ARIH1 or the indicated mutants were induced for 24 hr with 0.5 μg/mL DOX and were treated with 1 μM MLN4924 for 1 or 2 hr as indicated. Cells were lysed in 50 mM Tris pH 7.5, 150 mM NaCl, 0.5% NP40 with protease inhibitors to generate whole cell lysates. Clarified lysates were filtered through 0.45 μm filters and immunoprecipitated with 30 μl anti-FLAG magnetic beads per replicate (Sigma). Complexes were washed 4x with lysis buffer and 2x with PBS and eluted with FLAG peptide at room temperature. Elutions were subjected to disulfide bond reduction using DTT and alkylation with iodoacetamide followed by TCA precipitation. TCA-precipitated proteins were trypsinized, purified with Empore C18 extraction media (3M) and analyzed via LC-MS/MS with a LTQ-Velos linear ion trap mass spectrometer (Thermo) with an 18 cm3 125 μm (ID) C18 column and a 50 min 8%–26% acetonitrile gradient. Complexes were analyzed twice by LC-MS to generate technical duplicates. Spectra were searched with Sequest against a target-decoy human tryptic UNIPROT-based peptide database, and these results were loaded into the Comparative Proteomics Analysis Software Suite (CompPASS) (Sowa et al., 2009), to identify high confidence candidate interacting proteins (HCIPs). Individual experiments were analyzed using a stats table derived from analogous AP-MS data for 41 unrelated proteins to determine normalized weighted D- scores (NWD-score) and Z-scores based on spectral counts. All CompPASS scores are provided in Table S1. To identify bait-associated proteins, proteins were filtered at a 2% false discovery rate for those with a NWD-score ≥ 1.0 and a Z-score ≥ 4.
Tandem Mass Tagging Interaction Proteomics
For analysis of interaction partners using Tandem Mass Tagging (TMT), we employed methods described in (Huttlin et al., 2015) with modifications. Eluted immune complexes were reduced, alkylated and TCA precipitated. TCA precipitations were digested overnight at room temperature with Lys-C followed by trypsin digest for six hours at 37°C in 200 mM EPPS pH 8.0. Samples were TMT labeled in a final acetonitrile concentration of 30% (v/v). Following room temperature incubation for 1 hr, the reaction was quenched with hydroxylamine to a final concentration of 0.3% (v/v). The TMT labeled samples were pooled, vacuum centrifuged, resuspended in 5% formic acid and subjected to C18 StageTip desalting. Samples were dried down, resuspended in 5% acetonitrile and 5% formic acid for LC-MS/MS processing. Mass spectrometry data were collected using an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, San Jose, CA) coupled to a Proxeon EASY-nLC II liquid chromatography (LC) pump (Thermo Fisher Scientific). Peptides were fractionated on a 75 μm inner diameter microcapillary column packed with ~0.5 cm of Magic C4 resin (5 μm, 100Å, Michrom-Bioresources) followed by ~35 cm of GP-18 resin (1.8 μm, 200Å, Sepax, Newark, DE). Samples were separated using a 120 min gradient of 5%–22% acetonitrile in 0.125% formic acid at a flow rate of 550 nL/min. Each analysis used the multi-notch MS3-based TMT method (McAlister et al., 2014). The scan sequence began with an MS1 spectrum (Orbitrap analysis; resolution 120000; mass range 350-1400 m/z; automatic gain control (AGC) target 5.0E5; maximum injection time 100 ms). Precursors for MS2/MS3 analysis were selected using a Top10 method. MS2 analysis consisted of collision-induced dissociation (quadrupole ion trap analysis; AGC 1.5E4; normalized collision energy (NCE) 35; maximum injection time 150ms). Following acquisition of each MS2 spectrum, we collected an MS3 spectrum using our recently described method in which multiple MS2 fragment ions were captured in the MS3 precursor population using isolation waveforms with multiple frequency notches (McAlister et al., 2014). MS3 precursors were fragmented by high-energy collision-induced dissociation (HCD) and analyzed using the Orbitrap (NCE 55; AGC 2E5; maximum injection time 150 ms, resolution was 15,000 at 200 Th).
