Abstract
The increasing incidence of resistance to current HIV-1 therapy underscores the need to develop antiretroviral agents with new mechanisms of action. Integrase, one of three viral enzymes essential for HIV-1 replication, presents an important yet unexploited opportunity for drug development. We describe here the identification and characterization of L-870,810, a small-molecule inhibitor of HIV-1 integrase with potent antiviral activity in cell culture and good pharmacokinetic properties. L-870,810 is an inhibitor with an 8-hydroxy-(1,6)-naphthyridine-7-carboxamide pharmacophore. The compound inhibits HIV-1 integrase-mediated strand transfer, and its antiviral activity in vitro is a direct consequence of this ascribed effect on integration. L-870,810 is mechanistically identical to previously described inhibitors from the diketo acid series; however, viruses selected for resistance to L-870,810 contain mutations (integrase residues 72, 121, and 125) that uniquely confer resistance to the naphthyridine. Conversely, mutations associated with resistance to the diketo acid do not engender naphthyridine resistance. Importantly, the mutations associated with resistance to each of these inhibitors map to distinct regions within the integrase active site. Therefore, we propose a model of the two inhibitors that is consistent with this observation and suggests specific interactions with discrete binding sites for each ligand. These studies provide a structural basis and rationale for developing integrase inhibitors with the potential for unique and nonoverlapping resistance profiles.
Agents for the treatment of HIV-1 infection target two of the three virally encoded enzymes and belong to three mechanistic classes known as nucleoside reverse transcriptase, nonnucleoside reverse transcriptase (NNRTI), and protease inhibitors. Although treatment regimens comprising combinations of these agents have significantly reduced AIDS-related morbidity and mortality, it is estimated that 78% of treatment-naive patients harbor viruses that have evolved resistance to at least one of these drug classes (1, 2). The emergence of HIV-1 strains resistant to reverse transcriptase and protease inhibitors highlights the need to develop antiviral agents with novel mechanisms of action.
Integrase (3, 4), one of the three virally encoded enzymes required for HIV-1 replication, catalyses the integration of viral DNA into the genome of the host cell. The integration reaction requires three discrete steps: assembly of a stable preintegration complex at the termini of the viral DNA and two sequential transesterification reactions. In the first reaction, 3′-end processing, endonucleolytic cleavage of the two 3′ nucleotides at each DNA end generates 3′-hydroxyl groups that function as nucleophiles in the second reaction. The strand breakage of the cellular DNA and concomitant covalent linkage to the viral DNA is a consequence of the second transesterification reaction, strand transfer.
The discovery of a series of diketo acids containing HIV-1 integrase inhibitors (5, 6), exemplified by L-731,988 (1), (Fig. 1) that prevent integration and viral replication in cell culture provided the first proof of concept for HIV-1 integrase inhibitors as antiviral agents. These compounds were shown to act specifically as inhibitors of the integrase strand transfer reaction by virtue of their ability to bind selectively to the enzyme complexed with the viral (or donor) DNA and compete with the host (or target) DNA substrate (7, 8). The critical diketo carboxylate pharmacophore interacts with critical divalent metal ions in the active site (8) and mutations that engender resistance to these prototype inhibitors (5, 9), and closely related analogs map to the integrase active site proximal to residues that coordinate divalent metals (D66, D116, and E152). These observations are consistent with the proposed mechanism of action and provide further validation for integrase as the target of their antiviral effect (5). Subsequently, a series of rational structure activity relationships have afforded a variety of such analogs with improved activities (6, 10–13).
Fig. 1.
Evolution of L-870,810 from the diketo acids L-731,988 1 (5) and 2 (6).
Herein we describe the identification and characterization of an inhibitor, L-870,810 that contains a structurally distinct 8-hydroxy-(1,6)-naphthyridine-7-carboxamide pharmacophore. L-870,810 has a mechanism of action that is indistinguishable from the diketo acids, displays potent antiviral activity, and has pharmacokinetic properties suitable for clinical development. Although both the biochemical and antiviral activities of L-870,810 are analogous to the diketo acids, we provide evidence that these inhibitors exhibit discordant sensitivity to resistance mutations. Exploration of the structural basis for this unexpected result provides insights into this class of antiviral agents and suggests an approach to the development of integrase inhibitors with unique resistance profiles.
