Understanding plant response to salinity, one of the major abiotic stresses, provides insights into the improvement of tomato salt tolerance. This work focuses on the responses of tomato cultivars to salt stress. Genotypes, representative of content and enzyme activities. QPCR analysis of WRKY, ERF, LeNHX and HKT genes was also performed. A high K+, Ca2+ and proline accumulation as well as a decrease in Na+ concentration mediated salt tolerance. Concomitant with a pattern of high antioxidant enzyme activities, tolerant genotypes also displayed differential patterns of gene expression.
Keywords: Antioxidant enzymes, gene expression, ion contents, qPCR, salt stress, Tunisian tomato genotypes
Abstract
Salinity is a constraint limiting plant growth and productivity of crops throughout the world. Understanding the mechanism underlying plant response to salinity provides new insights into the improvement of salt tolerance-crops of importance. In the present study, we report on the responses of twenty cultivars of tomato. We have clustered genotypes into scale classes according to their response to increased NaCl levels. Three local tomato genotypes, representative of different saline scale classes, were selected for further investigation. During early (0 h, 6 h and 12 h) and later (7 days) stages of the response to salt treatment, ion concentrations (Na+, K+ and Ca2+), proline content, enzyme activities (catalase, ascorbate peroxidase and guiacol peroxidase) were recorded. qPCR analysis of candidate genes WRKY (8, 31and 39), ERF (9, 16 and 80), LeNHX (1, 3 and 4) and HKT (class I) were performed. A high K+, Ca2 +and proline accumulation as well as a decrease of Na+ concentration-mediated salt tolerance. Concomitant with a pattern of high-antioxidant enzyme activities, tolerant genotypes also displayed differential patterns of gene expression during the response to salt stress.
Introduction
High salinity is an important abiotic stress that reduces crop productivity in arid and semi-arid regions of the world (Foolad 2007). It is estimated that, worldwide, 800 million ha of land and 32 million ha of agricultural land are salt-affected (FAO 2015). In order to enhance productivity, improving salt tolerance of crop plants has the potential to make marginal areas agriculturally productive (Foolad 2007, Karan and Subudhi 2012). To achieve this goal, it is crucial to understand the physiological, biochemical and molecular mechanisms evolved by plants to cope with salt stress.
Soil salinization inhibits water uptake by the plants, causes ionic imbalance leading to ionic toxicity and osmotic stress (Munns and Tester 2008). To withstand salt stress, plants accumulate compatible solutes such as proline, which decreases the cytoplasmic osmotic potential, facilitating water absorption, and scavenges reactive oxygen species (ROS) molecules (Qureshi et al. 2013; Pottosin et al. 2014).
Multiple signalling pathways lead to the expression of genes that in turn allow the activation of the proteins that determine plant phenotype under salt stress (Marco et al. 2015). Data on signalling pathways have increased in recent years. Analysis of this data will not only elucidate the function and regulation of complex plant responses to salt stress but also the identification of genes whose function is unknown and which may have important roles in salt tolerance. These downstream signalling pathways comprise several active components including second messengers, phytohormones and phosphoprotein cascades. The Ca2+ is a second messenger in signalling network coupling the perception of a stressful environment to a significant plant adaptability (Tuteja and Mahajan 2007; Marco et al. 2015). Ca2+ acts at the crossroads of various signalling pathways (Gill and Tuteja 2010; Rany et al. 2016). High-salinity stress initiates the calcium signalling network (Tuteja 2009), inducing membrane depolarization, and may activate sensitive Ca2+ channels to generate a Ca2+ signature (Tester and Davenport 2003; Zhu 2003). Increases in Ca2+ concentrations and stimulus-induced enhancement in Ca2+ sensitivity (Young et al. 2006) function as an effective signal which modulates calcium-binding proteins thus transmitting signals in signal transduction pathways (Uozumi and Schroeder 2010).
Phytohormones such as abscisic acid (ABA), salicylic acid (SA), ethylene (ET) and jasmonic acid (JA) activate pathways that may act independently or synergistically with others triggered by stress (Marco et al. 2015). Protein kinases and phosphatases play a fundamental role in the coordination of the activity of many known signalling pathways (Marco et al. 2015). Transcriptome studies reveal that genes induced by these signalling cascades triggered by salt stress can be divided into two categories depending on the features of their products (Bohnert et al. 2001; Fowler and Thomashow 2002; Seki et al. 2002). The first, composed of functional proteins such as membrane proteins, protects cells against stress effects by restoring cellular homeostasis.
Ion channels in plant cells play crucial functions in adapting and overcoming salt stress (Uozumi and Schroeder 2010). Cation transporters as HKT and LeNHX enhance salt tolerance by regulation internal concentrations of Na+ in tissues. The expression level of HKT1-like transporters has been reported to be directly related to salt tolerance and Na+-specific tissue distribution depending to the plant source. HKT1;1 and HKT1;2 are two tomato Na+- selective transporters that contribute to Na+ and K+ homeostasis (Hauser and Horie 2010; Pardo and Rubio 2011). Salt tolerance is achieved by retrieval of Na+ from the xylem vessels to xylem parenchyma cells, promoting vacuolar accumulation and thus protecting photosynthetic leaf tissues from the adverse effect of Na+(Davenport et al. 2007; Plett et al. 2010, Xue et al. 2011; Munns et al. 2012). Several studies reported that HKT-I like transporters are associated with QTL on chromosome 7 in two populations of F(8) lines, derived from a salt sensitive genotype of Solanum lycopersicum cv. Cerasiforme, as female parent and two salt tolerant lines, as male parents, from Solanum pimpinellifolium and Solanum cheesmaniae. HKT-I like transporters seem to be involved in Na+ and K+ homeostasis in aerial parts of the plant (Ren et al. 2005; Villalta et al. 2008). NHX (Na+/H + Antiporters) and HKT (Histidine Kinase Transporter) genes, encoding K+ transporters and channels, have been implicated in multiple biological responses in various plant species (Gupta and Huang 2014). The two LeNHX1 and LeNHX2 isoforms localized in the tonoplast are essential for active K+ uptake, for stomatal function and for turgor regulation (Barragán et al. 2012) while LeNHX3 and LeNHX4 isoforms are involved in Na+, K+, and H+ homeostasis (Gàlvez et al. 2012). The HKT family improves salt tolerance by regulating ion transportation (Gupta and Huang 2014). In tomato, HKT1;1 and HKT1;2 are responsible for the major QTL involved in Na+ and K+ homeostasis (Asins et al. 2012). In Arabidopsis, HKT transporters protect the plant from the adverse effects of salinity by preventing excess Na+ accumulation in leaves. Experiments carried out on rice by Schroeder et al. (2013) suggest that HKT class I transporters remove excess Na+ from xylem, protecting the photosynthetic leaf tissues from the toxic effect of Na+. This first category also includes biosynthetic enzymes for metabolites acting in osmotic adjustment or protection as well as ROS detoxification enzymes.
High salinity has been reported to induce ROS formation and accumulation in plant cells (Chawla et al. 2013). Oxidative stress defenses occur through enzymatic antioxidant mechanism including catalase (CAT), superoxide dismutase (SOD), peroxidase (POX) and enzymes of the ascorbate-glutathione cycle as ascorbate peroxydase (APX), monodehydroascorbate dehydrogenase (MDHAR), dehydroascorbate reductase (DHAR) (Foyer and Noctor 2011; Chawla et al. 2013) and non-enzymatic antioxidants as phenolics, flavonoids (Munné-Bosch 2005; Gupta and Huang 2014; Rakhmankulova et al. 2015; Talbi et al. 2015). CAT is involved in scavenging of H2O2 during salt stress and other abiotic stress conditions (Willekens et al. 1997) and is considered as a major enzyme detoxifying H2O2 in tomato fruits (Murshed et al. 2014). Although APX performs the same general function as catalase, it catalyzes removal of H2O2 by using ascorbate as a reductant. APX is a family of isozymes widely involved in regulation of intracellular level of H2O2 in higher plants (Van Breusegem et al. 2001; Shigeoka et al. 2002). GPOX enzymes protect cells against oxidative damage generated by ROS. They catalyze the reduction of H2O2 or organic hydroperoxides to H2O or alcohols. The second category comprises a series of regulatory proteins (transcription factors, protein kinases) involved in the regulation of the signalling cascade that controls the expression of additional genes whose products could belong, in turn, to either of the two groups (Agarwal et al. 2006; Shinozaki and Yamaguchi-Shinozaki 2007).
The main stress-related transcription factors include members of APETALA2/Ethylene Responsive Factor (AP2/ERF), basic helix-loop-helix (bHLH), and basic leucine zipper (bZIP) proteins, the homeodomain-leucine zipper (HD-Zip), myelocytomatosis (MYC), myeloblastosis (MYB) and NAC families and members of the WRKY family (Lindemose et al. 2013). Previous studies revealed a significant induction of 18 different tomato SlWRKY genes under conditions of salt, drought or pathogen challenge, implying that they are regulators of plant responses to various biotic and abiotic stresses (Huang et al. 2012). Involvement of WRKY factors in plant salt adaptation was shown for WRKY15; 18; 20; 25; 33; 39; 40; 45; 60 and WRKY82 which increased salt tolerance in many plant species (Bakshi and Oelmüller 2014; Huang et al. 2012; Jiang and Deyholos 2009; Liu et al. 2011; Peng et al. 2012; Sun et al. 2014). These transcription factors are well interconnected with other complex signalling pathways corresponding to cell homeostasis, photosynthesis, oxidative pathway and enzyme activity (Baniwal et al. 2007; Eulgem and Somssich 2007). In Arabidopsis, AtWRKY25 and AtWRKY33 transcription level was increased by drought or NaCl treatment (Chen et al. 2012). Sharma et al. (2010) reported the induction of ERF family genes in response to various stress treatments (salt, cold, heat, dehydration, mechanical stress, oxidative stress, and submergence stress) suggesting a crosstalk between different stress-signalling pathways. All these elements interact with each other, forming a complex network which finally results in the modification of target proteins that may have enzymatic or structural function, leading to cellular responses at the physiological, biochemical and molecular levels.
Because of the extensive salinization of Tunisian soils, considerable efforts are being invested to improve salt tolerance in tomato. Achieving this goal depends on elucidation of mechanisms by which tomato plants are able to perceive stress and to activate appropriate cellular responses. In the present study, we explore the behaviour of tomato genotypes subjected to saline treatment to discover whether genotypic differences in response to salt stress exist. We explored the modulation of some physiological traits involved in the response to salt stress, mainly Na+, K+, Ca2+, and proline content, as well as expression of the antioxidant enzymes APX, CAT and GPOX. We also investigated the expression of a panel of genes (WRKY, LeNHX, HKT and ERF) involved in ion accumulation, encoding ion transporters or channels. Analyses were performed in leaves and roots at early (0, 6 and 12 h) and late (7 days) stages of the saline treatment. Our findings will help identifying potential candidate genes for local tomato genetic improvement to salt stress in Tunisia.
Methods
Plant material
Twenty tomato genotypes commonly cultivated by Tunisian growers were used in this investigation. They correspond to three TYLCV-tolerant lines (San Miguel, Ilanero and Romelia; Scott et al. 1995; Vidavsky and Czosnek 1998; Mejía et al. 2002) and 17 local Tunisian genotypes. Seeds were surface sterilized with 0.5 % NaCl solution, rinsed with water and incubated in Petri dishes on moist sterile filter paper at 27 °C in darkness until emergence of the radicle. Two days later, tomato seedlings were transferred into hydroponic tanks, each containing 10 L of half-strength modified Hoagland solution (Epstein 1972). The hydroponic solutions were vigorously aerated and renewed every 2 days during the growing period. The experiment was carried out with three replicates [see Supporting Information—Figure S1]. Plants were grown in an environmentally controlled chamber at 25 °C/18 °C, day/night and a 16-h light/8-h dark cycle with 40–50 % relative humidity (Asins et al. 2012).