TMT Total Proteome Analysis
For total proteome analysis using TMT, we employed methods described in (Huttlin et al., 2015) with modifications. 100 μg of whole cell lysate (lysed in 8M Urea, 50 mM NaCl, 50 mM EPPS pH 8.0, protease inhibitors) per replicate were reduced with 10 mM TCEP, alkylated with 15 mM Iodoacetamide. Excess iodoacetamide was quenched with 15 mM DTT. Samples were methanol-chloroform precipitated, resuspended in 200 mM EPPS pH 8.0 followed by sequential digestion with Lys-C (room temperature for 16 hr) and Trypsin (37°C for 6 hr). Samples were labeled with TMT reagent and pooled. We employed off-line basic pH reversed-phase fractionation on an Agilent 1260 HPLC prior to MS analysis. Approximately 0.6 mg of total peptide was loaded onto a 4.6 × 250 mM Aeris Peptide XB-C18 column with 2.6 micron core shell particles (Phenomenex, Torrance CA). Peptides were loaded in 100% solvent A (10mM ammonium bicarbonate, 5% (v/v) acetonitrile) and eluted over the course of a 60 min linear increase in solvent B (10mM ammonium bicarbonate, 90% (v/v) to 30%, followed by a short ramp to 100%. The fractionation was performed at a flow rate of 0.8 mL/min at 50°C. Ninety-six fractions (at 0.5 ml per fraction) were collected over the course of the gradient, and concatenated non-consecutively for a total of 12 fractions. Peptides were vacuum centrifuged to dryness prior to resuspension in 5% formic acid and C-18 stage tip clean up.
Peptides were separated using a 120 min gradient of 4%–22% acetonitrile in 0.125% formic acid at a flow rate of ~550 nL/min. Each analysis used the multi-notch MS3-based TMT method (McAlister et al., 2014). The scan sequence began with an MS1 spectrum (Orbitrap analysis; resolution 120000; mass range 400-1400 m/z; automatic gain control (AGC) target 5.0E5; maximum injection time 100 ms). Precursors for MS2/MS3 analysis were selected using a Top10 method. MS2 analysis consisted of collision-induced dissociation (quadrupole ion trap analysis; AGC 1.5E4; normalized collision energy (NCE) 35; maximum injection time 150ms). Following acquisition of each MS2 spectrum, we collected an MS3 spectrum using a method in which multiple MS2 fragment ions were captured in the MS3 precursor population using isolation waveforms with multiple frequency notches (McAlister et al., 2014). MS3 precursors were fragmented by high energy collision-induced dissociation (HCD) and analyzed using the Orbitrap (NCE 55; AGC 2E5; maximum injection time 150 ms, resolution was 60,000 at 200 Th).
Biochemical Assays
The use of pulse-chase assays allowed comparing the paths of UB transfer starting from different E2s. Conditions for the chase portions of pulse-chase assays used in different figures are as follows. For all assays except in Figures 4, S4, S5, 6, and S6 chase reactions consisted of mixing the E2~*UB thioester conjugate (0.4 μM final concentration in chase reaction) with the indicated combinations of ARIH1 or indicated mutants, neddylated or non-neddylated CRL–RBX1 (or indicated mutant) complexes, SRs and/or substrates at room temperature in 25 mM HEPES, 100 mM NaCl, 50 mM EDTA, 0.5 mg/ml BSA, pH 7.5. For reactions containing CUL3, mixtures consisted of pre-incubated complexes of CUL3–RBX1 or NEDD8 CUL3–RBX1 alone (0.4 μM final concentration in chase reaction), with KLHL12 (0.4 μM final concentration in chase reaction), and/or SEC13–SEC31A (0.6 μM final concentration in chase reaction) with or without WT or catalytically inactive ARIH1 (0.3 μM final concentration in chase reaction). For reactions monitoring ubiquitylation of the phosphopeptide derived from Cyclin E (pCyE) described previously (Jubelin et al., 2010) the final concentrations of CUL1–RBX1 and variants, substrate receptor FBW7ΔD, and WT, or the active site Cys357Ser mutant versions of ARIH1 were the same for the corresponding CUL–RBX1, SR, and ARIH components used in CRL3-based assays, but the pCyE peptide substrate was present in a five-fold excess (2 μM final concentration in chase reaction). For CRY1 ubiquitylation assays, the final concentration in chase reactions of CUL1–RBX1 variants and the SKP1–FBXL3–CRY1ΔC complex (1:1:1 stoichiometry due to coexpression and copurification) were also 0.4 μM. For assays monitoring ubiquitylation of phospho-p27, the substrate was a mutant version of p27 (Ser10Ala plus kinase inhibitory domain mutant F62A:F64A) that facilitates phosphorylation, which was first phosphorylated by incubation with Cyclin A–CDK2 as previously described (Vlach et al., 1997; Duda et al., 2008). Briefly, phosphorylation reactions consisted of mixing stoichiometric amounts (4 μM each) of p27 or the indicated mutant versions of p27 and Cyclin A-CDK2 in 25 mM HEPES, 50 mM NaCl, 10 mM MgCl2, 1 mM ATP, pH 7.5 and incubated at 30°C for 20 min. Phosphorylation reactions were diluted 10-fold into chase reactions to establish a 1:1:1 stoichiometric mixture of NEDD8~CUL1–RBX1, SKP1–SKP2–CKS1, CyclinA– CDK2–phospho-p27 at a final concentration of 0.4 μM. Dual-color ubiquitylation assays were performed as described above for their single color counterparts except chase reactions contained, where indicated, mixtures of the differentially labeled E2~*UB thioester conjugates.