Materials and Methods
Preparation of L-870,810 and Analogs. The synthesis of L-870,810 and analogs is described in ref. 14. The compound may be prepared as an anhydrous white powder or as a yellow anhydrous crystalline sodium salt (mp 338°C). The ionization constant of the phenol group was determined by spectrophotometric titration in aqueous methanol. The aqueous pKa was estimated to be 7.3 by extrapolation to zero organic solvent content. Additionally, the octanol:water partition coefficient, log P, was determined by HPLC analysis to be 2.09.
HIV-1 Integrase Assays. HIV-1 and simian immunodeficiency virus (SIV) integrase were purified as described in ref. 15. Rous sarcoma virus integrase was the generous gift of Duane Grandgenett (Saint Louis University, St. Louis). Integrase-mediated strand transfer activity was determined as described in ref. 12. Biotinylated donor DNA was immobilized onto streptavidin-coated microtiter plates. Integrase was assembled onto the donor in 20 mM Hepes (pH 7.6)/40 mM NaCl/5 mM 2-mercaptoethanol/50 μg/ml BSA with 25 mM MnCl2. The complexes were washed before the addition of a target substrate 3′-end-labeled with fluorescein isothiocyanate. Strand transfer reactions were performed in 2.5 mM MgCl2 by using 0.5 or 5 nM target DNA. Fluorescein isothiocyanate-labeled products were detected by using an antifluorescein isothiocyanate antibody conjugated with alkaline phosphatase (Roche Molecular Biochemicals) and a chemiluminescence substrate (Sapphire II, Tropix, Bedford, MA). Inhibition of strand transfer was evaluated by adding compounds immediately before the addition of target substrate.
Competitive Binding. 3H-L-731,988 (1) {4-[1-(4-fluorobenzyl)pyrrole-2-yl-2,4-diketobutanoic acid} was prepared by catalytic tritiation of the corresponding iodopyrrole. Competition binding experiments were performed as described in ref. 8.
HIV-1 Replication Assays. Assays were performed in MT-4 human T-lymphoid cells or primary PBMCs as described in ref. 16. Cells were infected en masse at low multiplicity (0.01). HIV-1 replication was assessed by p24 core antigen ELISA. Single-cycle infection assays were performed as described in ref. 5.
Measurement of HIV-1 DNA Synthesis, Integration, and 2-LTR Circles. Viral DNA synthesis and 2-LTR circles were evaluated by using quantitative PCR (TaqMan) (5). SupT1 cells were preincubated in the presence or absence of 10 μM L-731,988, 1 μM L-870,810, or 10 μM L-697,661 (NNRTI) for 1 h at 37°C and then infected with HIV-1 by coculture with the chronic producer line Molt-IIIB. Low-molecular-weight DNA was harvested at various times to quantify each HIV-1 product. Late reverse transcripts were amplified with primers and probes specific for DNA made after the second template switch of reverse transcription. Quantification of reverse transcription products at 6 h postinfection was performed relative to standards by using the HIV-1 proviral plasmid pNL4–3. HIV-1 2-LTR circles were evaluated at 24 h postinfection with a 2-LTR primer/probe set and standards generated with a cloned DNA fragment corresponding to a 2-LTR circle junction of HIV-1 DNA (154 bp from the U5 region and 408 bp from the U3 region). To determine the fraction of total HIV-1 DNA molecules in the 2-LTR circular form, the number of 2-LTR circles was divided by the number of total HIV-1 DNA molecules. Extracts were normalized for overall DNA recovery by assessing mitochondrial DNA with a primer/probe set for cytochrome oxidase.
Quantitative PCR was also used to evaluation integration in the presence or absence of inhibitors at 24 h postinfection by assessing integration events that occur near the repetitive genomic DNA element Alu with an Alu-LTR primer/probe set (17). To quantify relative integration events, genomic DNA from infected control cultures was serially diluted with a constant amount of DNA from uninfected cells to establish a standard curve.
Animal Pharmacokinetic Studies. The pharmacokinetic parameters of L-870,810 were determined in rats, dogs, and rhesus macaques. Three male Sprague–Dawley rats received an i.v. dose of 2 mg·kg–1 as a solution in DMSO via an indwelling cannula implanted in the jugular vein. After an overnight fast, three additional animals received an oral dose of 10 mg·kg–1 as a suspension in 1% aqueous methylcellulose. Similar procedures were used to evaluate pharmacokinetic parameters in dogs and rhesus macaques with four animals per group receiving i.v. and oral doses in a crossover fashion. Following an overnight fast, each animal received an oral dose of L-870,810 at 1 mg·kg–1 as a suspension in 0.5% aqueous methylcellulose. After a washout period, the animal received an i.v. dose of L-870,810 (1 mg·kg–1) as a solution in DMSO (0.1 mg·kg–1). L-870,810 plasma concentrations were determined by liquid chromatography MS analysis, and absolute oral bioavailability was determined by comparing the mean area under the plasma concentration–time curve obtained from each group.