Evaluation of salt tolerance
Plants with four fully developed true leaves were individually transferred into plastic pots (30 cm of diameter) containing a mixture of peat and sand then irrigated with one-half Hoagland solution supplemented with 150 mM NaCl (15 dS/m, pH 7.5). Salt treatment was initiated with 50 mM of NaCl solution (6 dS/m), increased to 100 mM (12 dS/m) on day two and finally to 150 mM (15 dS/m) on day three. We used three biological replicates for each of the 20 varieties. Each replicate consisted of a pool of 10 plants. A set of three plants for each genotype was grown in non-saline conditions and watered with the nutrient solution. Three weeks later, salt-treated plants were evaluated for salt tolerance, based on their visual phenotypes compared to control plants. Plants were rated for severity of salt susceptibility by on a 1–5 scale (Dasgan et al. 2002) [see Supporting Information—Figure S2].
Ten plants of each genotype were tested at 0 h, 6 h, 12 h and 7 days post-saline treatment. Fresh Root and leaf tissues were harvested, rinsed with demineralized water and weighted for immediate use. Half of the plants were dried in a forced air oven at 70 ºC to determine the dry weight (DW) [see Supporting Information—Figure S3]. The remaining plants were immediately stored at −80 ºC.
RNA isolation and quantitative real-time PCR analysis
Total RNA was isolated from root and leaf tissues using the TRIzol® LS Reagent (Trizol RNA stabilization solution, Invitrogen; Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. RNA was quantified by ND-1000 spectrophotometer (Nanodrop Technologies, USA). First-strand cDNA was synthesized from 2 µg of total RNA with oligo(dT) and MMLV reverse transcriptase (200 U/μl, Invitrogen) according to the manufacturer’s instructions. ABI A Prism 7000 sequence detection system (Applied Biosystems, USA) was used for quantitative real-time PCR (qPCR) under the following cycle conditions: 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C, 1 min at 60 °C. The Actin tomato gene (ACT) was used as internal reference gene (Lovdal and Lillo 2009 [see Supporting Information—Figures S3 and S4]. Genes and their corresponding primers are shown in Table 1. PCR reactions were carried out in 96-well optical reaction plates (Applied Biosystems, USA). Reaction included 50 ng of cDNA sample as a template, 400nM forward and reverse primers, and Igreen qPCR master Mix-Rox (BIOMATIK, USA). Relative quantification was performed by applying the 2−ΔΔCt method (Livak and Schmittgen 2001).
Table 1.
List of primers used for qPCR analysis.
| Genes names | Primers | References |
|---|---|---|
| SIWRKY 8 |
|
Huang et al. (2012) |
| SIWRKY 31 |
|
|
| SIWRKY 39 |
|
|
| SIERF 9 |
|
Sharma et al. (2010) |
| SIERF 16 |
|
|
| SIERF 80 |
|
|
| LeNHX1 |
|
Gálvez et al. (2012) |
| LeNHX3 |
|
|
| LeNHX4 |
|
|
| HKT1.1 |
|
Asins et al. (2012) |
| HKT1.2 |
|
|
| ACTIN |
|
Lovdal and Lillo (2009) |
Na+, K+ and Ca2+ measurements
Dried plant materials consisting to roots and leaves were digested with HNO3/HClO4 solution (2:1). Na+ and K+ ions were quantified using a flame photometer (Jenway Model PEP7, USA). Ca2+ concentrations were determined by atomic absorption spectrometry (Perkin-Elmer 5 5500, Waltham, MA, USA).
Proline concentration
Proline content in root and leaf tissues was measured via reaction with ninhydrin (Bates et al. 1973). For colorimetric determinations, a solution of proline, ninhydrin acid and glacial acetic acid (1:1:1) was incubated at 90 ºC for 1 h. Then, the reaction was cooled in an iced bath. The chromophore was extracted using 2 ml of toluene and its absorbance at 520 nm was determined by a BioMate spectrophotometer (ThermoSpectronic, USA).
Antioxidant enzyme activity measurements
One gram of either fresh leaf or root material was weighted individually and immediately homogenized in 5 ml of 50 mM K–phosphate buffer (pH 7.0), brought to 5 mM Na–ascorbate and 0.2 mM EDTA by the addition of concentrated stocks. The homogenate was centrifuged at 10 000g for 15 min at 4 °C. The resulting supernatant was used for enzyme assays. The extraction was carried out at 4 °C. Proteins were quantified according to Bradford (1976) using albumin bovine serum as a standard.
Activity of CAT was determined by monitoring the disappearance of H2O2 at 240 nm (extinction coefficient of 0.036 mM − 1 cm − 1) (Aebi 1984). One unit of activity was defined as the amount of enzyme required to decompose 1 µmol H2O2 per min at 25 °C. Activity of GPOX was measured by monitoring the increase in absorbance at 470 nm (ε = 26.6 mM − 1 cm − 1) during polymerization of guaiacol (Fielding and Hall 1978). One unit of activity was defined as the amount of enzyme producing 1 µmol of tetraguaiacol per min at 25 °C. Activity of APX was determined based on the decrease in absorbance at 290 nm (absorbance coefficient 2.8 mM−1 cm−1) as ascorbate was oxidized according to Nakano and Asada (1981). One unit of enzyme was defined as the amount necessary to decompose 1 µmol of ascorbate per min at 25 °C.
Statistical analysis
Data were analyzed using two-way ANOVAs with times and varieties as the two predictor variables. Differences at Tukey’s test HSDp = 0.05 were considered statistically significant. Analyses were performed using GraphPad Software (version 6.0, CA, USA). A heat map and a signal correlation were performed to visualize the correlation of the expression of candidate genes during salt treatment based on Pearson’s correlation. Only the comparisons with P < 0.05 were regarded as showing differential expression. The neutral/middle expression was set as the median of all the ΔCt values from tested varieties, red colour was used to indicate an increase with a ΔCt value below the median and the green indicated a decrease with ΔCt above the median. The ΔCt set of each considered variety is plotted on a scatter graph where the two axes are the Pearson correlation coefficients against two different query ΔCt sets. Analyses were performed with DataAssist™ v3.0 Software (Applied Biosystems, USA).
Results
Evaluation of tomato genotypes under salt stress
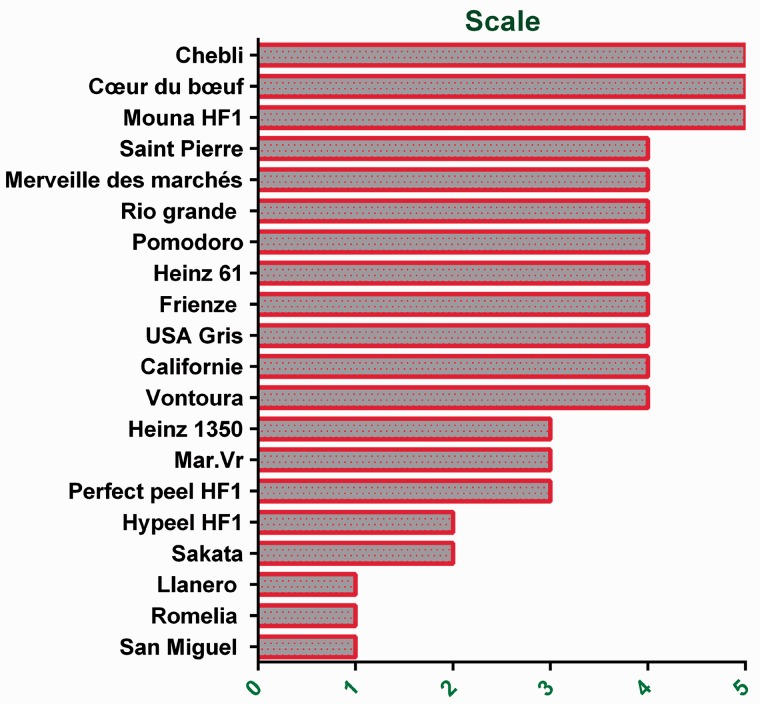
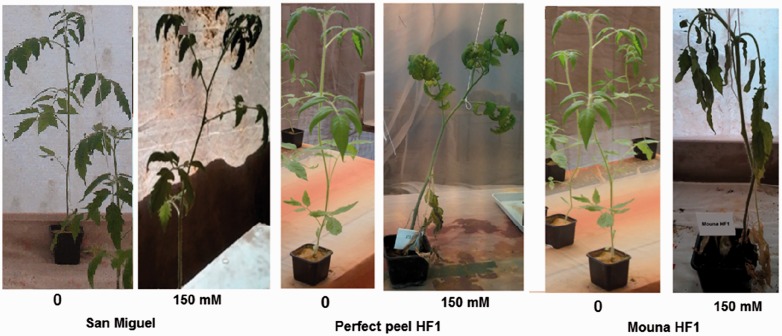
Based on symptoms and visual phenotypes, tomato varieties were screened for their response to the salt stress treatment according to Dasgan's (Dasgan et al. 2002) 1–5 scale classes. Three of the evaluated varieties (San Miguel, Ilanero and Romelia) showed no visual symptoms of dehydration and were clustered to the Class 1. Remaining genotypes were assigned to scale-classes as follow: 10 % (Sakata and Hypeel HF1) to Class 2, 15 % (Perfect peel HF1, Mar.Brand Heinz 1350) to class 3, 45 % (Ventura, California, USA gris, Frienze, Heinz 61, Pomodoro, Rio grande, Merveille des marchés and Saint Pierre) to Class 4 % and 15 % (Mouna HF1, Coeur du boeuf and Chebli) to Class 5 (Fig. 1). Salt treatment seems not to affect the maintenance of the growth of Class 1 genotypes while leading to reduced stem growth of those belonging to Classes 4 and 5. Subsequent analyses focused on three contrasting varieties. San Miguel represented the tolerant scale-Class 1 without any symptoms of salt damage; Perfect peel HF1 represented the mildly tolerant scale-Class 3 with curly and moderate dry leaves and the sensitive Mouna HF1 belonged to the scale-Class 5 showing all leaves with drying damage (Fig. 2).
Figure 1.
Clustering of Tunisian tomato genotypes based on their phenotypes according to Dasgan' scale classes (Dasgan et al. 2002).
Figure 2.
Visual appearance of the three selected genotypes growing in saline and non-saline conditions.