To characterize the RBR-CRL mechanism, it was necessary to identify a wide-range of defects in ARIH1 mutants. To facilitate identifying mutants with altered activities based on both the time dependent consumption of *UB~UBCH7 and appearance of pCyE-~*UB, the pulse-chase assays monitoring *UB transfer to pCyE by WT and mutant versions of ARIH1 (excluding OPEN Mutant variants of ARIH1, see below) in Figures 4, S4, 5, S5, and 6 were slowed by performing the reactions with the protein concentrations described above, but on ice in 25 mM MES, 100 mM NaCl, 50 mM EDTA, 0.5 mg/ml BSA, pH 6.5.
For ARIH1 ubiquitylation assays in the absence of NEDD8~CUL1–RBX1 (Assay 4; Figures 5, S5, and 6), the chase reaction consisted of the UBCH7~*UB thioester conjugate at a final concentration of 0.08 μM and Open Mutant B (R420A/N423A/E503A) version of ARIH1, or the indicated mutants that additionally carry the open mutant B mutations that bypass the need for a neddylated CRL to relieve autoinhibition, at a final concentration of 0.125 μM. Chase reactions were performed on ice in 25 mM MES, 100 mM NaCl, 50 mM EDTA, 0.5 mg/ml BSA, pH 6.5.
To monitor CRL client substrate ubiquitylation with Open Mutant B alone or combined with other ARIH1 mutations to investigate their effects while bypassing the need for a neddylated CRL to relieve autoinhibition (Assay 5; Figures 5, S5, and 6), the chase reaction consisted of the UBCH7~*UB thioester conjugate at a final concentration of 0.08 μM and the indicated variants of CUL1–RBX1 or NEDD8~CUL1–RBX1 alone (0.1 μM final concentration in chase reaction), with or without the substrate receptor FBW7ΔD (0.1 μM final concentration in chase reaction), with or without the pCyE substrate (0.5 μM final concentration in chase reaction), with ARIH1 Open Mutant B or the indicated ARIH1 mutants that additionally carry the open mutant B mutations (0.06 μM final concentration in chase reaction). Chase reactions were performed on ice in 25 mM MES, 100 mM NaCl, 50 mM EDTA, 0.5 mg/ml BSA, pH 6.5.
Assays to measure ARIH1 kinetic parameters utilized a modified pulse-chase protocol. Chase reactions consisted of the UBCH7~*UB thioester conjugate at a fixed final concentration of 8 μM with increasing concentrations of CUL1-RBX1-SKP1-FBW7ΔD (pCyE substrate was maintained at a five-fold molar excess throughout the titration) as indicated and ARIH1 at a final concentration of 0.01 μM. Chase reactions were performed at room temperature in 25 mM HEPES, 100 mM NaCl, 50 mM EDTA, 0.5 mg/ml BSA, pH 7.5. Reactions were quenched at the indicated times by mixing with SDS sample buffer, separated by SDS-PAGE and analyzed based on fluorescent signals of *UB using a Typhoon FLA9500 Phosphoimager (GE Healthcare). To generate a standard curve for calculating rates of product formation, serial dilutions of the UBCH7 *UB “pulse” load reaction were run on SDS-PAGE gels as quantification standards. An aliquot of the “pulse” load reaction was also subjected to SDS-PAGE and stained with Coomassie in order to estimate the percentage of UBCH7 converted to *UB~UBCH7 during the loading reaction to properly correlate fluorescent *UB signal to concentration of UBCH7~*UB for standard curve generation (typically 70%–80% of the UBCH7 in the load reaction was converted to UBCH7~*UB). Product formation of pCyE-*UB was quantified utilizing the *UB standard curve and reaction rates calculated in Prism. The resulting rates were fit to a Michaelis-Menten model to determine the kinetic parameters Km and Vmax. The results in Figure 2E are representative phosphoimager scans from one experiment. Data fit in Figure 2F are from the average of three independent experiments.