Molecular Modeling. Fifty conformers of L-870,810 and the diketo acid (2) were generated by using a proprietary distance geometry program based on the theory and algorithms in refs. 18 and 19. The conformers were subsequently energy-minimized with the MMFF force field (20–24) by using a distance-dependent dielectric constant (2r). The potential surface for the diketo acid (2) was quite flat, exhibiting a conformational energy range of only ≈2 kcal/mol. The potential surface for L-870,810 was more varied with a total conformational energy range of ≈15 kcal/mol. However, the energy distribution was uneven, with five conformers within a 1 kcal/mol range and the remainder in the range of 5–15 kcal/mol above the computed minimum. Thus, a representative low-energy conformer of each inhibitor was selected to illustrate the proposed overlays of the metal chelating pharmacophore elements as depicted in Fig. 4. The selection was supported by strong agreement between the selected low-energy conformer of L-870,810 and a small-molecule x-ray structure (data not shown). The selected conformer of each inhibitor was then manually docked into the active site of the C monomer of the 1BL3 HIV-1 integrase crystal structure (25). The keto and enol oxygens of 2 and the keto and 8-hydroxy oxygens of L-870,810 were aligned with two of the Mg-coordinated water molecules (11 and 149) in the crystal structure. Additionally, the carboxylate oxygen in 2 and nitrogen of the L-870,810 naphthyridine ring were positioned to interact with a putative second Mg positioned between D64 and E152. The positions of the pendant aromatic groups were modified slightly to optimize contacts with the active site surface.
Fig. 4.
Modeling of the diketo acid and 8-hydroxy-(1,6)-naphthyridine-7-carboxamide pharmacophores suggests two potential binding modes. Low-energy conformers of diketo acid 2 (green) and L-870,810 (magenta) with the pharmacophores aligned in the forward (A) and reverse (B) orientations. (C) The two inhibitors in the reverse orientation are docked in the active site, aligning the metal-binding pharmacophores with the two putative magnesium ions bound between positions D64 and D116 and between D116 and E152, respectively.
Results
L-870,810 Is a Structurally Unique, Orally Previously Bioavailable Inhibitor of Integrase-Mediated Strand Transfer. We reported the discovery of a series of diketo acid-containing HIV-1 integrase inhibitors (5, 6), exemplified by L-731,988 and the potent analog (2) (Fig. 1). These compounds bind selectively to the enzyme/viral DNA complex and act specifically as inhibitors of the strand transfer reaction. Structure–activity relationships for the diketo acid inhibitors have been discerned, and it was found that the carboxylate could be mimicked by a suitable heterocycle bearing a lone pair donor atom (26, 27), affording diketone-based inhibitors such as 3. Although the 1,3-diketone motif common to these compounds is a key element of the pharmacophore required for inhibitory activity, the chemical reactivity of this functionality provoked efforts to reduce its electrophilicity. In the 1,6-naphthyridine ketone (4) (12), one of the enolizable ketones is replaced with a phenolic-hydroxyl group. The remaining phenyl ketone was then replaced with a 4-fluorobenzyl carboxamide moiety. Further refinement with respect to in vitro antiviral potency (discussed below) as well as pharmacologic properties ultimately identified a six-membered cyclic sulfonamide as an optimal substituent at the 5-position of the 8-hydroxy-(1,6)-naphthyridine carboxamide core providing the inhibitor L-870,810 (14). L-870,810 has an oral bioavailability of 41%, 24%, and 51% and is characterized by low plasma clearance of 2.8, 2.0, and 6.6 ml·min–1·kg–1 in rats, dogs, and rhesus, respectively. Oral administration of L-870,810 can thus achieve 12 h through plasma concentrations that exceed the IC95 against HIV-1 measured in human serum.