Na+, K+ and Ca2+ contents
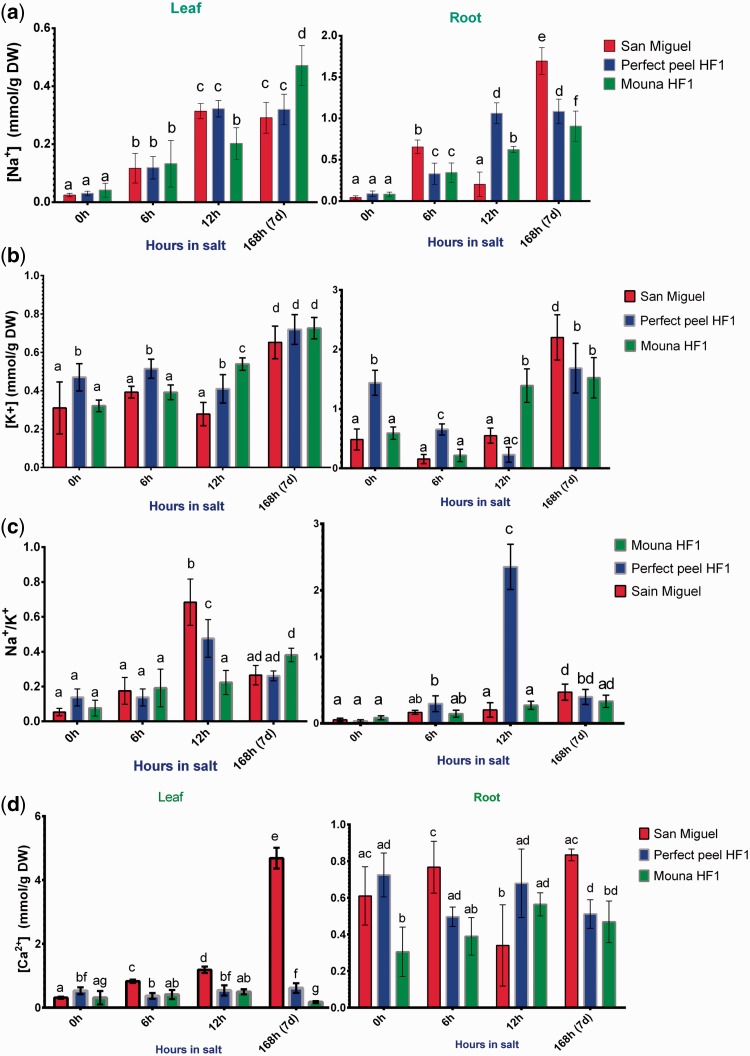
The distribution of Na+, K+ and the ratio Na+/K+ as well as Ca2+ in both leaves and roots were analyzed at 6 h, 12 h and 7 days post-stress application. Depending on the genotype, the increase in Na+ concentration followed a constant pattern in leaves (F2, 8 = 1.547, P = 0.2705) and varied significantly in roots (F2, 8 = 9.190, P = 0.0085). Na+ concentration also varied significantly according to the salt stress stage treatment in both tissues (Leaves: F3, 12 = 104.8, P < 0.0001; Roots: F3, 12 = 172.4, P < 0.0001). We also detected a significant interaction between genotypes and stages of stress treatment (F6, 24 = 11.75, P < 0.0001) and (F6, 24 = 67.87, P < 0.0001) in leaves and roots, respectively. In early stage of stress treatment (6 h), the three tested genotypes displayed similar Na+ concentrations in their leaves. By contrast, at 7 days post-treatment, the sensitive Mouna HF1 showed the highest Na+ concentration. Regardless of genotype, Na+ preferentially accumulated in roots rather than in leaves (Fig. 3A). Results showed the tolerant San Miguel accumulating the highest Na+ level in roots at the latest stage of salt treatment. Results from two-way ANOVA indicated that stages of treatment (leaves: F3, 12 = 77.31, P < 0.0001; roots: F3, 12 = 108.0, P < 0.0001) and genotypes (leaves: F2, 8 = 17.70, P = 0.0012; roots: F2, 8 = 5.674, P = 0.0292) had a significant overall effect on K+ concentration in tissues. In addition, we detected significant stages × genotypes interaction terms for both tissues (leaves: F6, 24 = 5.176, P = 0.0015; roots: F6, 24 = 20.67, P < 0.0001).K+ showed the same levels in leaves in all three genotypes (after 7 days of treatment). In roots, San Miguel and Mouna HF1 displayed similar K+ concentrations at early (6 h) stage of treatment whereas K+ content concentration increased within San Miguel at the latest stage (7 days) (Fig. 3B). As it appears to be a key determinant of salt tolerance, the Na+/K+ ratio was calculated Na+/K+ is statistically significant between genotypes (leaves: F2, 8 = 5.864, P = 0.0270; roots: F2, 8 = 341.2, P < 0.0001) and between stages of salt stress (leaves: F3, 12 = 145.8, P < 0.0001; roots: F3, 12 = 94.47, P < 0.0001). In addition, we detected significant stage × genotype interaction terms for both tissues (leaves: F6, 24 = 12.97, P < 0.0001; roots: F6, 24 = 120.7, P < 0.0001). In leaves, San Miguel and Perfect peel HF1 exhibited the highest leaf Na+/K+ ratio at 12 h post salt-treatment. All genotypes showed similar Na+/K+ ratio in both leaves and roots at the latest stage of treatment (Fig. 3C).
Figure 3.
Ion content in leaf and root tissues within San Miguel, Perfect peel HF1 and Mouna HF1 genotypes during 6 h, 12 h and 7 days post-NaCl treatment (15 dS/m, pH 7.5). (a) Na+, (b) K+, (c) Na+/K+ and (d) Ca2+. Values are expressed in mmole/g DW. The data reported are the mean ± SE of five biological replicas. Bars with different letters within each panel are significantly different at P > 0.05 according to Tukey's test.
In order to initiate defence–response mechanisms during saline stress, plants first need to perceive the stress, then activate the whole signalling cascade, starting with an increase of Ca2+ concentration. For this reason, Ca2+ concentration was also determined. Depending on the genotype, the increase in Ca2+ concentration followed a different pattern in leaves (F2, 8 = 520.4, P < 0.0001) and roots (F2, 8 = 25.15, P = 0.0004). Results revealed that stages of salt treatment had significant effect on Ca2+ concentration in leaves (F3, 12 = 214.5, P < 0.0001) but not in roots (F3, 12 = 0.9336, P = 0.4546). We also detected a significant interaction between these variables in tissues (leaves: F6, 24 = 403.4, P < 0.0001; roots: F6, 24 = 8.902, P < 0.0001). The highest Ca2+ concentration was recorded in San Miguel (the tolerant genotype) in both leaf and root tissues, at the latest stage of the treatment. By contrast, Perfect peel HF1 and Mouna HF1 genotypes showed reduced Ca2+ in leaves during first stage of salt stress. At 7 days post-salt imposition, both displayed the lowest lower Ca2+ concentrations in leaves and roots (Fig. 3D).
Effects of saline stress on proline accumulation
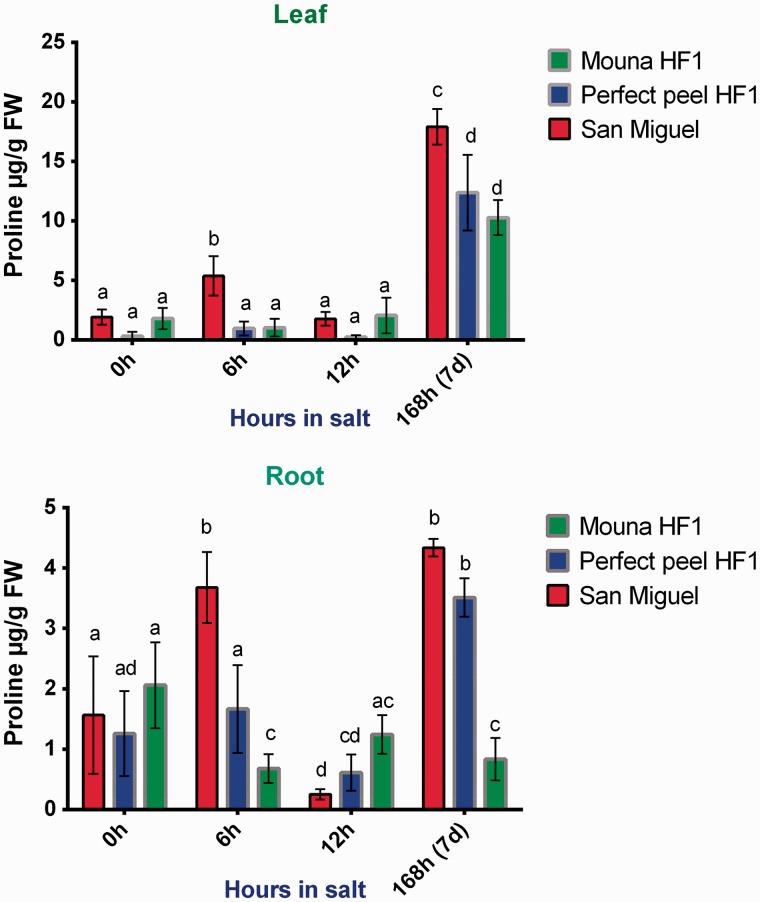
To combat osmotic stress imposed by high salinity, plants need to synthesize compatible organic solutes such as proline in the cytosol. Accumulation of proline was determined and seems to be linked to the scale class. Results from two-way ANOVA and Tukey's test indicated that the stage of salt treatment had a significant overall effect on proline concentration (leaves: F3, 12 = 260.2, P < 0.0001; roots: F3, 12 = 44.51, P < 0.0001). Similarly, results showed proline concentration varied significantly between genotypes in both tissues (leaves: F2, 8 = 48.22, P < 0.0001; roots: F2, 8 = 37.69, P < 0.0001). In addition, we detected significant interaction terms between genotypes and stages of treatment for leaves (F6, 24 = 8.949, P < 0.0001) and roots (F6, 24 = 23.20, P < 0.0001). In leaves, the tolerant San Miguel genotype showed a negligible content of proline during first stages of treatment with a peak observed at 6 h. However, proline concentration increased significantly at the late stage of the treatment, reaching 17 µg/g FW. Perfect Peel HF1 and Mouna HF1 genotypes displayed lower proline concentration at 7 days post-treatment reaching 13 µg/g FW and 10 μg/g FW, respectively (Fig. 4A). In roots, proline amount reached 4.33 µg/g FW, 3.50 µg/g FW and 0.82 µg/g FW in San Miguel, Perfect peel HF1 and Mouna HF1, respectively, at 7 days post-salt treatment (Fig. 4B). It is worth noting that the tolerant San Miguel genotype displayed similar proline concentration to Perfect peel HF1 at the latest stage of the treatment. Results indicate that proline is more abundant in leaves than roots within all the stressed genotypes especially at the end of the treatment.
Figure 4.
Proline accumulation in leaf and root tissues within San Miguel, Perfect peel HF1 and Mouna HF1 genotypes during 6 h, 12 h and 7 days post NaCl treatment (15 dS/m, pH 7.5). Data expressed as µg/g of fresh weight are the mean ± SE of 3 biological replicas. Bars with different letters within each panel are significantly different at P > 0.05 according to Tukey's test.
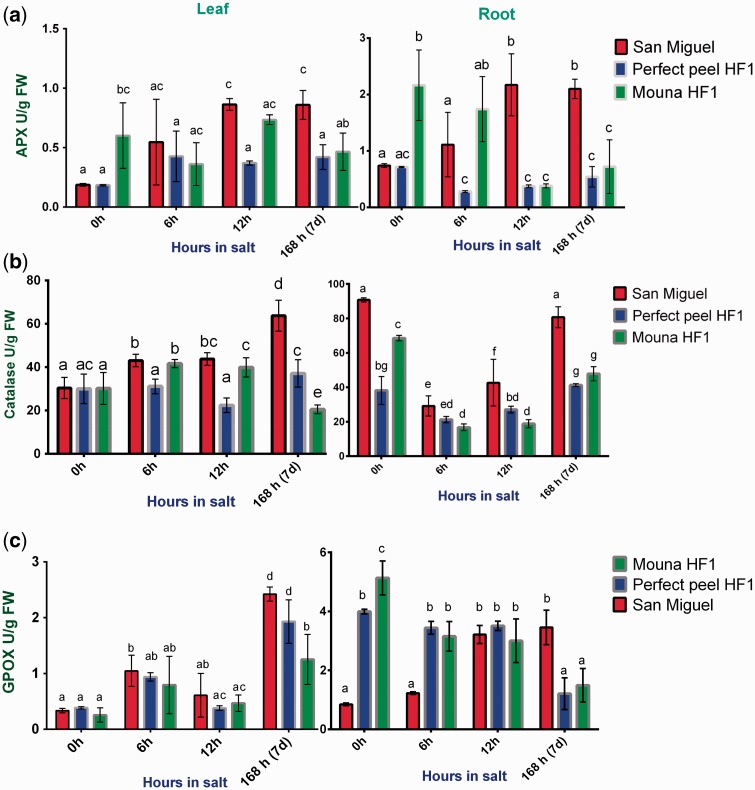
Antioxidant enzyme activities
To minimize the deleterious effects of ROS, plant cells suppress the buildup of harmful intracellular ROS concentrations. This is achieved by the action of the antioxidative defence systems as enzymatic ROS scavengers including APX, CAT and GPOX. In order to get further insight into the effect of salt stress on oxidative stress parameters, APX, CAT and GPOX activities were measured. Two way ANOVA followed by Tukey’s multiple comparisons test indicated that variation in APX activity is statistically significant between genotypes in roots (F2, 4 = 57.23, P = 0.0011) but not in leaves (F2, 4 = 6.382, P = 0.0569). By contrast, APX activity is significantly different between stages of stress treatment in leaves (F3, 6 = 6.252, P = 0.0282) but not in roots (F3, 6 = 0.3765, P = 0.7736). When considering interaction between genotypes and stages of stress treatment, results indicated that variation is statistically significant (leaves: F6, 12 = 3.976, P = 0.0201; roots: F6, 12 = 14.90, P < 0.0001). In both leaf and root tissues, APX activity gradually increased during all stages of the stress treatment within the San Miguel tolerant plant, whereas it was reduced with the remaining plants, especially in roots (Fig. 5A). CAT activity was not associated with significant changes at any stage of salt treatment in leaves (F3, 6 = 4.487, P = 0.0562) but not in roots (F3, 6 = 194.9, P <0.0001). CAT activity increased significantly among genotypes (leaves: F2, 4 = 25.48, P = 0.0053; roots: F2, 4 = 91.06, P = 0.0005). We also detected a significant interaction between genotypes and stages of stress treatment (leaves: F6, 12 = 29.81, P < 0.0001; roots: F6, 12 = 14.62, P < 0.0001). Leaf CAT activity displayed a significant increase within San Miguel at the latest stage post-salt treatment (Fig. 5B). Concerning GPOX activity, statistical results showed significant changes in both tissues either between genotypes (leaves: F2,4 = 13.06, P = 0.0176; roots: F2, 4 = 35.46, P = 0.0029) or between stages of salt treatment (leaves: F3, 6 = 31.75, P = 0.0004; roots: F3, 6 = 7.946, P = 0.0164). The interaction between these two variables is also significant (leaves: F6, 12 = 3.293, P = 0.0374; roots: F6, 12 = 52.82, P < 0.0001). GPOX leaf activity displayed the highest value within San Miguel and Perfect Peel HF1 genotypes at the end of the treatment (Fig. 5C). Otherwise, GPOX root activity showed a similar pattern within Perfect Peel HF1 and Mouna HF1 during the first and the last stage of treatment increasing within San Miguel at 7 days post-stress.