Reference Table of Concentrations of Components in Biochemical Assays
| Experiment (Fig Panel) | [E2~*UB] (μM) | [CRL] (μM) | [SR] (μM) | [Substrate] (μM) | [ARIH1] (μM) | pH | Temp (°C) |
|---|---|---|---|---|---|---|---|
| 2B, S2A, S2B, 3A | 0.4 | 0.4 | 0.4 | 0.6 | 0.3 | 7.5 | 23 |
| S2D | 0.4 | 0.4 | 0.4 | 0.6 | Titrate | 7.5 | 23 |
| 2C, 3B, S3C | 0.4 | 0.4 | 0.4 | 2.0 | 0.3 | 7.5 | 23 |
| 2E | 8.0 | Titrate | Titrate | Titrate | 0.01 | 7.5 | 23 |
| S2E, S2F, 3C, S3D | 0.4 | 0.4 | 0.4 | 0.4 | 0.3 | 7.5 | 23 |
| 3E, S3B | U7-0.4, 34-0.4 | 0.4 | 0.4 | 0.4 | 0.3 | 7.5 | 23 |
| S2G | U7-0.4, U5-8.0, 34-8.0 | 0.4 | 0.4 | 0.4 | 0.3 | 7.5 | 23 |
| S2H | U7-0.4, 34-Titrate | 0.4 | 0.4 | 0.4 | 0.3 | 7.5 | 23 |
| S3A | U7-0.4, 34-0.4 | 0.4 | 0.4 | 2.0 | 0.3 | 7.5 | 23 |
| “Assay 1” 4C, S4A-I, S5H, 6B, S6A, 6H | 0.4 | 0.4 | 0.4 | 2.0 | 0.3 | 6.5 | 0 |
| “Assay 4” S5F, S5K, S5O, S5S, S5T, 6E | 0.08 | - | - | - | 0.1 | 6.5 | 0 |
| “Assay 5” S5G, S5L, S5P, S5U, S5V, 6C, 6J | 0.08 | 0.1 | 0.1 | 0.5 | 0.06 | 6.5 | 0 |
QUANTIFICATION AND STATISTICAL ANALYSIS
TMT Data Analysis
For quantification, Mass spectra were processed using a Sequest-based in-house software pipeline (Huttlin et al., 2015). Searches were performed against all entries from the human UniProt database (March 11, 2014) including all protein sequences in reverse order. Precursor ion tolerance for total protein level analysis was set to 50 ppm, while the product ion tolerance was set to 1.0 Da. TMT tags on lysine residues and peptide N- termini (+229.163 Da) and carbamidomethylation of cysteine residues (+57.021 Da) were set as static modifications, while oxidation of methionine residues (+15.995 Da) was set as a variable modification. We extracted the signal-to-noise ratio for each TMT channel and found the closest matching centroid to the expected mass. Peptide- spectrum matches (PSMs) were adjusted to a 2% false discovery rate. PSM filtering was performed using linear discriminant analysis (Huttlin et al., 2015). Total signal to noise values for all peptides were summed for each TMT channel and all values were adjusted to account for variance in sample handling. For each peptide, a total minimum signal to noise value of 100 was required (Huttlin et al., 2015). Change in protein abundance was determined from reporter ion intensities averaged across all peptides for a given protein. Subsequent data analyses for TMT-IPs were carried out using the R statistical package (ver 3.2.3) and Bioconductor (ver 3.3). TMT channel intensities were normalized by bait peptide intensities and rank, then the data were log-transformed. The log-transformed data were analyzed using an empirical Bayes approach as implemented by limma. Differential interactors were determined using a fold change threshold of 2.
Statistical Analysis
For all tandem mass tagging experiments, N = 3 biological replicates for each condition examined. Statistical parameters including log fold-change, SEM and statistical significance are reported in the figures and supplemental tables. For kinetics of ubiquitylation with ARIH1, the values reported are the mean ± one SD from N = 3 independent experiments.