L-870,810 is a potent inhibitor of HIV-1 integrase and the highly homologous enzyme from SIV but is inactive against the more distantly related Rous sarcoma virus integrase (IC50 > 50 μM) (data not shown). Like the diketo acids, L-870,810 is a selective inhibitor of HIV-1 integrase-mediated strand transfer. When assayed with purified recombinant HIV-1 integrase, L-870,810 inhibits strand transfer with an apparent IC50 of 8 or 15 nM when using 0.5 nM and 5 nM target DNA, respectively. L-870,810 exhibits reduced activity with respect to assembly and 3′-end processing (IC50 of 85 and 250 nM in 0.5 and 5 nM target DNA). The preferential inhibition of strand transfer and the sensitivity of L-870,810 to the concentration of target substrate are consistent with studies (7) suggesting that these inhibitors bind to the target DNA site of the integration complex. In competition-binding experiments, L-870,810 displaces radiolabeled L-731,988 (1) from the integrase donor complex with a Ki of 3 nM (data not shown), indicating that these inhibitors bind to the assembled DNA complex within the same or overlapping regions of the active site.
L-870,810 Exhibits Broad-Spectrum Activity Against WT and Multidrug-Resistant HIV-1, HIV-2, and SIV. The antiviral activity of L-870,810 was profiled in viral-replication assays by using different cell types and a variety of M- and T-tropic isolates of HIV-1. In the presence of 10% FBS or 50% normal human serum, the compound inhibits the replication of the laboratory-adapted HIV-1 isolate H9/IIIB in MT-4 T lymphoid cells with a mean IC95 of 15 and 100 nM, respectively. L-870,810 also inhibits HIV-1 clinical isolates and exhibits comparable activity against nonsyncytia and syncytia viruses from clades A–D and F. As expected, L-870,810 is active against multidrug-resistant viruses such as MDRC4 (IC50 = 4 nM), which has multiple mutations in reverse transcriptase and protease and exhibits ≥5-fold resistance to most nucleoside reverse transcriptases, NNRTIs, and protease inhibitors (N. Parkin, personal communication). Consistent with the observation that L-870,810 inhibits SIV integrase, L-870,810 inhibits the replication of both SIV (IC95 of 7–15 nM in 10% FBS) and the highly homologous virus HIV-2 (IC95 of 8 nM in 10% FBS). The potential for efficacy in HIV-2 infection is of interest because most current antiretroviral agents are poorly active against this human pathogen.
Antiviral Activity of L-870,810 Is a Direct Consequence of Its Effect on Integration. The mechanism of action of L-870,810 on HIV-1 replication was evaluated by assessing DNA synthesis and integration in infected cells by quantitative PCR. SupT1 cells were acutely infected by coculturing the cells with the HIV-1 chronic producer Molt-IIIB cells in the presence or absence of L-870,810, the diketo acid L-731,988, and the NNRTI, L-697,661 (28). HIV-1-specific products were quantified and normalized relative to mitochondrial DNA. Neither L-870,810 nor the diketo acid affected reverse transcription at concentrations 10-fold greater than that required to inhibit replication (data not shown). In contrast, both L-870,810 and the diketo acid reduce the integrated HIV-1 DNA to undetectable levels and increase 2-LTR circles; at 24 h postinfection, a 5.9-fold increase in 2-LTR circles was observed with L-870,810 (Fig. 2). Neither integrated HIV-1 DNA nor unintegrated 2-LTR circle DNAs were detected in the presence of the NNRTI. As shown for the diketo acids, the absence of integration products and the accumulation of 2-LTR circles provide evidence that the antiviral activity of L-870,810 is a direct consequence of its effect on integration.
Fig. 2.
L-870,810 inhibits integration and increases the number of 2-LTR circles in HIV-1-infected cells. Integration products and 2-LTR circles in HIV-1-infected cells were evaluated in the presence or absence of an NNRTI, a diketo acid L-731,988, and L-870,810 by RT-PCR. All samples were standardized to a mitochondrial DNA control and quantified relative to known standards as described in Materials and Methods.
L-870,810-Resistant Variants Contain Unique Active-Site Mutations and Are Not Cross-Resistant to Diketo Acids. Like the diketo acid inhibitors, serial passage of HIV-1 in cell culture in the presence of L-870,810 selects viruses that exhibit reduced susceptibility to the inhibitor and accumulate mutations in integrase. Population sequencing of the integrase coding region in multiple clones intermittently during selection with L-870,810 identified mutations that were acquired sequentially over several months: F121Y/T125K (6 months) and V72I/F121Y/T125K and V72I/F121Y/T125K/V151I (three of eight and five of eight clones, respectively, after 9 months). Although the L-870,810 mutations map within the same region of integrase (see below), they are distinct from the mutations observed with diketo acid analogs [e.g., L-731,988 and L-708,906, T66I/S153Y, T66I/M154I (5); or 2, T66I/S153Y and N155S] (M.V.W. and D.J.H., unpublished data).