Figure 5.
Antioxidative enzyme activities in leaf and root tissues within San Miguel, Perfect peel HF1 and Mouna HF1 genotypes during 6 h, 12 h and 7 days post-NaCl treatment. (a) APX, (b) Cat and (c) GPOX. Values are the mean ± SE of 3 biological replicas. Bars with different letters within each panel are significantly different at P > 0.05 according to Tukey's test.
Analysis of differentially expressed (WRKY, ERF, LeNHX and HKT) genes by qRT-PCR
The expression profiles of tomato genes were analyzed in both leaf and root tissue. Three ERF family genes (ERF9, 16 and 80), three WRKY family genes (WRKY8, 31 and 39), 2 HKT class I gene transporters (HKT1;1 and 1;2) and three LeNHX genes (LeNHX1, 3 and 4) were selected and subjected to a qRT-PCR analysis for samples corresponding to the first stage (0 h, 6 h and 12 h) and a last stage (7 days) of the stress imposition.
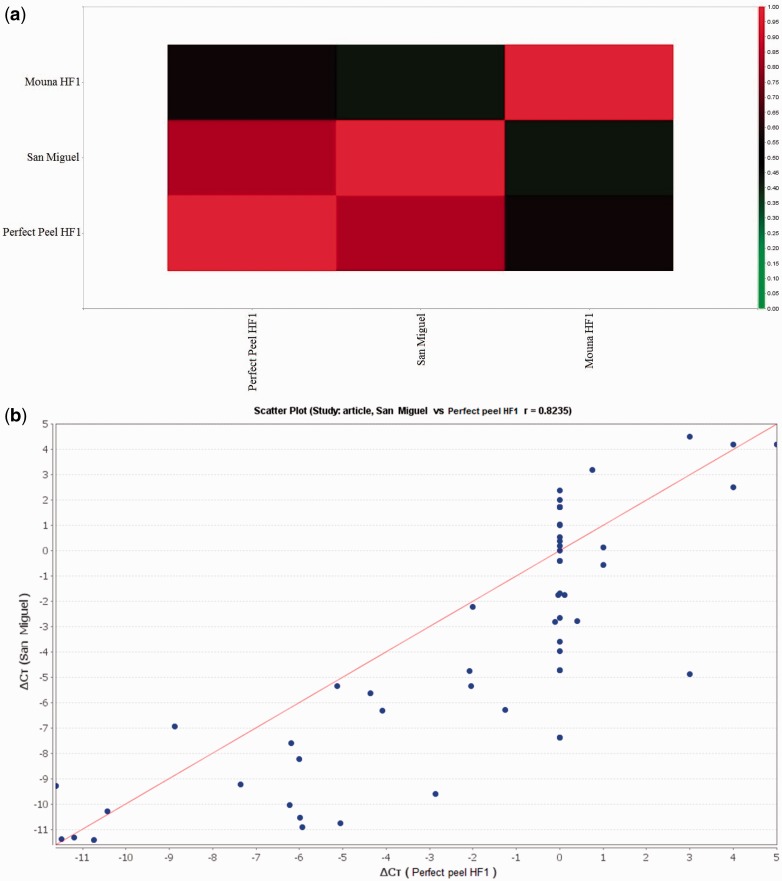
Heat maps of transcript expression were constructed [see Supporting Information—Figure S5] and genotype correlation analyses conducted. The correlation signal showed that the gene expression profile of San Miguel was very similar to that of Perfect peel HF1. In contrast, the expression profiles of San Miguel and Perfect peel HF1 were quite distinct from that of Mouna HF1 (Fig. 6A). In order to compare mRNA expression profiles in leaf and root tissues of the examined genotypes, correlation coefficients were calculated and showed in scatter plots. Analysis revealed a high-correlation coefficient (r = 0.82) between San Miguel and Perfect peel HF1, indicating that these two varieties are highly correlated with regard to the selected genes (Fig. 6B). By contrast, comparison of gene expression profiles pointed to a low correlation between San Miguel vs. Mouna HF1 and Perfect peel HF1 vs. Mouna HF1 (r = 0.42 and r = 0.56, respectively; Fig. 6C and D).
Figure 6.
(A) Signal correlation of San Miguel F1, Perfect peel HF1 and Mouna HF1 genotypes. The colour scale represents relative expression levels with red as increased transcript abundance and green as decreased transcript abundance. (B), (C) and (D) represent the scatter plot for global expression between San Miguel, Perfect Peel HF1and Mouna HF1, repectively. The Pearson correlation coefficient “r” is shown.
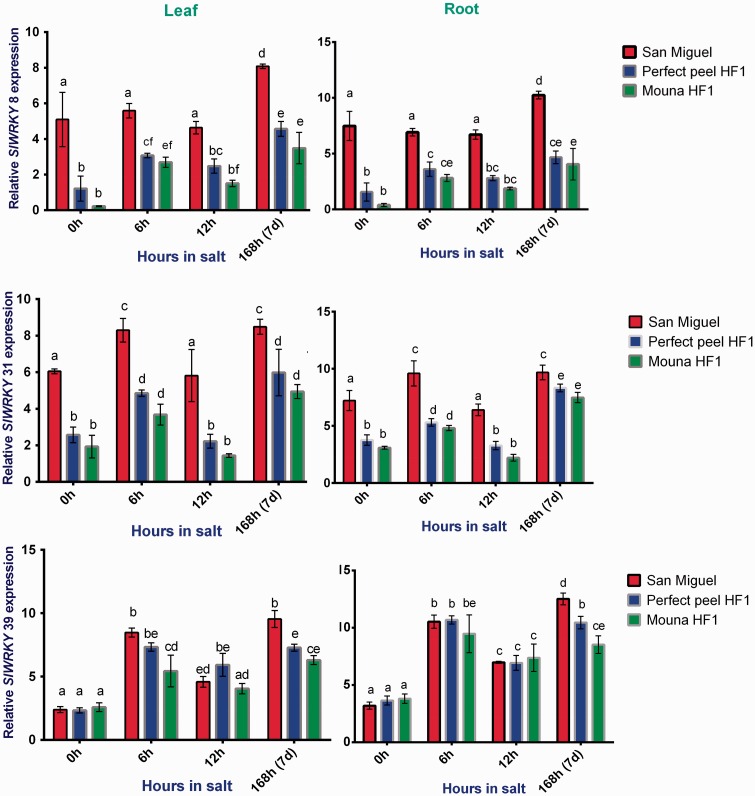
WRKY genes
In response to salt stress, WRKY genes showed up-regulated expression and displayed distinct patterns. Two-way ANOVA followed by Tukey’s multiple comparisons test indicated that relative SIWRKY8 and 31 expression is statistically significant between genotypes (leaves: F2, 4 = 316.5, P < 0.0001; roots: F2, 4 = 195.7, P= 0.0001) and (leaves: F2, 4 = 228.4, P < 0.0001; roots: F 2, 4 = 399.7, P < 0.0001), respectively. SIWRKY39 expression was significantly different in leaves (F 2, 4 = 44.80, P = 0.0018) but not in roots (F2, 4 = 6.077, P = 0.0613). Expression of SIWRKY8, 31 and 39 varied significantly between stages of salt stress (leaves: F3, 6 = 53.74, P < 0.0001; roots: F3, 6 = 69.92, P< 0.0001), (leaves: F3, 6 = 105.2, P < 0.0001; roots: F3, 6 = 245.8, P < 0.0001) and (leaves: F3, 6 = 198.0, P < 0.0001; roots: F3, 6 = 223.3, P < 0.0001). Results also indicated that interaction between genotypes and stages of treatment are not statistically different for SIWRKY8 (leaves: F6, 12 = 2.351, P = 0.0978; roots: F6,12 = 2.821, P = 0.0596) and for SIWRKY31 in leaves (F6, 12 = 0.6028, P = 0.7238). By cons, such interaction is supported statistically for SIWRKY31 in roots (F6, 12 = 4.068, P = 0.0185) and for SIWRKY39 in both tissues (leaves: F6, 12 = 7.349, P = 0.0018; roots: F6, 12 = 6.680, P = 0.0027). Within San Miguel, transcription of SlWRKY8 (Group II-d) was enhanced with saline treatment, particularly at 7 days whereas that of SlWRKY31 (Group I) showed similar expression pattern in both leaves and roots SlWRKY39 gene was expressed similarly between San Miguel and Perfect Peel HF1 genotypes during first stage of the treatment but showed increased expression relative to the tolerant one at the end of the treatment (Fig. 7).
Figure 7.
Relative gene expression of tomato WRKY8, WRKY31 and WRKY39 in response to salt stress in leaves and roots of San Miguel, Perfect peel HF1 and Mouna HF1 genotypes. Total RNA was purified from tissues of tomato plants treated with 150 mM NaCl for 0 h, 6 h, 12 h and 7 days. Transcript level was analyzed by qRT-PCR using primers indicated in Table 1. Tomato Actin gene was used as reference gene. Error bars show the standard error between three replicates performed. Bars with different letters within each panel are significantly different at P > 0.05 according to Tukey's test.
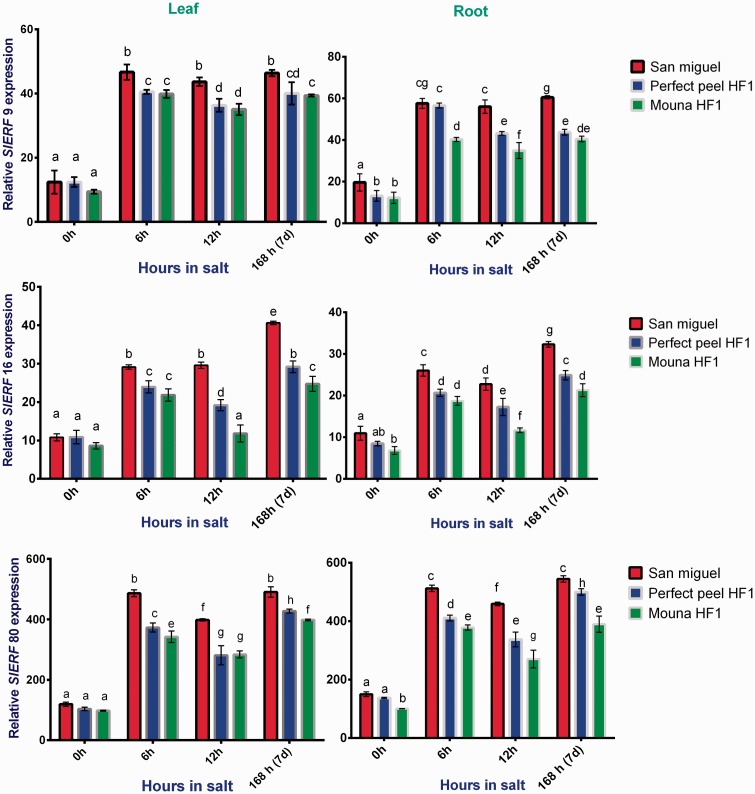
ERF genes
Under salt stress, gene expression of SIERF family genes belonging to Group VI (SIERF9 and 16) exhibited the same pattern, increasing significantly in both leaves and roots and in all three genotypes. Two-way ANOVA followed by Tukey’s multiple comparisons test indicated that the relative SIERF9, 16 and 80 expression varied significantly among genotypes (leaves: F2, 4 = 17.93, P = 0.0101; roots: F2, 4 = 120.6, P = 0.0003); (leaves: F2, 4 = 310.6, P < 0.0001; roots: F2, 4 = 116.0, P = 0.0003) and (leaves: F2, 4 = 88.56, P = 0.0005; roots: F2, 4 = 241.3, P < 0.0001), respectively. Expression also changed significantly between stages of salt stress for SIERF9, 16 and 80 (leaves: F3, 6 = 744.2, P < 0.0001; roots: F3, 6 = 487.3, P < 0.0001), (leaves: F3, 6 = 300.8, P < 0.0001; roots: F3, 6 = 339.1, P < 0.0001) and (leaves: F3, 6 = 1335, P < 0.0001; roots: F3, 6 = 492.2, P < 0.0001), respectively. In addition, we observed significant genotypes × stages interaction terms (leaves: F6, 12= 3.388, P = 0.0342; roots: F6, 12 = 21.27, P < 0.0001), (leaves: F6, 12 = 23.22, P < 0.0001; roots: F6, 12 = 7.022, P = 0.0022) and (leaves: F6, 12= 19.26, P < 0.0001; roots: F6, 12 = 18.39, P < 0.0001) for SIERF9,16 and 80, respectively. Expression was high overall in the tolerant genotype compared with the other two genotypes. Transcripts of SIERF80 accumulated in San Miguel at the first stage of treatment (6 h) to the same degree as in the latest stage (7 days) regardless of the tissue. These transcripts also increased significantly with Perfect Peel HF1 and Mouna HF1 genotypes (Fig. 8).
