Abstract
Background and Aims The broomrapes, Orobanche sensu lato (Orobanchaceae), are common root parasites found across Eurasia, Africa and the Americas. All species native to the western hemisphere, recognized as Orobanche sections Gymnocaulis and Nothaphyllon, form a clade that has a centre of diversity in western North America, but also includes four disjunct species in central and southern South America. The wide ecological distribution coupled with moderate taxonomic diversity make this clade a valuable model system for studying the role, if any, of host-switching in driving the diversification of plant parasites.
Methods Two spacer regions of ribosomal nuclear DNA (ITS + ETS), three plastid regions and one low-copy nuclear gene were sampled from 163 exemplars of Orobanche from across the native geographic range in order to infer a detailed phylogeny. Together with comprehensive data on the parasites’ native host ranges, associations between phylogenetic lineages and host specificity are tested.
Key Results Within the two currently recognized species of O. sect. Gymnocaulis, seven strongly supported clades were found. While commonly sympatric, members of these clades each had unique host associations. Strong support for cryptic host-specific diversity was also found in sect. Nothaphyllon, while other taxonomic species were well supported. We also find strong evidence for multiple amphitropical dispersals from central North America into South America.
Conclusions Host-switching is an important driver of diversification in western hemisphere broomrapes, where host specificity has been grossly underestimated. More broadly, host specificity and host-switching probably play fundamental roles in the speciation of parasitic plants.
Keywords: Amphitropical disjunction, cryptic speciation, holoparasite, host-switching, Orobanche, Orobanchaceae, parasite, phylogeny
INTRODUCTION
Parasitism is a highly successful life strategy that has evolved independently >60 times among animals, at least 12 times among angiosperms, and repeatedly in protozoans and prokaryotes (Poulin and Morand, 2000; Westwood et al., 2010). While the evolutionary significance of host–parasite associations has long been recognized (Kellogg, 1913), the main evolutionary mechanisms involved in the generation and maintenance of such ecological and phylogenetic diversity are still poorly understood, especially among parasitic flowering plants (de Vienne et al., 2013; Joel et al., 2013).
The parasitic broomrapes, Orobanche sensu lato (s.l.) [alternatively circumscribed as the genera Aphyllon and Myzorrhiza in the New World, and Boulardia, Orobanche sensu stricto (s.s.) and Phelipanche in the Old World: Schneeweiss (2013)], have attracted significant attention as an important system for understanding the evolutionary consequences of parasitism. This attention is in part a result of their extensive worldwide diversity (at least 170 species; Ulrich et al., 1995), a detailed and well-supported understanding of their placement within the family Orobanchaceae as well as the relationships among major clades (Schneeweiss et al., 2004a; Park et al., 2008; McNeal et al., 2013; Schneeweiss, 2013), and the significant economic damage caused by several Eurasian species to major agricultural systems worldwide (Joel et al., 2013).
Despite the interest in this group, relatively little is known about the role of host specificity in broomrape diversification. Understanding host specificity of parasites is predicated on a comprehensive understanding of lineage boundaries in the host (e.g. Labrousse et al., 2001; Timko et al., 2012) and, more importantly for Orobanche, the parasite. That is, failure to recognize evolutionary diversity in the parasite results in an overestimation of host breadth and may limit the ability to understand the evolutionary processes responsible for speciation in plant parasites (Refrégier et al., 2008). Therefore, it is important to distinguish true host generalists from taxa that comprise several cryptic lineages artificially united on the basis of superficial similarity but distinguished genetically and ecologically. Host specificity to the family or genus level has been cited as a key factor in the differentiation and genetic isolation of three subspecies of the European O. minor (Thorogood et al., 2008, 2009), but this has not been broadly tested across other Orobanche lineages. Several recently described species of Orobanche in North America also have unique host preferences in the Asteraceae: Orobanche riparia parasitizes Helianthieae sub-tribe Ambrosiinae and O. arizonica parasitizes Gutierrezia spp. However, neither these species concepts nor those of the other American Orobanche species has ever been tested phylogenetically.
Inclusion of western hemisphere Orobanche (sections Gymnocaulis and Nothaphyllon) in phylogenetic studies has been limited to several exemplars included in larger genus- or family-level analyses. These studies, supported by karyological and morphological evidence, have shown that these two sections are sister groups and together are sister to an Old World clade corresponding to Orobanche sect. Trionychon (Schneeweiss et al., 2004a; Park et al., 2008), more recently treated as the genus Phelipanche (Schneeweiss, 2013). This larger clade is supported by a shared base chromosome base number of x = 12 (Heckard and Chuang, 1973; Schneeweiss et al., 2004b).
Ecologically, Orobanche sections Gymnocaulis and Nothaphyllon parasitize a wide range of eudicot hosts, but most commonly perennial Asteraceae. Taxonomic diversity is concentrated in the California Floristic Province; however, species can be found across the Americas, as far north as the Alaska Peninsula and the Yukon Territory, east to Newfoundland, and south to central Mexico. Four poorly known species are found in South America. Affinities between South American Orobanche chilensis and North American O. ludoviciana have long been recognized (Beck, 1890), but explicit biogeographic hypotheses for this or other such relationships within the clade have yet to be proposed.
The wide ecological and host diversity among western hemisphere Orobanche, as well as its tractable taxonomic diversity, make it a valuable model system for understanding the main ecological and evolutionary processes affecting parasite diversification and speciation. Such investigations, however, are requisite for a robust understanding of evolutionary lineages, their host breadths and their relationships. Specifically our goals were to (1) reconstruct a well-resolved phylogeny of western hemisphere Orobanche that could be used to develop a revised, natural classification for the group; (2) evaluate the evolutionary significance of host-switching in Orobanche sect. Gymnocaulis by comprehensively sampling across the geographic and host ranges of each taxon; (3) test the monophyly of longstanding taxa as well as recently described segregates; and (4) infer biogeographical relationships between North American and South American Orobanche spp.
MATERIALS AND METHODS
Taxon and population sampling
163 Orobanche populations were sampled either from fresh collected tissue or from herbarium collections: 57 from sect. Gymnocaulis and 106 from sect. Nothaphyllon (voucher and host information is provided in Supplementary Data Table S1). This data set includes at least one exemplar of all taxa of Orobanche recognized within the last 75 years except for O. weberbaueri, a poorly known taxon from southern coastal Peru, perhaps known only from the type. Denser population sampling across sect. Gymnocaulis enabled more comprehensive geographic and host range sampling in the two currently recognized species of this section, O. fasciculata and O. uniflora (Fig. 1). Identifying the host breadth for each taxon was challenging, as many collectors note the nearest living plant as the host species without confirming a haustorial connection, resulting in a proliferation of dubious records. Our criteria for accepting a host was that a host taxon must have been independently reported at several populations by more than one collector, or a haustorial connection to an identifiable fragment of host must be present on the herbarium voucher. Host associations for sampled populations are listed in Table S1. For molecular phylogenetic analyses, one individual each of O. gracilis and O. hederae were used as the outgroup (Park et al., 2008; McNeal et al., 2013). Sequence data for the waxy locus were not available for these outgroups, so instead two more distantly related outgroup taxa were used, Castilleja ambigua and Triphysaria versicolor.