A panel of viruses containing the diketo acid resistance mutations and the L-870,810 mutations were constructed and evaluated for their susceptibility to L-870,810 and diketo acid (2) (Table 1). Viruses containing mutations selected by L-870,810 were 4- to 100-fold less sensitive to the inhibitor, and resistance was enhanced with the addition of those mutations accumulated during selection. However, in every case, the L-870,810 resistant viruses were sensitive to the diketo acid. Conversely, with the exception of N155S, the mutations selected with the diketo acid engendered resistance to this inhibitor without affecting L-870,810 activity. Similarly discordant profiles were also observed with other diketo acid and naphthyridine analogs (data not shown).
Table 1. Activity of L-870,810 and diketo acid (2) in single-cycle infection assays with HIV-1 containing site-directed mutations in integrase.
| Mutation susceptibility, fold
|
|||
|---|---|---|---|
| Mutations | Diketo acid (2) | L-870,810 | Sustiva |
| L-870,810-selected mutations | |||
| F121Y | 1 | 4 | 1 |
| T125K | 1 | 1 | 1 |
| V151I | 1 | 1 | 1 |
| F121Y/T125K | 1 | 16 | 1 |
| V72I/F121Y/T125K | 1 | 20 | 1 |
| V72I/F121Y/T125K/V151I | 3 | 100 | 1 |
| Diketo acid-selected mutations | |||
| T66I | 1 | 2 | 1 |
| S153Y | 4 | 1 | 1 |
| M154I | 1 | 1 | 1 |
| T66I/M154I | 1 | 2 | 1 |
| T66I/S153Y | 8 | 2 | 1 |
| N155S | 20 | 12 | 1 |
A ≥4-fold value indicates a significant loss of susceptibility vs. WT HXB2, shown in bold.
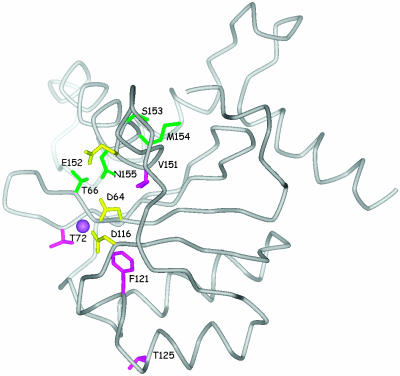
The lack of overt cross-resistance observed with the 8-hydroxy-(1,6)-naphthyridine carboxamides and diketo acids, although surprising, is entirely consistent with selection of distinct resistance mutations. Mapping of these residues within the integrase crystal structure is shown in Fig. 3. The active-site residues (D64, D116, and E152 in yellow in fig. 3) are believed to coordinate two magnesium ions, which interact with the pharmacophores in these inhibitors. Both the diketo acid and L-870,810 mutations surround the metal-binding site, consistent with the proposed mechanism of action. However, residues that are uniquely associated with either L-870,810 (Fig. 3, magenta) or diketo acid (Fig. 3, green) resistance cluster on distinct sides of the active site. Interestingly, N155S, the only mutation that conferred significant cross-resistance, points directly into the active site and hydrogen bonds with the critical metal-binding residue D116. A mutation of N155 could directly affect the interaction with both inhibitors by disrupting metal binding. This finding is consistent with the observation that the N155S mutant is impaired for enzymatic function and viral replication (<30% WT HIV in infectivity assays) (data not shown).
Fig. 3.
Mutations associated with diketo acid and L-870,810 resistance map to the integrase active site. The 3D structure (25) of HIV-1 integrase is depicted as a α-carbon pipe. The active site residues (D64, D116, and E152), highlighted in yellow, are believed to coordinate two divalent metals, although only one, shown as a purple ball, is evident in this structure. Residues associated with resistance to diketo acids 1 and 2 are shown in green; those associated with resistance to L-870,810 are shown in magenta.