Figure 8.
Relative gene expression of tomato ERF9, ERF16 and ERF80 in response to salt stress in leaves and roots San Miguel, Perfect peel HF1 and Mouna HF1genotypes. Total RNA was purified from tissues of tomato plants treated with 150 mM NaCl for 0 h, 6 h, 12 h and 7 days. Transcript level was analyzed by qRT-PCR using primers indicated in Table 1. Tomato Actin gene was used as reference gene. Error bars show the standard error between three replicates performed. Bars with different letters within each panel are significantly different at P > 0.05 according to Tukey's test.
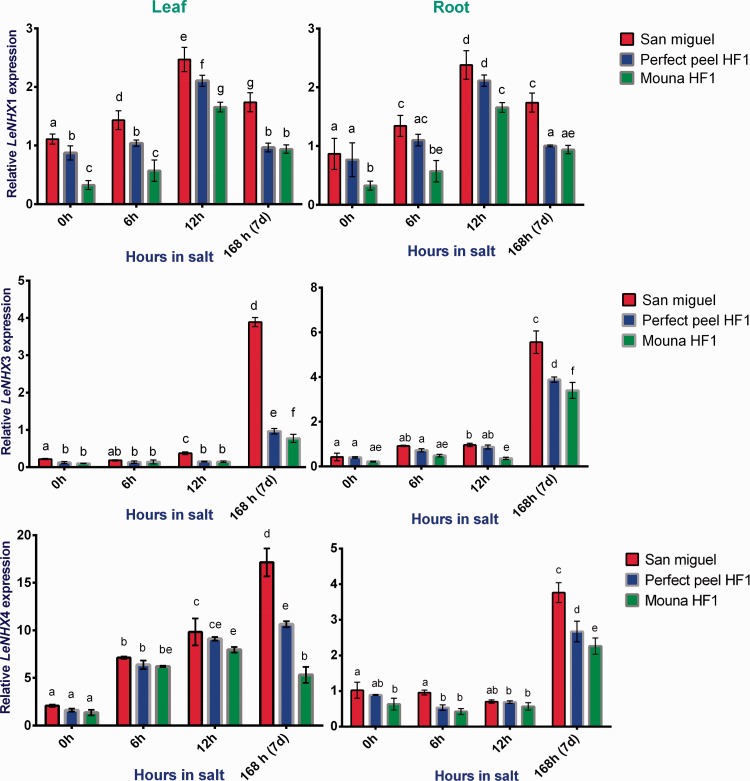
LeNHX genes
Results from two-way ANOVA indicated that stages of treatment had a significant overall effect on LeNHX1,3 and 4 transcripts abundance (leaves: F3, 6 = 667.5, P < 0.0001; roots: F3, 6 = 106.8, P < 0.0001), (leaves: F3, 6 = 779.8, P < 0.0001; roots: F3, 6 = 1347, P < 0.0001) and (leaves: F3, 6 = 308.2, P < 0.0001; roots: F3, 6 = 613.6, P < 0.0001). Similarly results supported significant change in relative LeNHX1, 3 and 4 expression among genotypes (leaves: F2, 4 = 63.11, P = 0.0009; roots: F2, 4 = 236.3, P < 0.0001), (leaves: F2, 4 = 1542, P < 0.0001; Roots: F2, 4 = 74.41, P = 0.0007) and (leaves: F2, 4 = 232.3, P < 0.0001; roots: F2, 4 = 34.91, P = 0.0029), respectively. We detected significant genotypes × stages interaction terms (leaves: F6,12 = 3092, P < 0.0001; roots: F6, 12 = 15.02, P < 0.0001) and (leaves: F6, 12 = 38.01, P < 0.0001; roots: F6, 12 = 12.95, P = 0.0001) for LeNHX3 and 4, respectively, as well as for LeNHX1 expression in leaves (F6, 12 = 5.219, P = 0.0074) but not in roots (F6, 12 = 2.090, P = 0.1306). In spite of a difference in the expression pattern of LeNHX1, 3 and 4, the tolerant San Miguel genotype exhibited the highest expression of all isoforms at the end of the saline stress. LeNHX1 showed an enhanced expression in both leaves and roots of stressed genotypes at 12 h post-salt treatment. LeNHX3 transcripts were negligible during the first stage of salt treatment, whereas they increase significantly at the end. Salinity significantly enhanced LeNHX4 expression in leaves and roots in all genotypes at 7 days of the stress imposition being highest in the tolerant genotype (Fig. 9).
Figure 9.
Relative gene expression of tomato LeNHX1, LeNHX3 and LeNHX4 in response to salt stress in in leaves and roots of San Miguel, Perfect peel HF1 and Mouna HF1 genotypes. Total RNA was purified from tissues of tomato plants treated with 150 mM NaCl for 0 h, 6 h, 12 h and 7 days. Transcript level was analyzed by qRT-PCR using primers indicated in Table 1. Tomato Actin gene was used as reference gene. Error bars show the standard error between three replicates performed. Bars with different letters within each panel are significantly different at P > 0.05 according to Tukey's test.
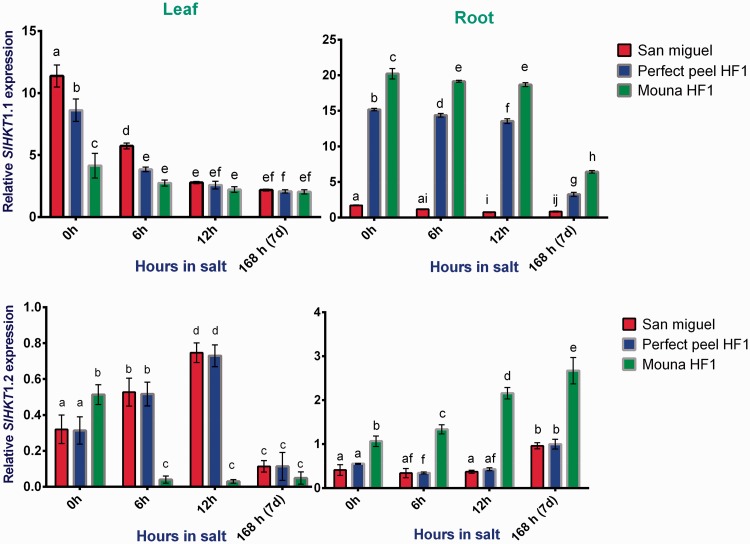
HKT Class I genes
Two way ANOVA followed by Tukey’s multiple comparisons test indicated that the relative SIHKT expression varied significantly between genotypes (leaves: F2, 4 = 124.1, P = 0.0003; roots: F2, 4 = 26 489, P < 0.0001) and (leaves: F2, 4 = 82.86, P = 0.0006; roots: F2, 4 = 221.4, P < 0.0001) and between stages of salt stress (leaves: F3, 6 = 343.7, P < 0.0001; roots: F3, 6 = 3520, P < 0.0001) and (leaves: F3, 6 = 55.95, P < 0.0001; roots: F3, 6 = 66.26, P < 0.0001) for SIHKT1;1 and 1;2, respectively. In addition, we observed significant interaction terms for these variables (leaves: F6, 12 = 29.57, P < 0.0001; roots: F6, 12 = 346.6, P < 0.0001) and (leaves: F6, 12 = 52.65, P < 0.0001; roots: F6, 12 = 60.86, P < 0.0001) for SIHKT1;1 and 1;2, respectively. HKT1;1 was gradually down regulated in leaves during all stages of the salt stress treatment. Transcript level was negligible in roots of San Miguel whereas it seems significantly higher within Mouna HF1. Similarly, high salinity increased the level of HKT1;2 transcripts in the roots of the sensitive genotype while remaining reduced during all the stress period within the tolerant and the mildly tolerant genotypes. In leaves, salinity clearly decreases HKT1;2 expression at 7 days with all tested genotypes (Fig. 10).
Figure 10.
Relative gene expression of tomato HKT1;1 and HKT1;2 in response to salt stress in leaves and roots San Miguel, Perfect peel HF1 and Mouna HF1 genotypes. Total RNA was purified from tissues of tomato plants treated with 150 mM NaCl for 0 h, 6 h, 12 h and 7 days. Transcript level was analyzed by qRT-PCR using primers indicated in Table 1. Tomato Actin gene was used as reference gene. Error bars show the standard error between three replicates performed. Bars with different letters within each panel are significantly different at P > 0.05 according to Tukey's test.
Discussion
Tomato plants, as sessile organisms, have evolved mechanisms that allow them to monitor their changing environment, as well as systems and strategies to react and adapt to these changes. Differences in sensitivity towards salt stress led to the classification of 15 local genotypes out of 20 as sensitive (Classes 3, 4 and 5). Therefore, salt stress seems to negatively affect tomato growth and would be expected to cause significant crop yield losses. The effects of such a stress may become obvious over weeks. In Tunisia, 25 % of the total of lands are salt-affected (FAO 2006). Tomato reaction to salt stress is carried through activating a stress response signal transduction network comprising physiological, biochemical, molecular and genetic changes.
Na+, K+, Ca2+ and proline contents correlate with scale classes
Under salt stress, genotypes belonging to tolerant and mildly tolerant scale classes (1 and 3, respectively) displayed the lowest Na+ accumulation in leaves when compared to the sensitive class (5). San Miguel, the tolerant genotype, accumulated Na+ preferentially in roots while the sensitive Mouna HF1 accumulated more Na+ in leaves at the longest time post stress. This is likely due to the superior Na+ exclusion mechanism of genotypes clustered in either scale 1 or 3 classes (Dasgan et al. 2002). This finding is comparable with the way that potato cultivars respond to NaCl transport, since sensitive cultivars transport relatively more Na+ to leaves (Jaarsma et al. 2013). When exposed to saline conditions, plants show reduced uptake and lesser tissue retention of K+ (Munns et al. 2002; Chakraborty et al. 2012). Tolerant genotypes are able to maintain K+ homeostasis during all the salt stress stages (Shabala and Cuin 2008; Hauser and Horie 2010). Therefore, K+ is considered as a key regulatory element in plant metabolic process by promoting Na+ exclusion and osmotic adjustment (Chakraborty et al. 2016). In our experiments, leaves of tolerant and sensitive tomato genotypes behaved similarly, at the later stage of salt treatment, although the tolerant genotype accumulated significantly more K+ in its roots. Maintaining a low Na+/K+ ratio in tissues is critical for plant growth and metabolism under salty conditions (Wang et al. 2015). We found that leaf Na+/K+ ratios were correlated with salinity scale classes recorded during the first stage of salt stress. Indeed, tomato genotypes belonging to scale Classes 1 and 3 displayed higher Na+/K+ ratios than scale-Class 5 genotype at 12 h post-salt stress whereas this ratio did not fluctuate between genotypes either in leaves or in roots at the late stage of the stress. In roots, despite a peak recorded at 12 h post salt treatment for the mildly tolerant genotype, all genotypes showed slight variation for Na+/K+ ratios regardless of their scale class. Dasgan et al. (2002) reported that tomato genotypes with lower Na+/K+ ratios indicated lower scale classes with less salt damage. The long-term salt tolerance in tomato plants seems to be related to a lower leaf Na+ accumulation by reducing Na+ transport from root to shoot and a concomitant uniform cyotosolic K+ concentration maintaining thus leaf Na+/K+ homeostasis over time (García-Abellan et al. 2014; Wu et al. 2014). A comparison between wild type and mutant tomato plants showed the ability of the more tolerant salt plants to maintain their K+ content under moderate salt stress in roots. K+ level significantly declined leading to high Na+/K+ ratio in mutant tomato plants. Thus, the mutants were more sensitive to salt stress than the wild type. These changes can be attributed to a stronger ionic stress due to K+ loss from the root tissues (Poóra et al. 2015). Villalta et al. (2008) suggested that several genes located in chromosome 7 of tolerant tomato plants are responsible for governing the active mechanism of Na+/K + regulation.