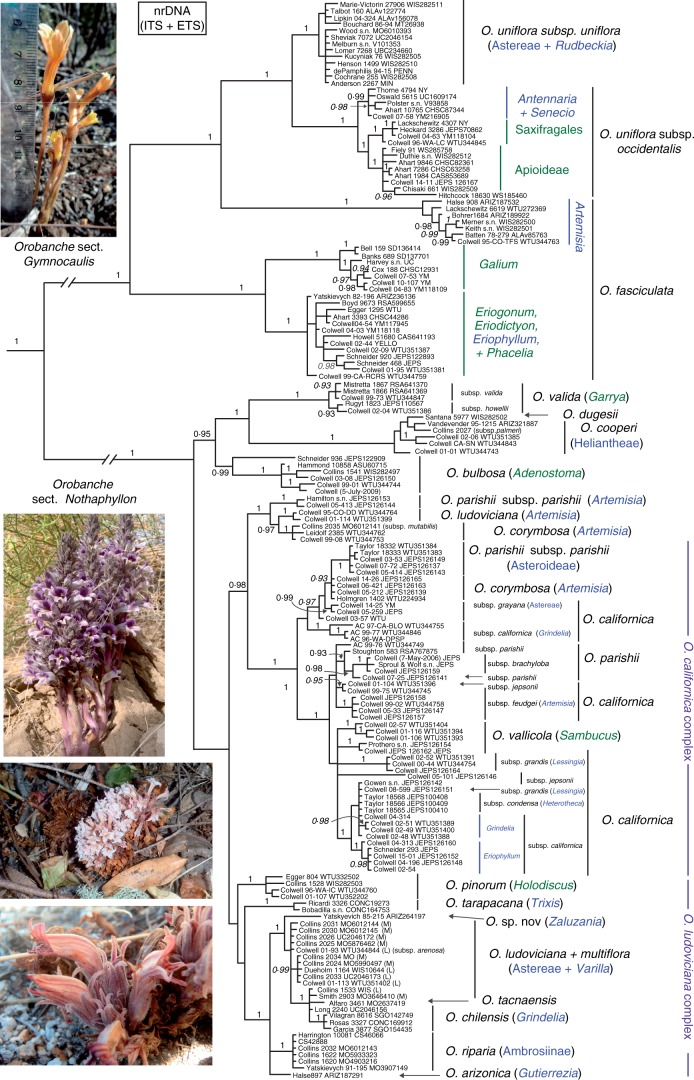
Fig. 1.
Bayesian inference majority-rule consensus tree of 162 Orobanche populations inferred from nrDNA (ITS + ETS). Tip labels include the collection number followed by the herbarium accession number, if available. Posterior probabilities >0·9 are shown in bold for nodes with >70 % maximum likelihood bootstrap (BS) support and in italics if BS support is < 70 %. The internal branches leading to section Gymnocaulis and section Nothaphyllon have been shortened by a factor of 1/2. Host associations are indicated in blue (Asteraceae) or green (other) to the genus or higher taxonomic level. Informally named clades are in purple. Outgroup taxa are not shown. Photographs, from top to bottom: O. fasciculata parasitizing Eriodicyton sp. (Schneider 606); O. cooperi parasitizing Hymenoclea salsola (Schneider 415); O. vallicola parasitizing Sambucus mexicana (Schneider 316); O. corymbosa parasitizing Artemisia tridentata (Colwell 14–26).
DNA extraction, amplification and sequencing
DNA was extracted from dried floral tissue using a DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA), or using a CTAB (cetyltrimethylammonium bromide) protocol (Doyle and Doyle, 1987). A total of six regions from the nuclear and plastid genomes were used to estimate the phylogeny: internal and external transcribed spacers of nuclear ribosomal DNA (ITS and ETS, respectively), introns 9–11 of the nuclear low-copy gene waxy, as well as the plastid trnL-trnF region (comprising the trnLUAA intron and the trnLUAA-trnFGAA intergenic spacer) and the matK and rps2 genes. ITS, matK, and rps2 were selected based on their prior use in genus- and family-level phylogenetic studies of Orobanche (Schneeweiss et al., 2004a; McNeal et al., 2013), and waxy for its use in the related (hemi-)parasitic genus Castilleja (Tank and Olmstead, 2008). The remaining two regions, ETS and trnL-trnF, were selected to provide additional rapidly evolving characters from the nuclear and plastid compartments, respectively. Due to difficulty assessing homology within some species of sect. Nothaphyllon, the waxy locus was mainly used to assess monophyly of sect. Nothaphyllon and to infer relationships within sect. Gymnocaulis.
Polymerase chain reaction (PCR) amplifications were performed using AccuPower PCR PreMix kits (Bioneer, Alameda, CA, USA) or by generating a master mix of 10 μL of 5× Promega buffer, 4 μL of 25 mm MgCl2, 1·25 μL of 10 mm dNTPs, 1 μL of 20 μm of each primer and 0·25 μL of Go-Taq DNA Polymerase (Promega, Madison, WI, USA) diluted to 50 μL. Complete information about primers, cycling parameters and amplicon sizes are provided in Table 1. PCR products were purified using ExoSAP (USB Products, Cleveland, OH, USA), and both DNA strands were sequenced using an ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA). GenBank accession numbers can be found in Table S1.
Table 1.
Molecular regions used in the phylogenetic analyses of Orobanche sections Gymnocaulis and Nothaphyllon, approximate lengths of complete ingroup sequences, PCR primers (5′–3′) and thermocycling parameters
| Gene region | Approximate amplicon length | Primer sequences | Reference | Thermocycling parameters |
|---|---|---|---|---|
| ITS | 590 bp | AB_101: TGG TCC CGT GAA GTG TTC G | Schneeweiss et al. (2004a) | 94 °C, 4 min; 35 × (95 °C, 1 min; 48 °C, 1 min; 72 °C, 1 min); 72 °C, 10 min. |
| AB_102: CCG GTT CGC TG CCG TAA C | Schneeweiss et al. (2004a) | |||
| ETS | 430 bp | ETS_B: ATA GAG CGC GTG AGT GGT G | Beardsley and Olmstead, 2002) | 96 °C, 2 min; 35 × (94 °C, 30 s; 56 °C, 30 s; 72 °C, 45 s); 72 °C, 3 min. |
| ETS_seq: (C) TGG CAG GAT CAA CCA GGT A | This study | |||
| waxy (introns 9–11) | 585–630 bp | waxy_9F-ORO: GAT GCT AAG CCW TTG TTG A | This study | 92 °C, 5 min; 40 × (94 °C, 45 s; 53·5 °C, 45 s; 72 °C, 1 min); 72 °C, 5 min. |
| waxy_11R: CCA TRT GGA ASC CAG TRT A | Tank and Olmstead (2009) | |||
| matK 3′ intron | 680–760 bp | matK 8: CTT CGA CTT TCT TGT GCT | Steele and Vilgalys (1994) | 94 °C, 5 min; 40 × (92 °C, 1 min; 51 °C, 40 s; 72 °C, 1 min); 72 °C, 10 min. |
| matK_psbA5′R: AAC CAT CCA ATG TAA AGA CGG TTT | Shaw et al. (2005) | |||
| rps2 | 675 bp | rps2_2F: AAA TGG AAT CCT AAA ATG GC | This study | 94 °C, 2 min 30 s; 35 × (94 °C, 1 min; 50 °C, 1 min; 72 °C, 1 min); 72 °C, 7 min. |
| rps2_18F: GGR KAR AAA TGA CAA GAA GAT ATT GG | dePamphilis et al. (1997) | |||
| rps2_661R: ACC CTC ACA AAT GCG AAT ACC AA | dePamphilis et al. (1997) | |||
| trnL-trnF spacer | 710–810 bp | trL ‘c’: CGA AAT CGG TAG ACG CTA CG | Taberlet et al. (1991) | 94 °C, 5 min; 40 × (92 °C, 1 min; 51·5 °C, 1 min; 72 °C, 1 min); 72 °C, 5 min. |
| trnF ‘f': ATT TGA ACT GGT GAC ACG AG | Taberlet et al. (1991) |
Two different forward primers for rps2 were used.