The majority of mutations are uniquely associated with resistance to one inhibitor and map more distal to the metal-binding residues (Fig. 3). Moreover, the mutations associated with each inhibitor map to discrete sites in the crystal structure, indicating that their respective interactions extend beyond the metal-binding center in opposing directions within the active site and suggest that the two compounds may bind in opposite orientations. Assuming that the diketo acid and 8-hydroxy-(1,6)-naphthyridine carboxamide moieties form a metal-chelating pharmacophore, low-energy models of L-870,810 and diketo acid (2) can be overlaid in a “forward” or “reverse” direction, as illustrated in Fig. 4 A and B. In the reverse overlay, there is overlap of the metal-binding pharmacophores but not of the pendant substituents. The two molecules were docked in the active site such that the two pharmacophores could engage magnesium ions bound between D64 and D116 and between D116 and E152, respectively (Fig. 4C). In this model the overlay, which places the two inhibitors in the reverse orientation, is the most consistent with both a similar binding mode for the two inhibitors and a divergent pattern of resistant mutations.
Discussion
We have described the identification and characterization of L-870,810, an orally bioavailable inhibitor of integrase that inhibits HIV-1 replication through its effect on viral integration. Based on the compound's potent activity against diverse clinical isolates and drug-resistant HIV-1 in vitro and a promising preclinical pharmacokinetic profile, L-870,810 exemplifies a class of agents that target HIV-1 integrase with the potential for clinical utility. L-870,810 is also the prototype of a structural class of 8-hydroxy-1,6-naphthyridine-7-carboxamides, which provide a previously uncharacterized pharmacophore for inhibitor design. Like previously described diketo acids, the 8-hydroxy-1,6-naphthyridine-7-carboxamides selectively inhibit integrase-mediated strand transfer (5, 7). Although these inhibitors are mechanistically indistinguishable, we have shown that distinct resistance profiles can be obtained with homologous naphthyridine and diketo acid analogs, providing the proof of concept and rationale for designing integrase inhibitors with the potential for unique and nonoverlapping resistance.
We have explored the molecular basis for the discordant resistance profiles of the naphthyridine and diketo acid, and we suggest that, although the two pharmacophores engage divalent metals within the integrase active site, they may bind in opposite orientations. The 8-hydroxy-(1,6)-naphthyridine carboxamide conserves the spatial orientation of the chelating moieties in the diketo acid that have been shown to be essential for activity, and this spacing is consistent with the distance between the two active-site metals observed in the crystal structure of ASV integrase and other divalent metal-dependent phosphotransferases (8). Although in some cases resistance may be mediated by mutations that can affect the metal-coordinating residues in integrase and alter the geometry of the active site (e.g., mutations at residues 155 or 66), other residues associated with resistance are more distal to the metal-binding site. The observation that the latter mutations can be selective for different inhibitors and map to distinct regions within the integrase active site suggests that these inhibitor–enzyme interactions may extend beyond the pharmacophore in different directions.
Molecular modeling studies of low-energy conformers indicate the essential elements of the diketo acid and 8-hydroxy-(1,6)-naphthyridine-7-carboxamide pharmacophores can align equally well in either direction. However, the distinct resistance profiles of the compounds would be most consistent with the two pharmacophores binding in reverse orientations, fixing the respective substituents in opposing directions. The proposed model, which places the two inhibitors in the reverse orientation, is also consistent with the structure–activity relationships observed for structurally related analogs in both inhibitor series (data not shown). The precise orientation of each inhibitor in the active site therefore may be the result of establishing the most favorable interactions of the specific substituents and accommodating the increased rigidity and bulkiness of the naphthyridine relative to the more flexible diketo acid. The suggestion that affinity and specificity is driven by the pendant groups in these molecules is consistent with studies that have shown that diketone analogs, which lack a carboxylate, bind with relatively high affinity even though they do not inhibit integrase enzymatic activity (8), and with the observation that pharmacophores in which substituents are extended in both directions can exhibit enhanced affinity (data not shown). In addition, although the respective structure–activity relationships are quite distinct, the adaptable diketo acid pharmacophore can be exploited as a template for inhibitors of a variety of divalent metal-dependent phosphotransferases, such as HIV-1 RNase H (29) and hepatitis C virus polymerase (30). In contrast, the more rigid naphthyridines appear to be particularly restricted to integrase (S.D.Y. and D.J.H., unpublished data). These observations are interesting in the context of published molecular dynamics simulations with a diketo tetrazole inhibitor, 5CITEP, that have suggested the potential for the existence of two conformations of this flexible inhibitor in the docked structure and two possible ligand-binding regions adjacent to the integrase active site (31).