Supplementary Material
Highlights.
● The RBR E3 ARIH1 assembles with numerous cullin-RING E3s via NEDD8 in mammalian cells
● ARIH1 ligates ubiquitin to CRL substrates, enabling CDC34-dependent chain elongation
● Two-step ARIH1 reactions involve multiple complementary interaction surfaces with CRLs
● Several CRL substrates accumulate with ARIH1 dominant negative or depletion
ACKNOWLEDGMENTS
We thank C. McGourty, M. Rape, D. van Dalen, A. Ordureau, J. Lee, and D. Miller. Funding was supplied by NIH R37NS083524 and AG011085 (to J.W.H.); ALSAC, HHMI, and NIH R37GM069530, P30CA021765 (to B.A.S.); NIH K01 DK098285 (to J.A.P.); ERC grant 281699 (to H.O.); and SCILLS (to A.F.A.). B.A.S. is a Howard Hughes Medical Institute Investigator.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, D.C.S., D.Y.R., D.M.D., J.W.H., and B.A.S.; Investigation, D.C.S., D.Y.R., D.M.D., I.R.K., J.L.O., J.A.P., and A.F.A.; Formal Analysis: D.C.S., D.Y.R., and J.A.P.; Resources, D.C.S., D.Y.R., D.M.D., I.R.K., J.L.O., A.d.J., and A.F.A. Funding Acquisition and Supervision, H.O., A.F.A., J.W.H., and B.A.S.; Visualization and Writing, D.C.S., D.Y.R., A.F.A., J.W.H., and B.A.S.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2016.07.027.
REFERENCES
- Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, et al. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell. 2011;145:1075–1087. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dove KK, Stieglitz B, Duncan ED, Rittinger K, Klevit RE. Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO Rep. 2016;17:1221–1235. doi: 10.15252/embr.201642641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Olszewski JL, Schuermann JP, Kurinov I, Miller DJ, Nourse A, Alpi AF, Schulman BA. Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure. 2013;21:1030–1041. doi: 10.1016/j.str.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Harper JW, Tan MK. Understanding cullin-RING E3 biology through proteomics-based substrate identification. Mol. Cell. Proteomics. 2012;11:1541–1550. doi: 10.1074/mcp.R112.021154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. High-resolution CRISPR screens reveal fitness genes and genotypespecific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. The BioPlex Network: A systematic exploration of the human interactome. Cell. 2015;162:425–440. doi: 10.1016/j.cell.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CS, Shemorry A, Auerbach D, Varshavsky A. The N-end rule pathway is mediated by a complex of the RING-type Ubr1 and HECT-type Ufd4 ubiquitin ligases. Nat. Cell Biol. 2010;12:1177–1185. doi: 10.1038/ncb2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgärtel C, Schekman R, Rape M. Ubiquitin-dependent regulation of COPII coat size and function. Nature. 2012;482:495–500. doi: 10.1038/nature10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubelin G, Taieb F, Duda DM, Hsu Y, Samba-Louaka A, Nobe R, Penary M, Watrin C, Nougayrède JP, Schulman BA, et al. Pathogenic bacteria target NEDD8-conjugated cullins to hijack host-cell signaling pathways. PLoS Pathog. 2010;6:e1001128. doi: 10.1371/journal.ppat.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsall IR, Duda DM, Olszewski JL, Hofmann K, Knebel A, Langevin F, Wood N, Wightman M, Schulman BA, Alpi AF. TRIAD1 and HHARI bind to and are activated by distinct neddylated Cullin-RING ligase complexes. EMBO J. 2013;32:2848–2860. doi: 10.1038/emboj.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- Lechtenberg BC, Rajput A, Sanishvili R, Dobaczewska MK, Ware CF, Mace PD, Riedl SJ. Structure of a HOIP/E2 ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature. 2016;529:546–550. doi: 10.1038/nature16511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Sweredoski MJ, Graham RL, Kolawa NJ, Smith GT, Hess S, Deshaies RJ. The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Mol. Cell Proteomics. 2011;10:M110.006460. doi: 10.1074/mcp.M110.006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Schulman BA, Harper JW. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013;14:1050–1061. doi: 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte R, Sayad A, Brown KR, Sanchez-Garcia F, Reimand J, Haider M, Virtanen C, Bradner JE, Bader GD, Mills GB, et al. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell. 2016;164:293–309. doi: 10.1016/j.cell.2015.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín I. RBR ubiquitin ligases: Diversification and streamlining in animal lineages. J. Mol. Evol. 2009;69:54–64. doi: 10.1007/s00239-009-9252-3. [DOI] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wühr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. MultiNotch MS3 en ables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal. Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta. 2014;1843:47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan TP, Ardley HC, Nuber U, Rose SA, Jones PF, Markham AF, Scheffner M, Robinson PA. The ubiquitin-conjugating enzymes UbcH7 and UbcH8 interact with RING finger/IBR motif-containing domains of HHARI and H7-AP1. J. Biol. Chem. 1999;274:30963–30968. doi: 10.1074/jbc.274.43.30963. [DOI] [PubMed] [Google Scholar]