Sequence alignment and phylogenetic reconstruction
Sequences were checked for base-calling errors and assembled into contigs using Geneious v.6.1.7 (Biomatters, Auckland, New Zealand). Sequence alignments were generated using the MUSCLE plug-in with default settings. Maximum likelihood (ML) and Bayesian inference (BI) analyses were conducted separately on the concatenated chloroplast DNA matrix (cpDNA), the concatenated ribosomal spacers (nrDNA) and the waxy locus using the CIPRES Science Gateway (Miller et al., 2010). The ML analyses were performed with RAxML-HPC2 v.8.2.6 (Stamatakis, 2014) using the GTRCAT model with 25 rate categories and 1000 rapid bootstrap (BS) replicates. The BI analyses were performed using MrBayes v.3.2.6 (Ronquist et al., 2012). We used an AIC (Akaike information criterion) comparison implemented in jmodeltest2 (Darriba et al., 2012) to select a GTR + Γ substitution model (approximated using four rate categories). The estimated substitution rates for the nrDNA, cpDNA and waxy alignments were then used as priors in the MrBayes analysis. Default settings were used for other priors. Three independent runs of four chains each (one cold, three heated) were sampled every 1000 generations for 2 500 000 generations. The first 20 % of samples were discarded as burn-in. Convergence was assessed in several ways: the average standard deviation of split frequencies was <0·01, the potential scale reduction factor was close to 1·00 for all parameters, and the effective sample sizes (ESS) were >800.
RESULTS
nrDNA
Strongly supported clades in the Bayesian ITS/ETS analysis (Fig. 1) were consistent with those identified by ML (data not shown). Orobanche sect. Gymnocaulis and sect. Nothaphyllon were both resolved as monophyletic [posterior probability (PP) = 1·0, BS = 100] and sister to each other. Within sect. Gymnocaulis, seven major clades were resolved (PP = 1·0, BS ≥ 80). Under the current classification, three of these together correspond to a paraphyletic O. fasciculata. Plants from each of these clades showed unique host preferences: plants in two of these groups parasitize hosts of single genera, Artemisia (Asteraceae) and Galium (Rubiaceae). The third group of plants form a clade of generalists that parasitize numerous species within Eriogonum (Polygonaceae), Eriophyllum (Asteraceae), and Eriodictyon and Phacelia (Hydrophyllaceae). The remaining four clades constituted a monophyletic O. uniflora (PP = 1·0, BS = 100). Three of these clades include parasites specific to hosts in the genera Antennaria and Senecio (Asteraceae), on members of Saxifragaceae and Crassulaceae (Saxifragales s.s.) and on Apioideae (Apiaceae), respectively. These clades together are currently recognized as O. uniflora subsp. occidentalis and were resolved sister to the fourth clade corresponding to subsp. uniflora. Members of this clade parasitize Rudbeckia and several genera of Astereae in the Asteroideae.
Populations of the remaining American Orobanche species, representing sect. Nothaphyllon were generally resolved in one of eight major clades (PP > 0·95, BS > 90): (1) a clade of populations from the western USA parasitic on Artemisia previously determined as one of three taxa: O. parishii subsp. parishii, O. ludoviciana or O. corymbosa; (2) a taxonomically and ecologically diverse clade, the O. californica complex, which included O. californica and O. vallicola, as well as the remainder of O. parishii and O. corymbosa populations; (3) O. pinorum; (4) O. tarapacana; (5) the O. ludoviciana complex, including O. multiflora, O riparia, O. chilensis, O. tacnaensis, O arizonica, the remainder of O. ludoviciana and a collection from Hidalgo, Mexico (Yatskievych 85-215) that does not match the morphology of any described species; (6) O. valida; (7) O. cooperi and O. dugesii; and (8) O. bulbosa. Clades 6–8, found predominantly in south-western North America, constituted a monophyletic group (PP = 0·95, BS = 77) that was sister to the rest of the section (clades 1–5). Resolution at the subspecific level of the paraphyletic O. californica was variable. For example, populations of subsp. californica along the central California coast parasitizing Grindelia stricta and those in far northern California and Washington parasitizing Grindelia integrifolia were resolved in separate strongly supported sub-clades within the O. californica complex (clade 2, above). Other subspecies, such as subsp. grandis and subsp. condensa, formed a polytomy. The polyploid O. parishii subsp. brachyloba was nested within one of three separate clades of O. parishii subsp. parishii.