High-affinity interaction of strand transfer inhibitors with integrase requires a specific DNA bound conformation (7), and obtaining a structure of the inhibitor-bound integrase complex with DNA has not been achieved to date. Whereas crystal structures of various integrase subdomains have been published (25, 32, 33), the active site has proven highly flexible, and the structure in the region most relevant for the interaction with these inhibitors is not precisely known. The suggestion that the active site adopts a unique conformation upon binding the DNA is consistent with the observation that in the proposed model the residues associated with resistance are located proximal to the respective inhibitors but do not make direct contact with each molecule. The lack of structural information with sufficient detail to facilitate the design of integrase inhibitors therefore continues to present a substantial challenge. Although we do not have accurate structural information, resistance profiling and the association of selected mutations with specific inhibitors provide valuable insights into how these inhibitors may interact with the integrase active site. The observation that different classes of integrase inhibitors both select for and are influenced by unique mutations indicates that high-affinity molecules can be obtained, wherein the critical interactions are established in different regions of the active site, and suggests a molecular basis for developing novel integrase inhibitors with discordant resistance profiles.
Acknowledgments
We thank C. Petropolous and N. Parkin for assistance in evaluating multidrug-resistant variants.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NNRTI, nonnucleoside reverse transcriptase; SIV, simian immunodeficiency virus.
References
- 1.Richman, D. D. (2001) Nature 410, 995–1001. [DOI] [PubMed] [Google Scholar]
- 2.Little, S. J., Holte, S., Routy, J.-P., Daar, E. S., Markowitz, M., Collier, A. C., Koup, R. A., Mellors, J. W., Connick, E., Conway, B., et al. (2002) N. Engl. J. Med. 347, 385–394. [DOI] [PubMed] [Google Scholar]
- 3.Esposito, D. & Craigie, R. (1999) Adv. Virus Res. 52, 319–333. [DOI] [PubMed] [Google Scholar]
- 4.Asante-Appiah, E. & Skalka, A. M. (1999) Adv. Virus Res. 52, 351–369. [DOI] [PubMed] [Google Scholar]
- 5.Hazuda, D. J., Felock, P., Witmar, M., Wolfe, A., Stillmock, K., Grobler, J. A., Espeseth, A., Gabryelski, L., Schleif, W., Blau, C. & Miller, M. D. (2000) Science 287, 646–650. [DOI] [PubMed] [Google Scholar]
- 6.Wai, J. S., Egbertson, M. S., Payne, L. S., Fisher, T. E., Embrey, M. W., Tran, L. O., Melamed, J. Y., Langford, H. M., Guare, J. P., Jr., Zhuang, L., et al. (2000) J. Med. Chem. 43, 4923–4926. [DOI] [PubMed] [Google Scholar]
- 7.Espeseth, A. S., Felock, P., Wolfe, A., Witmer, M., Grobler, J., Anthony, N., Egbertson, M., Melamed, J. Y., Young, S., Hamill, T., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 11244–11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grobler, J. A., Stillmock, K., Hu, B., Witmer, M., Felock, P., Espeseth, A. S., Wolfe, A., Egbertson, M., Bourgeois, M., Melamed, J., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 6661–6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fikkert, V., Van Maele, B., Vercammen, J., Hanston, A., Van Remoortel, B., Michiels, M., Gurnari, C., Pannecouque, C., De Maeyer, M., Engelborghs, Y., et al. (2003) J. Virol. 77, 11459–11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payne, L. S., Tran, L. O., Zhuang, L. H., Young, S. D., Egbertson, M. S., Wai, J. S., Embrey, M. W., Fisher, T. E., Guare, J. P., Langford, H. M., et al. (2001) PCT Int. Appl. 236, WO 01/0057A1.
- 11.Fujshita, T., Yoshinaga, T. & Sato, A. (2000) PCT Int. Appl. 554, WO 00/3086A1.