When submitted to salt stress, another striking difference was observed between tomato genotypes. Tolerant scale Class 1 genotype exhibited an enrichment Ca2+ in leaf tissues. In addition to its effect on preventing Na+ entry into cells, Ca2+ is the most important universal signal carrier used by plants to convey information in many different cellular processes. Ca2+ seems to be necessary for maintenance of an appropriate K+ concentration in tissues (Subbarao et al. 1990). In addition, high Ca2+ has a beneficial effect by contributing to the maintenance of K+ uptake enhancing salt tolerant in tomato plants (Bacha et al. 2015).
The accumulation of ions requires the accumulation of solutes in the cytosol playing a role in both osmoprotection and osmotic adjustment under abiotic stress (Hasegawa et al. 2000; Flowers and Colmer 2008; Munns and Tester 2008). This accumulation of osmolytes, especially that of proline, is a common phenomenon in plants. Besides its role as an osmolyte, proline contributes to scavenging ROS, stabilizing subcellular structures, modulating cell redox homeostasis, supplying energy and functioning as a signal (Kavi-Kishor et al. 2005; Verbruggen and Hermans 2008; Szabados and Savouré 2010; Sharma et al. 2011). Although proline accumulation is a common response to salt stress in tomato, the extent of its accumulation varies between tolerant and sensitive genotypes. Indeed, our findings revealed that proline accumulation increases greatly within the tolerant genotype, mainly in leaves and when compared to the most sensitive genotype. Proline is accumulated preferentially in leaves in order to maintain chlorophyll level and cell turgor to protect photosynthetic activity under salt stress (Silva-Ortega et al. 2008). Proline has also a potential role in scavenging ROS products (Soshinkova et al. 2013).The accumulation of proline in plants under stress is caused either by the induction of expression of proline biosynthesis genes (P5CS, P5CR) or by the repression of the genes of its degradation pathway (PDH silencing) (Marco et al. 2015).
Salt stress-induced up-regulation of antioxidant enzymes
The increases in CAT, APX and GPOX activities are an adaptive trait to overcome salt damage by reducing toxic levels of H2O2 and provide protection against oxidative stress (Sudhakar et al. 2001; Bor et al. 2003; Mittova et al. 2003; Chawla et al. 2013). In our study, salt stress modulates the responses of antioxidative enzymes in both leaves and roots according to the tested genotype and the period of stress imposition. Oxidative stress defence occurs in the tolerant San Miguel genotype through an increase in APX and GPOX activities especially in roots and leaves. CAT, APX and GPOX have been reported as antioxidant enzymes in different plant tissues (Chawla et al. 2013). CAT is often related to an enhanced tolerance to salt stress (Mittova et al. 2004; Gao et al. 2008). Similarly, APX activity under salinity stress increases (Gossett et al. 1994; Hernàndez et al. 2000; Lee et al. 2001; Mittova et al. 2004). Within root organelles of salt-tolerant genotypes of tomato, the increase in APX activity was higher than that of SOD under salt stress. This finding indicates that under salinity, the rate of H2O2 detoxification is higher than that of its production leading to alleviation of oxidative stress. Similarly, decreased H2O2 and lipid peroxidation levels were found in peroxisomes of salt-treated tolerant plants. These responses to salinity were the result of differentially increased activities of APX and CAT over that of SOD (Mittova et al. 2004). An improved stress tolerance has been observed in several transgenic plants over-expressing antioxidant enzymes such as GPOX and APX (John et al. 2010). Kim et al. (2014) reported a positive response of GPOX to environmental stimuli such as salt stress enhancing tolerance of Panax ginseng plants against abiotic stresses. APX activity was clearly enhanced in the salt-tolerant L. pennellii (Mittova et al. 2015). Overall, our findings demonstrate that inherent activities of the isozymes are present in tomato leaf and root but are expressed differentially between genotypes. These variations reflect differences in both tissues- and species-dependent expression of these isozymes (Mittova et al. 2015).
Expression analysis of transporters (HKT Class 1 and LeNHX)
Many plants have developed an efficient method to keep Na+ concentration in the cytoplasm at a low level (Gupta and Huang 2014). The data in this work showed that leaf expression of HKT1.2 was high and similar between San Miguel and Perfect peel HF1 during the first stage of salt treatment before reducing drastically and similarly in both genotypes at the later stage. At the same time, leaf expression of HKT1;1 decreased gradually in leaves regardless of the tested genotype. Overall, root expression of HKT1;1 and HKT1;2 was significantly reduced in the tolerant genotype compared with the most sensitive one. The differences observed in the expression levels of HKT1 genes in tomato genotypes are probably linked to the contribution of each allele to Na+ movement and tissue content. These findings are in accord with those published by Almeida et al. (2013) since they found a positive relation between HKT1;1 and HKT1;2 expression and Na+ content in leaves but not in roots. Interestingly, in Arabidopsis, the salt tolerance of ecotypes adapted to coastal and saline soils is associated with high leaf Na+ concentration due to a weak expression of the AtHKT1;1 allele in roots (Rus et al. 2006; Baxter et al. 2010).
In addition to HKT, LeNHX cation/H+ antiporters contribute to the sequestration of Na+ in vacuoles, the regulation of the homeostasis of K+ and endosomal pH regulation under normal and saline conditions (Barragán et al. 2012; Bassil et al. 2012; Leidi et al. 2010). We involved in our study three different isoforms corresponding to LeNHX1, LeNHX3 and LeNHX4. LeNHX1 is a tonoplast localized protein mediating K+ uptake at the tonoplast, for turgor regulation and stomatal function (Barragán et al. 2012). LeNHX3 and LeNHX4 are involved in Na+, K+ and H+ homeostasis (Gàlvez et al. 2012). LeNHX3 seems also to be linked to a QTL for Na+ leaf concentration (Villalta et al. 2007; 2008). In our study, these three isoforms were differentially expressed allowing the discrimination between tolerant and sensitive genotypes. LeNHX1 showed a low expression in normal conditions and rapidly increases during the early stage of stress imposition, then it decreased at the latest stage (at 7 days). Expression of LeNHX1 seems to be correlated with low accumulation of Na+ in leaves and high accumulation in roots of the tolerant genotype. LeNHX3 and LeNHX4 were highly expressed within San Miguel, especially during the latest period of the treatment. This pattern is associated with a reduced Na+ content in leaf tissue and a high accumulation in roots in line with previous studies (Venema et al. 2003; Almeida et al. 2014). Either in the absence of stress or during the early stage of salt stress, LeNHX3 expression remained drastically reduced in both leaf and root tissues. This expression increased later, especially in the tolerant genotype. This may be due to enhanced cellular Na+ concentration as described by Gàlvez et al. (2012). Apart from LeNHX3, LeNHX4 showed also basal expression level in leaves in the absence of salt stress that increased 7 days post-treatment particularly in the tolerant genotype. This agreed with other-published data showing that LeNHX4 and closely related isoforms in Arabidopsis are rapidly induced by salt stress in roots and especially in leaves (Gàlvez et al. 2012; Pardo et al. 2006).
Expression analysis of WRKY and ERF genes
In response to salinity stress, a large number of salt-responsive transcription factors and genes, being either upregulated or downregulated, have been identified and characterized using transcriptomic and genomic approaches (Bakshi and Oelmüller 2014). In this present work, transcription of SIWRKY8, SIWRKY31 and SIWRKY39 was induced with a similar a pattern of expression between leaf and root tissues. SIWRKY8 and SIWRKY31 showed abundant transcripts accumulation particularly within the tolerant genotype at all times of the treatment and even in the absence of the salt stress. In contrast, SIWRKY39 was highly and similarly expressed within sensitive and tolerant genotypes at the beginning of the stress (12 h) then showing a significant increase in the tolerant genotype (7 days). These transcription factors were reported to be up-regulated by salt stress (Huang et al. 2012). Thus, 81 WRKY genes were reported to display constitutive or induced expression patterns which are tomato tissue-specific. The majority of the WRKY gene family as well as their orthologs in Arabidopsis showed up-regulation under stress (Jiang and Deyholos 2009; Liu et al. 2011).
Besides members of WRKY gene family, ERF transcription factors are the most important regulators modulating gene expressions (Sharma et al. 2010). Our study indicated that rapid and high expression of SIERF9, 16 and 80 is closely connected to salt tolerance during all the stages of the salt stress treatment. ln line with such findings, Sharma et al. (2010) showed that SIERF80 was 400-foldes up-regulated during salt stress. They also provided evidence that over expressing of SlERF5 in transgenic tomato plants leads to an increased resistance to salt and drought stress.
Conclusion
Salinity is one of the major abiotic stresses world-wide, particularly in Tunisia due to the soil salinization and the poor quality of water irrigation. Salinity severely limits yields, threatening land productivity in arid and semi-arid areas leading to food imbalance of these regions. The ability to face abiotic challenges involves a complex of responses at the whole plant level. Responses are themselves part of effective ways to improve and protect tomato crops from the adverse effects of soil salinization. This is the first study investigating phenotypical, physiological and molecular responses of Tunisian tomato genotypes to salt stress. Associations were pointed out between exhibited phenotypes, ion and proline accumulation, APX, CAT and GPX activities and gene expression. Salt tolerance seems to be related to a lower leaf accumulation in the long term by reducing Na+ transport from root to leaves. Besides, accumulation of proline was found to be linked to tolerance being much higher within tolerant genotype. As production of ROS generated by salt stress is always enhanced, APX, Cat and GPOX activities were stimulated mainly in the tolerant genotype at the later stage of treatment. The described expression pattern of allelic genes belonging to WRKY (8, 31 and 39), ERF (9, 16 and 80), LeNHX (1, 3 and 4) and HKT (Class 1) families support the view that they are involved in mechanisms associated with a response to salt stress and can be considered as markers to be used in discriminating tomato genotypes. Data generated from this study will be helpful in selecting candidate genotypes to be used by growers in coastal areas or as progenitors in breeding programmes.
Sources of Funding
This work was partially supported by the Ministry of Higher Education and Scientific Research of Tunisia and carried out within the USAID-MERC Project TA-MOU-08-M28-048.
Contributions by the Authors
C.G., H.F. and F.G. conceived and designed the experiments. C.G. performed the experiments. C.G., H.F. and F.G. analysed the data. F.G. wrote the paper with assistance from C.G and D.G.
Conflict of Interest Statement
None declared.
Supplementary Material
Acknowledgements
We thank the Tunisian Ministry of Agriculture for support, Pr M BOUSSAID (INSAT institution, Carthage University), Pr A CHAOUI (Faculty of Sciences of Bizerte, Carthage University), Pr H Ben HMAD and Pr M DACHRAOUI (Faculty of Sciences of Tunis, El Manar University) from Tunisia for assistance. We also thank Pr G ANFOKA (Faculty of Agricultural Technology Al-Balqa, Applied University, Jordan) for constant support.
Supporting Information
The following additional information is available in the online version of this article —
Figure S1. Seedlings grown in Hoagland solution to half-strength in jars with continuous aeration. Plants with four fully developed true leaves were transferred into plastic pots containing a mixture of peat and sand, then irrigated with one-half Hoagland solution added with 150 mM NaCl (15 dS/m, pH 7.5).