cpDNA
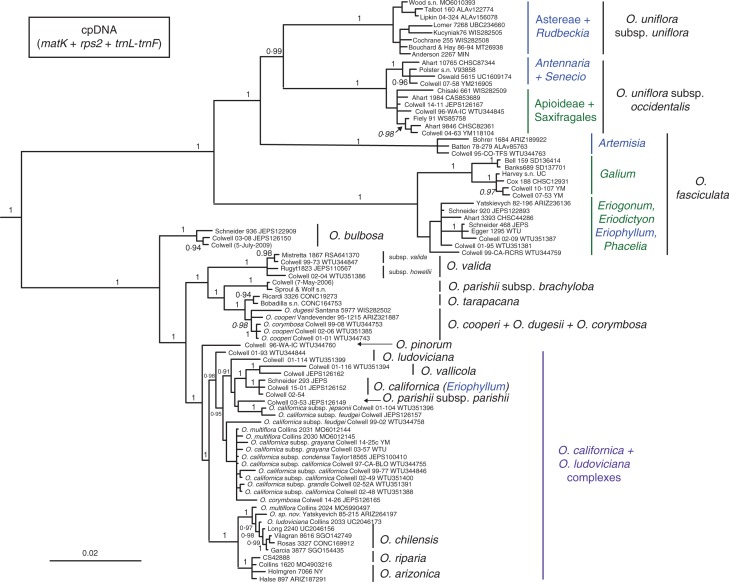
Strongly supported clades from the Bayesian analysis of three plastid regions (Fig. 2) were consistent with those identified by ML (data not shown). Orobanche sect. Gymnocaulis was resolved as monophyletic (PP = 1·0, BS = 100). Within sect. Gymnocaulis, six host-specific clades were resolved, congruent with the nrDNA results. Three of these were sub-clades of the monophyletic O. uniflora (PP = 0·99, BS = 97): a clade of plants parasitizing Antennaria and Senecio (PP = 1·0, BS = 93) and a less supported clade of plants parasitizing Apioideae (Apiaceae), Saxifragaceae and Crassulaceae (PP = 0·71, BS = 88), together corresponding to subsp. occidentalis (PP = 1·0, BS = 100) and sister to a clade of plants that parasitize several genera of Asteroideae corresponding to subsp. uniflora (PP = 1, BS = 100). Orobanche fasciculata was found to be paraphyletic: a strongly supported clade parasitizing Artemisia (PP = 1·0, BS = 100) was resolved sister to O. uniflora. The remaining two clades of O. fasciculata were resolved as sister groups, one strongly supported and parasitizing Galium spp. in California and Oregon (PP = 1·0, BS = 100), and the other weakly supported and parasitizing a variety of distantly related core eudicot genera (PP = 0·65, BS < 50).
Fig. 2.
Bayesian inference majority-rule consensus tree of 86 Orobanche populations inferred from three concatenated cpDNA regions (matK, rps2 and trnL-trnF). Tip labels consist of the taxon name (if not included in a sidebar), the collection number and the herbarium accession numbers if available. Posterior probabilities >0·9 are shown in bold for nodes with >70 % maximum likelihood bootstrap (BS) support and in italics if BS support is < 70 %. Some host associations are indicated in blue (Asteraceae) or green (other) to the genus or higher taxonomic level; for others, see Fig. 1. Informally named groups are given in purple. Outgroups are not shown.
Deep relationships within Orobanche sect. Nothaphyllon were generally well resolved, albeit with variable support at the species and subspecies level. Populations of O. bulbosa formed a clade (PP = 1·0, BS = 96) that was sister to the remainder of the section, which in turn comprised two well-supported sub-clades (PP = 1·0, BS > 95). The first included strongly supported clades corresponding to single taxa that diverged from the remainder of the sub-clade in succession: O. valida (PP = 1·0, BS = 100), O. parishii (PP = 1·0, BS = 100) and finally O. tarapacana (PP = 0·94, BS = 72), which was sister to a clade of O. cooperi, O. dugesii and one accession of O. corymbosa (PP = 0·98, BS = 68). The second well-supported sub-clade included the only sampled population of O. pinorum sister to the O. californica and O. ludoviciana complexes. Relationships within this sub-clade were poorly resolved, except for strong support of O. riparia + O. arizonica, O. vallicola, a clade of O. californica subsp. californica parasitic on Eriophyllum staechadifolium, and O. chilensis + several populations from central North America (PP = 1·0, BS > 97).
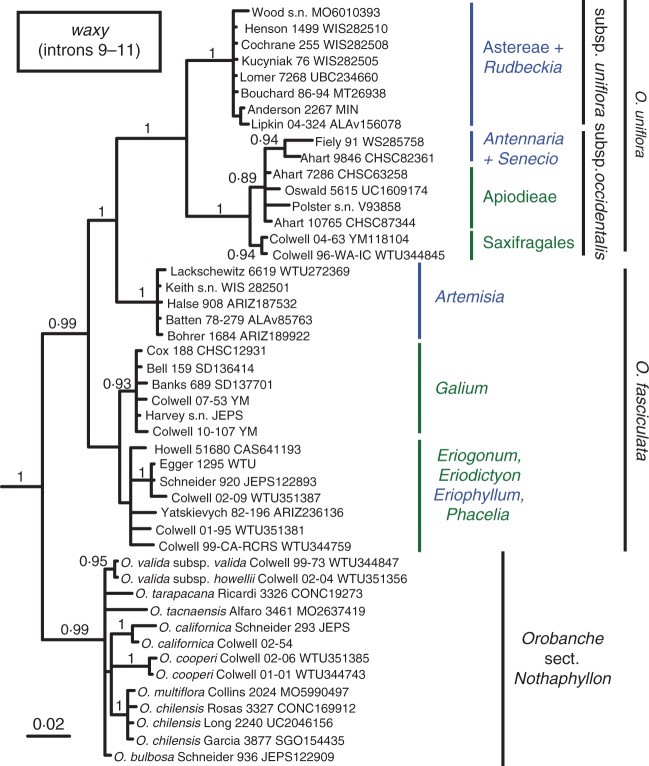
waxy
Orobanche sect. Gymnocaulis and sect. Nothaphyllon were each resolved as monophyletic (PP = 0·99, BS > 75). Within sect. Gymnocaulis, five host-specific clades were resolved with strong support (PP > 0·92, BS > 73), congruent with both nrDNA and cpDNA results. These included a clade of plants parasitizing several genera in the Asteroideae corresponding to O. uniflora subsp. uniflora, as well as two clades together corresponding to O. uniflora subsp. occidentalis – the first, which was comprised of plants parasitizing Saxifragaceae and Crassulaceae (Saxifragales s.s), and another that included a sub-clade of parasites on Antennaria and Senecio (Asteraceae) united in a moderately supported polytomy with several populations that parasitize Apioideae (PP = 0·89, BS = 0·74). The remaining two strongly supported clades include plants currently recognized as O. fasciculata: one was sister to O. uniflora and parasitizes Artemisia; the other parasitizes Galium and was sister to the remaining populations of O. fasciculata, which formed a third, weakly supported clade (PP = 0·74, BS = 67) including parasites on a variety of core eudicot hosts. In contrast to Orobanche sect. Gymnocaulis, infraspecific sampling density and phylogenetic resolution within O. sect. Nothaphyllon was limited, although conspecific populations of O. valida, O. californica subsp. californica and O. cooperi, as well as O. chilensis + O. multiflora were each resolved as monophyletic (PP > 0·94, BS > 90). Tree files were uploaded to Open Tree of Life (http://www.opentreeoflife.org), study ID ot_732.