- Pierce NW, Lee JE, Liu X, Sweredoski MJ, Graham RL, Larimore EA, Rome M, Zheng N, Clurman BE, Hess S, et al. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell. 2013;153:206–215. doi: 10.1016/j.cell.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley SR, Kuzmanov A, Kuang J, Karpel J, Lažetić V, Karina EI, Veo BL, Fay DS. Implicating SCF complexes in organogenesis in Caenorhabditis elegans. Genetics. 2014;196:211–223. doi: 10.1534/genetics.113.158485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–139. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol. Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Sviderskiy VO, Monda JK, Lydeard JR, Cho SE, Harper JW, Schulman BA. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell. 2014;157:1671–1684. doi: 10.1016/j.cell.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane M, Harumiya Y, Ishida N, Hirai A, Miyamoto C, Hatakeyama S, Nakayama K, Kitagawa M. Down-regulation of p27(Kip1) by two mechanisms, ubiquitin-mediated degradation and proteolytic processing. J. Biol. Chem. 1999;274:13886–13893. doi: 10.1074/jbc.274.20.13886. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz B, Rana RR, Koliopoulos MG, Morris-Davies AC, Schaeffer V, Christodoulou E, Howell S, Brown NR, Dikic I, Rittinger K. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature. 2013;503:422–426. doi: 10.1038/nature12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streich FC, Jr., Lima CD. Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 2014;43:357–379. doi: 10.1146/annurev-biophys-051013-022958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittal V, Stewart MD, Brzovic PS, Klevit RE. Regulating the regulators: recent revelations in the control of E3 ubiquitin ligases. J. Biol. Chem. 2015;290:21244–21251. doi: 10.1074/jbc.R115.675165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlach J, Hennecke S, Amati B. Phosphorylation-dependent degradation of the cyclin-dependent kinase inhibitor p27. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Kovacev J, Pan ZQ. Priming and extending: a UbcH5/ Cdc34 E2 handoff mechanism for polyubiquitination on a SCF substrate. Mol. Cell. 2010;37:784–796. doi: 10.1016/j.molcel.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhu W, Nhan T, Toth JI, Petroski MD, Wolf DA. CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat. Commun. 2013;4:1642. doi: 10.1038/ncomms2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W, Busino L, Hinds TR, Marionni ST, Saifee NH, Bush MF, Pagano M, Zheng N. SCF(FBXL3) ubiquitin ligase targets cryptochromes at their cofactor pocket. Nature. 2013;496:64–68. doi: 10.1038/nature11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1's C-terminal tail. Proc. Natl. Acad. Sci. USA. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–929. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- Yoo SH, Mohawk JA, Siepka SM, Shan Y, Huh SK, Hong HK, Korn-blum I, Kumar V, Koike N, Xu M, et al. Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell. 2013;152:1091–1105. doi: 10.1016/j.cell.2013.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Saeki Y, Murakami A, Kawawaki J, Tsuchiya H, Yoshihara H, Shindo M, Tanaka K. A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc. Natl. Acad. Sci. USA. 2015;112:4630–4635. doi: 10.1073/pnas.1422313112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Mao H, Novitsky EJ, Tang X, Rychnovsky SD, Zheng N, Huang L. Gln40 deamidation blocks structural reconfiguration and activation of SCF ubiquitin ligase complex by Nedd8. Nat. Commun. 2015;6:10053. doi: 10.1038/ncomms10053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemla A, Thomas Y, Kedziora S, Knebel A, Wood NT, Rabut G, Kurz T. CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat. Commun. 2013;4:1641. doi: 10.1038/ncomms2628. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr. Opin. Struct. Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.