- 12.Zhuang, L., Wai, J. S., Embrey, M. W., Fisher, T. E., Egbertson, M. S., Payne, L. S., Guare J. P., Jr., Vacca, J. P., Hazuda, D. J., Felock, P. J., et al. (2003) J. Med. Chem. 46, 453–456. [DOI] [PubMed] [Google Scholar]
- 13.Pluymers, W., Paism, G., Pannecouque, C., Fikkert, V., Pommier, Y., Burke, T. R., De Clercq, E., Witvrouw, M., Neamati, N. & DeBeyser, Z. (2002) Antimicrob. Agents Chemother. 46, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anthony, N. J., Gomez, R. P., Young, S. D., Egbertson, M., Wai, J. S., Zhuang, L., Embrey, M., Tran, L., Melamed, J. Y., Langford, H. M., et al. (2002) PCT Int. Appl. 97, WO 02/30931-A2.
- 15.Hazuda, D. J., Felock, P., Hastings, J. C., Pramanik, B. & Wolfe, A. L. (1997) J. Virol. 71, 7005–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vacca, J. P., Dorsey, B. D., Schleif, W. A., Levin, R. B., McDaniel, S. L., Darke, P. L., Zugay, J., Quintero, J. C., Blahy, O. M., Roth, E., et al. (1994) Proc. Natl. Acad. Sci. USA 91, 4096–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler, S. L., Hansen, M. S. T. & Bushman, F. D. (2001) Nat. Med. 7, 631–633. [DOI] [PubMed] [Google Scholar]
- 18.Crippen, C. M. & Havel, T. F. (1988) Distance Geometry and Molecular Conformation, ed. D. Bawden, (Wiley, New York).
- 19.Kuszewski, J., Nilges, M. & Brunger, A. T. (1992) J. Biomol. NMR 2, 33–56. [DOI] [PubMed] [Google Scholar]
- 20.Halgren, T. A. (1996) J. Comput. Chem. 17, 490–586. [Google Scholar]
- 21.Halgren, T. A. (1996) J. Comput. Chem. 17, 520–552. [Google Scholar]
- 22.Halgren, T. A. (1996) J. Comput. Chem. 17, 553–586. [Google Scholar]
- 23.Halgren, T. A. & Nachbar, R. B. (1996) J. Comput. Chem. 17, 587–615. [Google Scholar]
- 24.Halgren, T. A. (1996) J. Comput. Chem. 17, 616–641. [Google Scholar]
- 25.Maignan, S., Guilloteau, J. P., Zhou-Liu, Q., Clement-Mella, C. & Mikol, V. (1998) J. Mol. Biol. 282, 359–368. [DOI] [PubMed] [Google Scholar]
- 26.Young, S. D. (2001) Curr. Opin. Drug Discov. Dev. 4, 402–410. [PubMed] [Google Scholar]
- 27.Neamati, N. (2002) Expert Opin. Ther. Pat. 12, 709–724. [Google Scholar]
- 28.Saag, M. S., Emini, E. A., Laskin, O. L., Douglas, J., Lapidus, W. I., Schleif, W. A., Whitley, R. J., Byrmes, V. W., Hildebrand, C., Kappes, J. C., et al. (1993) N. Engl. J. Med. 329, 1065–1072. [DOI] [PubMed] [Google Scholar]
- 29.Shaw-Reid, C. A., Munshi, V., Graham, P., Wolfe, A., Witmer, M., Danzeisen, R., Olsen, D. B., Carroll, S. S., Embrey, M., Wai, J. S., et al. (2003) J. Biol. Chem. 278, 2777–2780. [DOI] [PubMed] [Google Scholar]
- 30.Summa, V., Petrocchi, A., Pace, P., Matassa, V. G., De Francesco, R., Altamura, S., Tomei, L., Koch, U. & Neuner, P. (2004) J. Med. Chem. 47, 14–17. [DOI] [PubMed] [Google Scholar]
- 31.Schames, J. R., Henchman, R. H., Siegel, J. S., Sotriffer, C. A., Ni, H. & McCammon, J. A. (2004) J. Med. Chem. 47, 1879–1881. [DOI] [PubMed] [Google Scholar]
- 32.Goldgur, Y., Craigie, R., Cohen, G. H., Fujiwara, T., Yoshinaga, T., Fujishita, T., Sugimoto, H., Endo, T., Murai, H. & Davies, D. R. (1999) Proc. Natl. Acad. Sci. USA 96, 13040–13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen, J. C.-H., Krucinski, J., Miercke, L. J. W., Finer-Moore, J. S., Tang, A. H., Leavitt, A. D. & Stroud, R. M. (2000) Proc. Natl. Acad. Sci. USA 97, 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]