DISCUSSION
Host specificity and speciation
Among extant western hemisphere Orobanche, we report many previously unrecognized, host-specific lineages in both sect. Gymnocaulis and sect. Nothaphyllon that are strongly supported by both plastid and nuclear DNA sequences (Figs 1–3). This cryptic diversity has two complementary implications – one evolutionary, the other ecological. First, biodiversity within western hemisphere Orobanche is substantially richer than recognized by current taxonomy, perhaps because extensive reduction of structural characters in these parasites has limited the potential for morphological diagnosis of recently diverged evolutionary lineages. Secondly, the host breadth of each evolutionary lineage is narrower than previously assumed, although some lineages with wide host ranges are still present (e.g. O. fasciculata p.p.). Host specificity in plant parasites has been correlated to various life history and other host traits such as weediness or perenniality (Schneweeis, 2007). Host-switching has been cited as a driver of speciation of numerous parasites across the tree of life (Ricklefs et al., 2004; de Vienne et al., 2013), including other lineages of parasitic plants (Norton and Carpenter, 1998; Norton and Lange, 1999; Bolin et al., 2011), as well as within the genus Orobanche (Thorogood et al., 2009). Our evidence strongly supports this hypothesis. The abundance of host-specific clades found here suggests that host-switching may be an even more important driver of evolutionary divergence in parasitic plants than previously recognized.
Fig. 3.
Bayesian inference majority-rule consensus tree of 47 Orobanche populations inferred from the waxy locus (introns 9–11). Tip labels consist of the taxon name (if not included in a sidebar), the collection number and the herbarium accession numbers if available. Posterior probabilities >0·8 are shown. All labelled nodes have maximum likelihood bootstrap scores ≥74 %. Host associations for sect. Gymnocaulis are indicated in blue (Asteraceae) or green (other) to the genus or higher taxonomic level; for sect. Nothaphyllon see Fig. 1 or Table S1. Outgroups are not shown.
Although some Orobanche taxa are specific to a single host species, most parasitize several closely related species that are unique and sometimes phylogenetically distant from the hosts of their nearest relatives. In many ways, Orobanche spp. occupy an ecological middle-ground between species such as Epifagus virginiana (Orobanchaceae), which can only grow on Fagus grandifolia, and true generalists such as dodders (Cuscuta spp., Convolvulaceae) in which a single individual may parasitize numerous distantly related hosts (Press and Graves, 1995). Therefore, it is unlikely that host–parasite co-speciation plays an appreciable role in driving diversification in western hemisphere Orobanche in contrast to some plant–animal, animal–animal or prokaryote–animal host–parasite systems (de Vienne et al., 2013). Instead, we argue that the more common mode – host-switching followed by physiological specialization and divergence – is dominant in this system.
Specialization and evolutionary divergence (cladogenesis) following host-switching is an expected outcome given the complex challenges of host detection, host invasion and evasion or neutralization of host defences, which may occur pre- or post-attachment. Pre-attachment host defences may include reduced germination stimulants (i.e. strigolactones, Cameron et al., 2006; Xie et al., 2010), increased germination inhibitory compounds (Fernández-Aparicio et al., 2011), chemical inhibition of haustorial development (Pérez-de-Luque et al., 2005a, b) or structural fortifications to serve as a mechanical barrier to invasion. Potential hosts can repel parasitic plants following attachment using a variety of mechanisms that disrupt the flow of nutrients or block vessel elements (Goldwasser et al., 1999, 2000; Pérez-de-Luque et al., 2005a), initiate programmed cell death (Gurney et al., 2006), increase lignification and suberization of cell walls (Labrousse et al., 2001; Pérez-de-Luque et al., 2008) or elicit chemical defence through increased peroxidases or the transfer of toxins from the host to the parasite (Gurney et al., 2003). These multiple layers of incompatibility must be overcome for a successful invasion of the host, and provide the physiological basis for host specificity in parasitic Orobanchaceae (Yoder, 1997; Yoshida and Shirasu, 2009; Thorogood and Hiscock, 2010). Consequently, distantly related hosts with more divergent physiologies probably require different invasion strategies. Various suites of host-specific traits may therefore represent different adaptive peaks for an Orobanche lineage.
Drès and Mallet (2002) cite a number of insect–plant systems to show how the formation of host-specific races may eventually lead to sympatric speciation of parasites through outbreeding depression, even in the presence of gene flow. The generalist clade of O. fasciculata shows poorly supported phylogenetic sub-structure and may provide the opportunity to explore this hypothesis in a plant–parasite system. Among the other host-specific clades of O. sect. Gymnocaulis, sympatric speciation following this model may already have occurred. The strong support for these clades by all three loci (nrDNA, cpDNA and waxy) suggest minimal, if any, continued gene flow among these lineages, even between geographically neighbouring populations. Isolation by host may also be reinforced by autogamy or apomixis, which is common in New World Orobanche species in contrast to more variable mating systems among species of Eurasian Orobanche and predominance of outcrossing among other lineages of parasitic angiosperms (Musselman et al., 1982; Jones, 1989; Bellot and Renner, 2013). Autogamy has been identified as the predominant mating system in O. pinorum, with occasional outcrossing by bees (Ellis et al., 1999), is common among O. fasciculata parasitizing Artemisia (Reuter, 1986), and has been anecdotally reported in O. uniflora subsp. occidentalis and Orobanche bulbosa (K. L. Chambers 2952, OSC198410; Butterwick 5434 & Parfitt, ASU, JEPS; Schneider 1032, JEPS (Parfitt and Butterwick, 1981)). Some populations of Orobanche uniflora subsp. uniflora are obligatorily parthenogenic, while other populations show a ‘wholly different…reproductive process’ (Jenson, 1951). As discussed previously, gene flow between different host races is expected to be detrimental if parent taxa are adapted to separate hosts, since a hybrid may be adapted to neither of them.
Geographic differentiation may play a subordinate role in lineage diversification, and may be restricted to cases where sister clades parasitize closely related hosts, such as between the subspecies of O. valida, which both parasitize Garrya. Much more commonly, ranges are at least partially overlapping, and closely related parasite lineages differing in their hosts can co-occur on a regional or even a local scale. This is particularly well pronounced in sect. Gymnocaulis, discussed in detail below.
Cryptic diversity in section Gymnocaulis
Cryptic lineages are found in both sections of New World Orobanche [e.g. a polyphyletic O. parishii subsp. parishii (Fig. 1)], but most extensively in O. sect. Gymnocaulis, in which we identified over twice as many host-specific clades as exist commonly recognized taxa. Moreover, these clades are often subtended by long stem branches relative to clades that represent different recognized species in sect. Nothaphyllon. This disparity, which is robust to the gene region(s) used (Figs 1–3), may be due to more extensive reduction of morphological and thus diagnostic features in sect. Gymnocaulis, as well as more limited systematic and taxonomic study of this section (Achey, 1933; Watson, 1975) relative to sect. Nothaphyllon (Munz, 1930; Collins, 1973; Heckard, 1973; Heckard and Chuang, 1975; Collins and Yatskievich, 2015). Similar levels of cryptic diversity may be found in other holoparasitic lineages, particular endoparasites such as Cytinus (Cytinaceae) that show even more extensive morphological reduction than Orobanche and a more intimate host–parasite relationship (De Vega et al., 2008).