Figure S2. The salinity scale classes used in the experiment according to Dasgan et al. (2002). 1/normal green plants with or without slight inward curly leaves; 2/green plants with complete inward curly leaves; 3/addition to complete curly leaves, dry leaves from moderate to severe damages; 4/most leaves with drying damages; 5/all leaves of the plant with drying damages.
Figure S3. Data relative to dry weight of leaves and roots. Data, expressed as g DW per plant, are means SD of three independent experiments, using 10 plants per genotype and per treatment. Bars with different letters within each panel are significantly different at P > 0.05 according to Tukey's test.
Figure S4. Stability of tomato Actin gene expression (expressed as cycle threshold- (Cts-)) in response to salt stress treatment in San Miguel, Perfect peel HF1 and Mouna HF1 genotypes. Total RNA was purified from leaf and root of tomato plants treated with 150 mM NaCl for 0 h, 6 h, 12 h and 7 days. Transcript level was analyzed by qRT-PCR using primers indicated in Table 1. The data reported are the mean ± SE of three values.
Figure S5. Expression profiles of candidate genes. The results of the relative expression levels (Δct) of candidate genes under salt stress treatments (6 h, 12 h and 7 days) compared to the controls were used for hierarchical cluster analysis with Data Assist v3.01. The colour scale represents relative expression levels, with red as increased transcript abundance and green as decreased transcript abundance.
Literature Cited
- Aebi H. 1984. Catalase in vitro. Methods of Enzymology 105:121–126. [DOI] [PubMed] [Google Scholar]
- Agarwal PK, Agarwa lP, Reddy MK, Sopory S. 2006. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Reporter 25:1263–1274. [DOI] [PubMed] [Google Scholar]
- Almeida P, de Boer GJ, de Boer AH. 2014. Differences in shoot Na+ accumulation between two tomato species are due to differences in ion affinity of HKT1;2. Journal of Plant Physiology 171:438–447. [DOI] [PubMed] [Google Scholar]
- Almeida P, Katsching D, de Boer AH. 2013. HKT transporters – state of the art. International Journal of Molecular Sciences 14:20359–20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asins MJ, Villalta I, Aly MM, Olías R, Alvarez DE, Morales P, Huertas R, Li J, Jaime-Pérez N, Haro R, Raga V, Carbonell EA, Belver A. 2012. Two closely linked tomato HKT coding genes are positional candidates for the major tomato QTL involved in Na+/K+ homeostasis. Plant Cell Environment 36:1171–1191. [DOI] [PubMed] [Google Scholar]
- Bacha H, Mansour E, Guasmi F, Triki T, Ferchichi A. 2015. Proline, glycine bétaïne et composition minérale des plantes de Solanum lycopersicum L. (var. Microtom) sous stress salin. Journal of New Sciences 22:1007–1013. [Google Scholar]
- Baniwal SK, Chan KY, Scharf KD, Nover L. 2007. Role of heat stress transcription factor HsfA5 as specific repressor of HsfA4. Journal of Biological Chemistry 282:3605–3613. [DOI] [PubMed] [Google Scholar]
- Bakshi M, Oelmüller R. 2014. WRKY transcription factors: Jack of many trades in plants. Plant Signaling and Behavior 9:e27700.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barragán V, Leidi EO, Andrés Z, Rubio L, De Luca A, Fernández JA, Cubero B, Pardo JM. 2012. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell Online 24:1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassil E, Coku A, Blumwald E. 2012. Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. Journal of Experimental Botany 63:5727–5740. [DOI] [PubMed] [Google Scholar]
- Bates L, Waldren RP, Teare ID. 1973. Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207. [Google Scholar]
- Baxter I, Brazelton JN, Yu D, Huang YS, Lahner B, Yakubova E, Li Y, Bergelson J, Borevitz JO, Nordborg M, Vitek O, Salt DE. 2010. A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1. PLoS Genetic 6:e1001193.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert HJ, Ayoubi P, Borchert C, Bressan RA, Burnap RL, Cushman JC, Cushman MA, Deyholos M, Fischer R, Galbaith DW, Hasegawa PM, Jenks M, Kawasaki S, Koiwa H, Kore-eda S, Lee BH, Michalowski CB, Misawa E, Nomura M, Ozturk N, Postier B, Prade R, Song CP, Tanaka Y, Wang H, Zhu JK. 2001. A genomics approach towards salt stress tolerance. Plant Physiology and Biochemistry 39:295–311. [Google Scholar]
- Bor M, Ozdemir F, Turkan I. 2003. The effect of salt stress on lipid peroxidation and antioxidants in leaves of sugar beet Beta vulgaris L. and wild beet Beta maritima L. Plant Science 164:77–84. [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantification of icrogram quantities of protein utilizing the principal of protein-dye binding. Analytical Biochemstry 72:248–254. [DOI] [PubMed] [Google Scholar]
- Chakraborty K, Bhaduri D, Meena HN, Kalariya K. 2016. External potassium (K+) application improves salinity tolerance by promoting Na+-exclusion, K+- accumulation and osmotic adjustment in contrasting peanut cultivars. Plant Physiology and Biochemistry 103:143–153. [DOI] [PubMed] [Google Scholar]
- Chakraborty K, Sairam RK, Bhattacharya RC. 2012. Differential expression of salt overly sensitive pathway genes determines salinity stress tolerance in Brassica genotypes. Plant Physiology and Biochemistry 51:90–101. [DOI] [PubMed] [Google Scholar]
- Chawla S, Jain S, Jain V. 2013. Salinity induced oxidative stress and antioxidant system in salt-tolerant and salt-sensitive cultivars of rice (Oryzasativa L.). Journal of Plant Biochemistry and Biotechnology 22:27–34. [Google Scholar]
- Chen L, Song Y, Li S, Zhang L, Zou C, Yu D. 2012. The role of WRKY transcription factors in plant abiotic stresses. Biochimica et Biophysica Acta 1819:120–128. [DOI] [PubMed] [Google Scholar]
- Dasgan HY, Aktas H, Abak K, Cakmak I. 2002. Determination of screening techniques to salinity tolerance in tomatoes and investigation of genotype responses. Plant Science 163:695–703. [Google Scholar]
- Davenport RJ, Munoz‐Mayor A, Jha D, Essah PA, Rus A, Tester M. 2007. The Na+ transporter AtHKT1; 1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell and Environment 30:497–507. [DOI] [PubMed] [Google Scholar]
- Epstein E. 1972. Mineral nutrition of plants: principles and perspectives ed. London: John Wiley and Sons, 412. [Google Scholar]
- Eulgem T, Somssich IE. 2007. Networks of WRKY transcription factors in defense signaling. Current Opinion in Plant Biology 10:366–371. [DOI] [PubMed] [Google Scholar]
- FAO. 2006. Conférence électronique sur la salinisation: Extension de la salinisation et stratégie de prévention et réhabilitation. www.dgroups.org/groups/fao/salinization.
- FAO. 2015. Technical issues of salt-affected soils.
- Fielding JL, Hall JL. 1978. A biochemical and cytochemical study of peroxidase activity in roots of Pisum sativum I. A comparison of DAB-peroxidase and guaiacol-peroxidase with particular emphasis on the properties of cell wall activity. Journal of Experimental Botany 29:969–981. [Google Scholar]
- Fowler S, Thomashow MF. 2002. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers TJ, Colmer TD. 2008. Salinity tolerance in halophytes. New Phytologist 179:945–963. [DOI] [PubMed] [Google Scholar]
- Foolad MR. 2007. Genome mapping and molecular breeding of tomato. International Journal of Plant Genomics ID 643581–52. http://dx.doi.org/10.1155/2007/64358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. 2011. Ascorbate and glutathione: the heart of the redox hub. Plant Physiology 155:2–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gàlvez FJ, Baghour M, Hao G, Cagnac O, Rodriguez-Rosales MP, Venema K. 2012. Expression of LeNHX isoforms in response to salt stress in salt sensitive and salt tolerant tomato species. Plant Physiology and Biochemistry 51:109–115. [DOI] [PubMed] [Google Scholar]
- Gao S, Ouyang C, Wang S, Xu Y, Tang L, Chen F. 2008. Effects of salt stress on growth, antioxidant enzyme and phenyalanine ammonia-lyase activities in Jatropha curcas L seedlings. Plant Soil Environment 54:374–381. [Google Scholar]
- García-Abellan JO, Egea I, Pineda B, Sanchez-Bel P, Belver A, Garcia-Sogo B, Flores FB, Atares A, Moreno V, Bola MC. 2014. Heterologous expression of the yeast HAL5 gene in tomato enhances salt tolerance by reducing shoot Na+ accumulation in the long term. Physiologia Plantarum 152:700–713. [DOI] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. 2010. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiology and Biochemistry 48:909–930. [DOI] [PubMed] [Google Scholar]
- Gossett DR, Millhollon EP, Lucas MC. 1994. Antioxidant response to NaCl stress in salt-tolerant and salt-sensitive cultivars of cotton. Crop Sciences 34:706–714. [Google Scholar]
- Gupta B, Huang B. 2014. Mechanism of salinity tolerance in plants: physiological, biochemical and molecular characterization. International Journal of Genomics ID 7015961–7015918. http://dx.doi.org/10.1155/2014/701596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. 2000. Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology Plant Molecular Biology 51:463–499. [DOI] [PubMed] [Google Scholar]
- Hauser F, Horie T. 2010. A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environment 33:552–565. [DOI] [PubMed] [Google Scholar]
- Hernàndez JA, Jiménez A, Mullineaux PM, Sevilla F. 2000. Tolerance of pea (Pisum sativum L.) to long-term salt stress is associated with induction of antioxidant defenses. Plant Cell Environment 23:853–862. [Google Scholar]
- Huang S, Gao Y, Liu J, Peng X, Niu X, Fei Z, Cao S, Liu Y. 2012. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Molecular Genetics and Genomics 287:495–513. [DOI] [PubMed] [Google Scholar]
- Jaarsma R, deVries RSM, de Boer AH. 2013. Effect of salt stress on growth, Na+ accumulation and proline metabolism in potato (Solanum tuberosum) cultivars. PloS One 8:e60183.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Deyholos MK. 2009. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Molecular Biology 69:91–105. [DOI] [PubMed] [Google Scholar]
- John R, Pandey R, Sopory SK, Rajam MV. 2010. Engineering antioxidant enzymes for abiotic stress tolerance in plants. Journal of Plant Biology 37:1–18. [Google Scholar]
- Karan R, Subudhi PK. 2012. A stress inducible SUMO conjugating enzyme gene of a grass halophyte Spartina alterniflora (SaSce9) enhances salinity and drought stress tolerance in Arabidopsis. BMC Plant Biology 12:187.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavi-Kishor PB, Sangam S, Amrutha RN, Sri Laxmi P, Naidu KR, RaoK RSS, Sreenath R, Reddy KJ, Theriappan P, Sreenivasulu N. 2005. Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: its implications in plant growth and abiotic stress tolerance. Current Science 88:424–438. [Google Scholar]
- Kim ST, Kim SG, Agrawal GK, Kikuchi S, Rakwal R. 2014. Rice proteomics: a model system for crop improvement and food security. Proteomics 14:593–610. [DOI] [PubMed] [Google Scholar]
- Lee DH, Kim YS, Lee CB. 2001. The inductive responses of the antioxidant enzymes by salt stress in rice (Oryza sativa L.). Journal of Plant Physiology 158:737–745. [Google Scholar]
- Leidi EO, Barragán V, Rubio L, El-Hamdaoui A, Ruiz MT, Cubero B, Fernández JA, Bressan RA, Hasegawa PM, Quintero FJ, Pardo JM. 2010. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant Journal 61:495–506. [DOI] [PubMed] [Google Scholar]
- Lindemose S, Shea C, Jensen M, Skriver K. 2013. Structure, function and networks of transcription factors involved in abiotic stress responses. International Journal of Molecular Sciences 14:5842–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yang W, Liu D, Han Y, Zhang A, Li S. 2011. Ectopic expression of a grapevine transcription factor VvWRKY11 contributes to osmotic stress tolerance in Arabidopsis. Molecular Biology Reporter 38:417–427. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC (T)) method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- Lovdal T, Lillo C. 2009. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold and light stress. Analytical Biochemistry 387:238–242. [DOI] [PubMed] [Google Scholar]