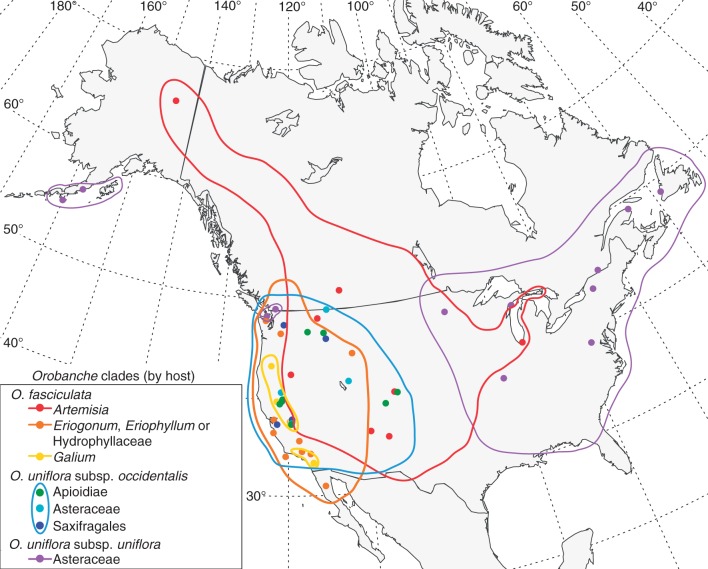
Each clade of Orobanche sect. Gymnocaulis shows at least partial range overlap with its sister group, with generally increasing overlap with decreasing phylogenetic distance (Fig. 4). The clade of O. fasciculata parasitic on Galium is entirely included within the range of its sister group, which is a generalist clade parasitic on various eudicot hosts. The clade of O. fasciculata parasitic on Artemisia grows coarsely sympatrically (i.e. sympatric at regional scales) with both subspecies of its sister group, O. uniflora. These subspecies, O. subsp. uniflora and O. subsp. occidentalis, once thought to be allopatric, are now known to co-occur based on a recent floristic discovery in southern British Columbia and subsequent reinterpretation of historic herbarium records (A. C. Schneider, unpubl. data). Most strikingly, the three closely related clades resolved within O. uniflora subsp. occidentalis, which parasitize species in the Asteraceae, Apiaceae, and Saxifragaceae plus Crassulaceae, respectively, share nearly entirely overlapping ranges at both coarse continental and local scales. For example, populations of all three clades can be found in Yosemite National Park and the adjacent Sierra National Forest.
Fig. 4.
Range map of host-specific clades of Orobanche sect. Gymnocaulis. Coloured circles represent individuals sampled in the phylogeny (Figs 1–3). Coloured lines show the approximate range of each clade. Further study is needed to determine the range of each of the three host-specific lineages of O. uniflora subsp. occidentalis, which in this figure are treated as one unit. Range maps should be considered tentative, particularly in northern Canada and west-central USA, pending a thorough taxonomic and phytogeographical study.
Relationships in section Nothaphyllon
In sect. Nothaphyllon, we also find strong support for host-specific species, including the recently described O. arizonica, O. riparia and a clade currently recognized as O. californica subsp. californica that parasitzes Eriophyllum stachaedifolium on the central California coast, which is currently being described by the second author and George Yatskievych. Most other clades have distinct host associations, generally with perennial Asteraceae, but usually not specific to the species level (Fig. 1).
Most of the taxonomic diversity in O. sect. Nothaphyllon is concentrated in a large clade supported by nrDNA and cpDNA, which is comprised of two sub-clades supported by nrDNA (Fig. 1) and morphological analysis (Collins 1973; Heckard, 1973). The first sub-clade corresponds to the O. californica complex, which includes O. californica and its subspecies, O. parishii, O. corymbosa and O. vallicola. The second clade represents the O. ludoviciana complex, which includes O. ludoviciana (except for populations parasitizing Artemisia), O. multiflora, the recently described O. arizonica, O. riparia, the disjunct South American species O. chilensis and O. tacnaensis, and a collection from Hidalgo, Mexico that does not fit the description of any described taxon (Yatskievych 85-215; ARIZ).
Several earlier diverging lineages native to western North America are also strongly supported as monophyletic by both nrDNA and cpDNA, including O. valida, O. bulbosa and the recently revised O. cooperi + O. dugesii complex (Figs 1 and 2; Collins and Yatskievych, 2015). However, relationships among these lineages are unclear: O. bulbosa is resolved either as sister to the rest of the section (nrDNA, Fig. 1) or as a grade with O. bulbosa diverging earliest (cpDNA, Fig. 2). The conflict among gene partitions is in most cases probably explained by incomplete lineage sorting, but in other cases may be a result of reticulate evolution. For example, based on its phylogenetic placement in two separate clades (Fig. 1), and morphological and host affinities (Artemisia, especially A. tridentata), O. corymbosa may represent a hybrid between O. californica and O. ludoviciana, both of which in part also parasitize Artemisia. In certain other cases, polyploidy may be a driver of speciation. Heckard and Chuang (1975) published detailed chromosome counts for most species of North American Orobanche. The octoploid O. parishii subsp. brachyloba forms a clade nested within O. parishii subsp. parishii (Fig. 1), its likely tetraploid progenitor (ploidy assignment based on a chromosome base number of x = 12; for a more detailed discussion, see Schneeweiss et al., 2004b), or, if an allopolyploid, one of two parental lineages. Octoploid lineages have also been reported in O. cooperi and O. corymbosa subsp. corymbosa (but not O. ludoviciana). A full discussion of the systematics and taxonomy of these and other individual species is needed, but is beyond the scope of this paper.
Repeated dispersal to South America
Based on the nrDNA phylogeny, we find support for the longstanding hypothesis that O. chilensis is closely related to O. ludoviciana and O. multiflora (Beck, 1890), thereby contributing to the broadly recognized pattern of amphitropical disjunction between the Great Plains of North America and northern Chile/southern Peru (Wen and Ickert-Bond, 2009). Of the two other sampled Orobanche species from South America, O. tacnaensis, was resolved with O. chilensis, but the two samples of O. tarapacana Phil. formed a separate, earlier diverging lineage resulting from north to south dispersal. Phylogenetic placement of O. tarapacana is uncertain due to conflict between the nrDNA and cpDNA trees; O. tarapacana is sister to either the O. ludoviciana complex, the O. cooperi complex or perhaps a hybrid between the two (Figs 1 and 2).
Conclusions and future directions
Parasitic Orobanchaceae is becoming a model system for understanding plant parasitism at various levels of biological organization and scale (Joel et al., 2013; McNeal et al., 2013; Wicke et al., 2013; Yang et al., 2015). Our results emphasize the importance of host specificity and host-switching as a driver of evolutionary divergence in obligate plant parasites. We find evidence for twice as many host-specific lineages in O. sect. Gymnocaulis as recognized taxa, and denser sampling in other clades such as O. sect. Nothaphyllon is likely to uncover more. This robust understanding of fine-scale evolutionary relationships provides the necessary phylogenetic framework to develop a more natural classification for this group, and understand genetic, ecological, functional and life-history consequences of host–parasite associations more broadly.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of Table S1: vouchers, host associations and GenBank accession numbers for 163 Orobanche populations and outgroup taxa.
ACKNOWLEDGEMENTS
We thank the curatorial staff at the following herbaria for providing access to specimens and/or providing tissue for DNA sequencing. ACAD, ALA, ARIZ, CAS, CHSC, CONC, JEPS, MO, MT, NY, OSC, RSA, SD, SGO, UBC, UC, V, WIS, WS and WTU. We also thank Rafael Casanova and Derek Anderson for assistance in procuring collections, Dave Tank for advice on using the waxy locus, Harold Chun and Amanda Seng for laboratory assistance, Turner Collins and George Yatskievych for fruitful discussion, and Jakub Těšitel and one anonymous reviewer for their comments on an earlier version of the manuscript. Funding was provided by grants to A.C.S. from the Lawrence R. Heckard Endowment Fund of the Jepson Herbarium, the California Native Plant Society (CNPS) Doc Burr Grant, and the CNPS Santa Clara Valley Chapter. A.C.S. was also supported by an internal Berkeley Graduate Fellowship.
LITERATURE CITED
- Achey DM. 1933. A revision of the section Gymnocaulis of the genus Orobanche. Bulletin of the Torrey Botanical Club 60: 441–451. [Google Scholar]
- Beardsley PM, Olmstead RG. 2002. Redefining Phrymaceae: the placement of Mimulus, tribe Mimuleae, and Phryma. American Journal of Botany 87: 1093–1102. [DOI] [PubMed] [Google Scholar]
- Beck G. 1890. Monographe der Gattung Orobanche. Biblitoheca Botanica 19. Cassel: Theoder Fischer. [Google Scholar]
- Bellot S, Renner SS. 2013. Pollination and mating systems of Apodanthaceae and the distribution of reproductive traits in parasitic angiosperms. American Journal of Botany 100: 1083–1094. [DOI] [PubMed] [Google Scholar]
- Bolin JF, Maass E, Mussellman LJ. 2011. A new species of Hydnora (Hydnoraceae) from South Africa. Systematic Botany 36: 255–260. [Google Scholar]
- Cameron DC, Coats AM, Seel WE. 2006. Differential resistance among host and non-host species underlies the variable success of the hemiparasitic plant Rhinanthus minor. Annals of Botany 98: 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins LT. 1973. Systematics of Orobanche sect. Myzorrhiza. PhD thesis, University of Wisconsin–Milwaukee.
- Collins L.T., Yatskievych G. 2015. Orobanche arizonica sp. nov. and nomenclatural changes in Orobanche cooperi (Orobanchaceae). Phytoneuron 2015-48: 1–19. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computer. Nature Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vega C, Berjano R, Arista M, Ortiz PL, Talavera S, Stuessy TF. 2008. Genetic races associated with the genera and sections of host species in the holoparasitic plant Cytinus (Cytinaceae) in the Western Mediterranean basin. New Phytologist 178: 875–887. [DOI] [PubMed] [Google Scholar]
- dePamphilis CW, Young ND, Wolfe AD. 1997. Evolution of plastid gene rps2 in a lineage of hemiparasitic and holoparasitic plants: many losses of photosynthesis and complex patterns of rate variation. Proceedings of the National Academy of Sciences, USA 94: 7367–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin of the Botanical Society of America 19: 11–15. [Google Scholar]
- Drès M, Mallet J. 2002. Host races in plant-feeding insects and their importance in sympatric speciation. Proceedings of the Royal Society B: Biological Sciences 357: 471–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MW, Taylor RJ, Harrod RJ. 1999. The reproductive biology and host specificity of Orobanche pinorum Geyer (Orobanchaceae). Madroño 46: 7–12. [Google Scholar]
- Fernández-Aparicio M, Yoneyama K, Rubiales D. 2011. The role of strigolactones in host specificity of Orobanche and Phelipanche seed germination. Seed Science Research 21: 55–61. [Google Scholar]
- Goldwasser Y, Hershenhorn J, Plakhine D, Kleifeld Y, Rubin B. 1999. Biochemical factors involved in vetch resistance to Orobanche aegyptiaca. Physiolgical and Molecular Plant Pathology 54: 87–96. [Google Scholar]
- Goldwasser Y, Plakhine D, Kleifeld Y, Zamski E, Rubin B. 2000. The differential susceptibility of vetch (Vicia spp.) to Orobanche aegyptiaca: anatomical studies. Annals of Botany 85:257–262. [Google Scholar]
- Gurney AL, Grimanelli D, Kanampiu F, Hoisington D, Scholes JD, Press MC. 2003. Novel sources of resistance to Striga hermonthica in Tripsacum dactyloides, a wild relative of maize. New Phytologist 160: 557–568. [DOI] [PubMed] [Google Scholar]
- Gurney AL, Slate J, Press MC, Scholes JD. 2006. A novel form of resistance in rice to the angiosperm parasite Striga hermonthica. New Phytologist 169: 199–208. [DOI] [PubMed] [Google Scholar]
- Heckard LR. 1973. A taxonomic re-interpretation of the Orobanche californica complex. Madroño 22: 41–70. [Google Scholar]
- Heckard LR, Chuang TI. 1975. Chromosome numbers and polyploidy in Orobanche (Orobanchaceae). Brittonia 27: 179–186. [Google Scholar]
- Jenson HW. 1951. The normal and parthenogenic forms of Orobanche uniflora in the eastern United States. Cellule 54: 135–142. [PubMed] [Google Scholar]
- Joel DM, Gressel J, Musselman LJ, eds. 2013. Parasitic Orobanchaceae. Heidelberg: Springer. [Google Scholar]
- Jones M. 1989. Studies on the pollination of Orobanche species in the British Isles. Progress in Orobanche Research Proceedings. Tubingen, 6–17.
- Kellogg VL. 1913. Distribution and species-forming of ecto-parasites. American Naturalist 47: 129–158. [Google Scholar]
- Labrousse P, Arnaud MC, Serieys H, Bervillé A, Thalouarn P. 2001. Several mechanisms are involved in resistance of Helianthus to Orobanche cunana Wallr. Annals of Botany 88: 859–868. [Google Scholar]
- McNeal JR, Bennett JR, Wolfe AD, Mathews S. 2013. Phylogeny and origins of holoparasitism in Orobanchaceae. American Journal of Botany 100: 971–983. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE). 14 November 2010, New Orleans, Louisiana, USA.