- Marco F, Bitrián M, Carrasco P, Rajam MV, Alcázar R, Antonio FT. 2015. Genetic engineering strategies for abiotic stress tolerance in plants. Plant Biology and Biotechnology 2:579–610. [Google Scholar]
- Mejía L, Teni RE, Czosnek H, Vidavski F, Bettilyon A, Nakhla MK, Maxwell DP. 2002. Field evaluation of tomato experimental lines and hybrids for resistance to Begomoviruses in Guatemala. Phytopathology 92:S54. [Google Scholar]
- Mittova V, Guy M, Tal M, Volokita M. 2004. Salinity upregulates the antioxidative system in root mitochondria and peroxisomes of the wild salt-tolerant tomato species Lycopersiconpennellii. Journal of Experimental Botany 55:1105–1113. [DOI] [PubMed] [Google Scholar]
- Mittova V, Tal M, Volokita M, Guy M. 2003. Up-regulation of the leaf mitochondrial and peroxisomalantioxidative systems in response to salt-induced oxidative stress in the wild salt-tolerant tomato species Lycopersicon pennellii. Plant Cell Environment 26:845–856. [DOI] [PubMed] [Google Scholar]
- Mittova V, Volokita M, Guy M. 2015. Antioxidative systems and stress tolerance: insight from wild and cultivated tomato species In: Gupta KJ, Igamberdiev AU, eds. Reactive oxygen and nitrogen species signaling and communication in plants. Switzerland: Springer International Publishing Inc, 89–131. [Google Scholar]
- Munné-Bosch S. 2005. The role of alpha-tocopherol in plant stress tolerance. Journal of Plant Physiology 162:743–748. [DOI] [PubMed] [Google Scholar]
- Munns R, Husain S, Rivelli AR, James RA, Condon AG, Lindsay MP, Lagudah ES, Schachtman DP, Hare RA. 2002. Avenues for increasing salt tolerance of crops, and the role of physiologically based selection traits. Plant & Soil 247:93–105. [Google Scholar]
- Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, Plett D, Gilliham M. 2012. Wheat grain yield on saline soils is improved by an ancestral Na transporter gene. Nature Biotechnology 30:360–366. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. 2008. Mechanisms of salinity tolerance. Annual Review of Plant Biology 59:651–681. [DOI] [PubMed] [Google Scholar]
- Murshed R, Lopez-Lauri F, Sallanon H. 2014. Effect of salt stress on tomato fruit antioxidant systems depends on fruit development stage. Physiology and Molecular Biology of Plants 20:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Asada K. 1981. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiology 22:867–880. [Google Scholar]
- Pardo JM, Cubero B, Leidi EO, Quintero FJ. 2006. Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. Journal of Experimental Botany 57:1181–1199. [DOI] [PubMed] [Google Scholar]
- Pardo JM, Rubio F. 2011. Na+ and K+ transporters in plant signalling In: Geisler M, Venema K, eds. Transporters and pumps in plant signaling. Berlin, Germany: Springer Verlag Inc, 65–99. [Google Scholar]
- Peng X, Hu Y, Tang X, Zhou P, Deng X, Wang H, Guo Z. 2012. Constitutive expression of rice WRKY 30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta 236:1485–1498. [DOI] [PubMed] [Google Scholar]
- Plett D, Safwat G, Gilliham M, Skrumsager Moller I, Roy S, Shirley N, Jacobs A, Johnson A, Tester M. 2010. Improved salinity tolerance of rice through cell type‐specific expression of AtHKT1;1. PLoS One 5:e12571.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poóra P, Kovácsa J, Borbélya P, Takácsa Z, Szepesia Á, Tari I. 2015. Salt stress-induced production of reactive oxygen-and nitrogen species and cell death in the ethylene receptor mutant never ripe and wild type tomato roots. Plant Physiology and Biochemistry 97:313–322. [DOI] [PubMed] [Google Scholar]
- Pottosin I, Velarde-Buendía AM, Bose J, Zepeda-Jazo I, Shabala S, Dobrovinskaya O. 2014. Cross-talk between reactive oxygen species and polyamines in regulation of ion transport across the plasma membrane: implications for plant adaptive responses. Journal of Experimental Botany doi:10.1093/jxb/ert423. [DOI] [PubMed] [Google Scholar]
- Rakhmankulova ZF, Shuyskaya EV, Shcherbakov AV, Fedyaev VV, Biktimerova GY, Khafisova RR, Usmanov IY. 2015. Content of proline and flavonoids in the shoots of halophytes inhabiting the South Urals. Russian Journal of Plant Physiology 62:71–79. [Google Scholar]
- Rany B, Aldon D, Cotelle V, Galaud JP, Thuleau P, Mazars C. 2016. Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Frontiers in Plant Science 7:327.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX. 2005. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nature Genetics 37:1141–1146. [DOI] [PubMed] [Google Scholar]
- Rus A, Baxter I, Muthukumar B, Gustin J, Lahner B, Yakubova E, Salt DE. 2006. Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genetics 2:1964–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi M, Abdin M, Ahmad J, Iqbal M. 2013. Effect of long-term salinity on cellular antioxidants, compatible solute and fatty acid profile of sweet Annie (Artemisia annua L.). Phytochemistry 95:215–223. [DOI] [PubMed] [Google Scholar]
- Schroeder JI, Delhaize E, Frommer WB, Guerinot ML, Harrison MJ, Herrera-Estrella L, Horie T, Kochian LV, Munns R, Nishizawa NK, Tsay YF, Sanders D. 2013. Using membrane transporters to improve crops for sustainable food production. Nature 497:60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Stevens MR, Barten JHM, Serra CA. 1995. Introgression of resistance to whitefly-transmitted geminiviruses from Lycopersicon chilense to tomato In: Gerling D, Mayer RT, eds. Bemisia: taxonomy, biology, damage, control and management. England: Intercept Inc Andover, 357–367. [Google Scholar]
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K. 2002. Monitoring the expression profiles of ca 7000Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant Journal 31:279–292. [DOI] [PubMed] [Google Scholar]
- Shabala S, Cuin TA. 2008. Potassium transport and plant salt tolerance. Physiologia Plantarum 133:651–669. [DOI] [PubMed] [Google Scholar]
- Sharma MK, Kumar R, Solanke AU, Sharma R, Tyagi AK, Sharma AK. 2010. Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Molecular Genetics and Genomics 284:455–475. [DOI] [PubMed] [Google Scholar]
- Sharma S, Villamor JG, Verslues PE. 2011. Essential role of tissue-specific proline synthesis and catabolism in growth and redox balance at low water potential. Plant Physiology 157:292–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K. 2002. Regulation and function of ascorbate peroxidase isoenzymes. Journal of Experimental Botany 53:1305–1319. [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. 2007. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany 58:221–227. [DOI] [PubMed] [Google Scholar]
- Silva-Ortega CO, Ochoa-Alfaro AE, Reyes-Agüerob JA, Aguado-Santacruz GA, Jimenez-Bremont JF. 2008. Salt stress increases the expression of P5CS gene and induces proline accumulation in cactus pear. Plant Physiology and Biochemistry 46:82–92. [DOI] [PubMed] [Google Scholar]
- Soshinkova TN, Radyukina NL, Korolkova DV, Nosov AV. 2013. Proline and functioning of the antioxidant system in Thellungiella salsuginea plants and cultured cells subjected to oxidative stress. Russian Journal of Plant Physiology 60:41–54. [Google Scholar]
- Subbarao GV, Johansen C, Jana MK, Kumar Rao JVDK. 1990. Effects of sodium/calcium ratio in modifying salinity response of pigeonpea (Cajanuscajan). Journal of Plant Physiology 136:439–443. [Google Scholar]
- Sudhakar C, Lakshmi A, Giridarakumar S. 2001. Changes in the antioxidant enzyme efficacy in two high yielding genotypes of mulberry (Morusalba L.) under NaCl salinity. Plant Sciences 161:613–619. [Google Scholar]
- Sun LJ, Huang L, Li DY, Zhang HJ, Song FM. 2014. Comprehensive expression analysis suggests overlapping of rice OsWRKY transcription factor genes during abiotic stress responses. Plant Physiology Journal 50:1651–1658. [Google Scholar]
- Szabados L, Savouré A. 2010. Proline: a multifunctional aminoacid. Trends Plant Sciences 15:89–97. [DOI] [PubMed] [Google Scholar]
- Talbi S, Romero-Puertas MC, Hernandez A, Terron L, Ferchichi A, Sandalio LM. 2015. Drought tolerance in a saharian plant oudneya africana: role of the antioxidant defenses. Environmental and Experimental Botany 111:114–126. [Google Scholar]
- Tester M, Davenport R. 2003. Na+ tolerance and Na+ transport in higher plants. Annals of Botany 91:503–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N. 2009. Cold, salinity, and drought stress In: Hirt H, eds. Plant stress biology, from genomics towards system biology. Weinheim, Germany: Wiley-Blackwell Inc, 137–159. [Google Scholar]
- Tuteja N, Mahajan S. 2007. Calcium signaling network in plants: an overview. Plant Signaling and Behavior 2:79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N, Schroeder JI. 2010. Ion channels and plant stress: past, present and future In: Demidchik V, Maathuis F, eds. Ion channels and plant stress responses. Berlin: Springer Inc, 1–22. [Google Scholar]
- Van Breusegem F, Vranova E, Dat JF, Inzé D. 2001. The role of active oxygen species in plant signal transduction. Plant Science 161:405–414. [Google Scholar]
- Venema K, Belver A, Marín-Manzano MC, Rodriguez Rosales MP, Donaire JP. 2003. A novel intracellular K+/H+ antiporter related to Na+/H+ antiporters is important for K+ ion homeostasis in plants. Journal of Biological Chemistry 278:22453–22459. [DOI] [PubMed] [Google Scholar]
- Verbruggen N, Hermans C. 2008. Proline accumulation in plants: a review. Amino Acids 35:753–759. [DOI] [PubMed] [Google Scholar]
- Vidavsky F, Czosnek H. 1998. Tomato breeding lines resistant and tolerant to Tomato yellow leaf curl virus issued from Lycopersicon hirsutum. Phytopathology 88:910–914. [DOI] [PubMed] [Google Scholar]
- Villalta I, Bernet GP, Carbonell EA, Asins MJ. 2007. Comparative QTL analysis of salinity tolerance in terms of fruit yield using two Solanum populations of F7 lines. Theoretical and Applied Genetics 114:1001–1017. [DOI] [PubMed] [Google Scholar]
- Villalta I, Reina-Sanchez A, Bolarin MC, Cuartero J, Belver A, Venema K, Carbonell EA, Asins MJ. 2008. Genetic analysis of Na (+) and K (+) concentrations in leaf and stem as physiological components of salt tolerance in Tomato. Theoretical and Applied Genetics 116:869–880. [DOI] [PubMed] [Google Scholar]
- Wang P, Guo Q, Wang Q, Zhou XR, Wang SM. 2015. PtAKT1 maintains selective absorption capacity for K+ over Na+ in halophyte Puccinellia tenuiflora under salt stress. Acta Physiologiae Plantarum 37:100. doi: 10.1007/s11738-015-1846-3. [Google Scholar]
- Willekens H, Chamnongpol S, Davey M, Schraudner M, Langebartels C, Van Montagu M. 1997. Catalase is a sink for H2O2 and is indispensable for stress defense in C3 plants. EMBO Journal 16:4806–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhu M, Shabala L, Zhou M, Shabala S. 2014. K+ retention in leaf mesophyll, an overlooked component of salinity tolerance mechanism: a case study for barley. Journal of Integrative Plant Biology 57:171–185. [DOI] [PubMed] [Google Scholar]
- Xue S, Yao X, Luo W, Jha D, Tester M, Horie T, Schroeder JI. 2011. AtHKT1;1 mediates nernstian sodium channel transport properties in Arabidopsis root stelar cells. PLoS One 6:e24725.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JJ, Mehta S, Israelsson M, Godoski J, Grill E, Schroeder JI. 2006. CO2 signaling in guard cells: calcium sensitivity response modulation, a Ca2+-independent phase, and CO2 insensitivity of the gca2 mutant. Proceedings of the National Academy of Sciences 103:7506–7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. 2003. Regulation of ion homeostasis under salt stress. Current Opinion of Plant Biology 6:441–445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.