- Munz PA. 1930. The North American species of Orobanche section Myzorrhiza. Bulletin of the Torrey Botanical Club 57: 611–624. [Google Scholar]
- Musselman LJ, Parker C, Dixon N. 1982. Notes on autogamy and flower structure in agronomically important species of Striga (Scrophulariaceae) and Orobanche (Orobanchaceae). Beiträge zur Biologie der Pflanzen 5: 329–343 [Google Scholar]
- Norton DA, Carpenter MA. 1998. Mistletoes as parasites: host specificity and speciation. Trends in Ecology and Evolution 13: 101–104. [DOI] [PubMed] [Google Scholar]
- Norton DA, De Lange PJ. 1999. Host specificity in parasitic mistletoes (Loranthaceae) in New Zealand. Functional Ecology 13: 552–559. [Google Scholar]
- Parfitt BD, Butterwick M. 1981. Noteworthy collections: Orobanche uniflora L. subsp. occidentalis. Madroño 28: 37–38. [Google Scholar]
- Park J-M, Manen J-F, Colwell AE, Schneeweiss GM. 2008. A plastid gene phylogeny of the non-photosynthetic parasitic Orobanche (Orobanchaceae) and related genera. Journal of Plant Research 121: 365–376. [DOI] [PubMed] [Google Scholar]
- Pérez-de-Luque A, Jorrín J, Cubero JI, Rubiales D. 2005a. Orobanche crenata resistance and avoidance in pea (Pisum spp.) operate at different developmental stages of the parasite. Weed Research 45: 379–387. [Google Scholar]
- Pérez-de-Luque A, Rubiales D, Cubero JI, et al. 2005b. Interaction between Orobanche crenata and its host legumes: unsuccessful haustorial penetration and necrosis of the developing parasite. Annals of Botany 95: 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-de-Luque A, Moreno MT, Rubiales D. 2008. Host plant resitance against broomrapes (Orobanche spp.): defence reactions and mechanisms of resistance. Annals of Applied Biology 152: 131–141. [Google Scholar]
- Poulin R, Morand S. 2000. The diversity of parasites. Quarterly Review of Biology 75: 277–293. [DOI] [PubMed] [Google Scholar]
- Press M, Graves J, eds. 1995. Parasitic plants. London: Chapman & Hall. [Google Scholar]
- Refrégier G, Le Gac M, Jabbour F, et al. 2008. Cophylogeny of the anther smut fungi and their caryophyllaceous hosts: prevalence of host shifts and importance of delimiting parasite species. BMC Evolutionary Biology 8: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter BC. 1986. The habitat, reproductive ecology, and host relations of Orobanche fasciculata Nutt. (Orobanchaceae) in Wisconsin. Bulletin of the Torrey Botanical Club 113: 110–117. [Google Scholar]
- Ricklefs RE, Fallon SM, Bermingham E. 2004. Evolutionary relationships, cospeciation, and host switching in avian malaria parasites. Systematic Biology 53: 111–119. [DOI] [PubMed] [Google Scholar]
- Ronquist FM, Teslenko P, van der Mark DL, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneeweiss GM. 2007. Correlated evolution of life history and host range in the nonphotosynthetic parasitic flowering plants Orobanche and Phelipanche (Orobanchaceae). Journal of Evolutionary Biology 20: 471–478. [DOI] [PubMed] [Google Scholar]
- Schneeweiss GM. 2013. Phylogenetic relationships and evolutionary trends in Orobanchaceae In: DM Joel, J Gressel, LJ Mussleman, eds. Parasitic Orobanchaceae. Berlin: Springer, 243–265. [Google Scholar]
- Schneeweiss GM, Colwell AEL, Park J-M, Jang C-G, Stuessy TF. 2004a. Phylogeny of holoparasistic Orobanche (Orobanchaceae) inferred from nuclear ITS sequences. Molecular Phylogenetics and Evolution 30: 465–478. [DOI] [PubMed] [Google Scholar]
- Schneeweiss GM, Palomeque T, Colwell AE, Weiss-Schneeweiss H. 2004b. Chromosome numbers and karyotype evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. American Journal of Botany 91: 439–448. [DOI] [PubMed] [Google Scholar]
- Shaw J, Lickey EB, Beck JT, et al. 2005. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. American Journal of Botany 92: 142–166. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogeneies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele KP, Vilgalys R. 1994. Phylogenetic analyses of Polemoniaceae using nucleotide sequences of the plastid gene matK. Systematic Botany 19: 126–142. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. 1991. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology 17: 1105–1109. [DOI] [PubMed] [Google Scholar]
- Tank DC, Olmstead RC. 2008. From annuals to perennials: phylogeny of subtribe Castillejinae (Orobanchaceae). American Journal of Botany 95: 608–625. [DOI] [PubMed] [Google Scholar]
- Tank DC, Olmstead RG. 2009. The evolutionary origin of a second radiation of annual Castilleja (Orobanchaceae) speceis in South America: the role of long distance dispersal and allopolyploidy. American Journal of Botany 96: 1907–1921. [DOI] [PubMed] [Google Scholar]
- Thorogood CJ, Hiscock SJ. 2010. Compatibility interactions at the cellular level provide the basis for host specificity in the parasitic plant Orobanche. New Phytologist 186: 571–575. [DOI] [PubMed] [Google Scholar]
- Thorogood CJ, Rumsey FJ, Harris SA, Hiscock J. 2008. Host-driven divergence in the parasitic plant O. minor. Molecular Ecology 17: 4289–4303. [DOI] [PubMed] [Google Scholar]
- Thorogood CJ, Rumsey FJ, Hiscock SJ. 2009. Host-specific races in the holoparasitic angiosperm Orobanche minor: implications for speciation in parasitic plants. Annals of Botany 103: 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timko MP, Huang K, Lis KE. 2012. Host resistance and parasite virulence in Striga–host plant interactions: a shifting balance of power. Weed Science 60: 307–315. [Google Scholar]
- Uhlich H, Pusch J, Barthel KJ. 1995. Die Sommerwurzarten Europas. Magdeburg: Westarp Wissenschaften. [Google Scholar]
- de Vienne DM, Refrégier G, López-Villavicencio M, Tellier A, Hood ME, Giraud T. 2013. Cospeciation vs host-shift speciation: methods for testing, evidence from natural associations and relation to coevolution. New Phytologist 198: 347–385. [DOI] [PubMed] [Google Scholar]
- Watson KC. 1975. Systematics of Orobanche section Gymnocaulis (Orobanchaceae). MS thesis, California State University Chico.
- Wen J, Ickert-Bond SM. 2009. Evolution of the Madrean–Tethyan disjunctions and the North American amphitropical disjunctions in plants. Journal of Systematics and Evolution 47: 331–348. [Google Scholar]
- Westwood JH, Yoder JI, Timko MP, dePamphilis CW. 2010. The evolution of parasitism in plants. Trends in Plant Science 15: 227–235. [DOI] [PubMed] [Google Scholar]
- Wicke S, Müller KF, de Pamphilis CW, et al. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants in the broomrape family. The Plant Cell 25: 3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Yoneyama K, Kisugi T. 2010. The strigolactone story. Annual Review of Phytopathology 48: 93–117. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wafula EK, Honaas LA, et al. 2015. Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Molecular Biology and Evolution 32: 767–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder J. 1997. A species-specific recognition system directs haustorium development in the parasitic plant Tryphysaria (Scrophulariaceae). Planta 202: 407–413. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Shirasu K. 2009. Multiple layers of incompatibility to the parasitic witchweed, Striga hermonthica. New Phytologist 183: 180–189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.