Abstract
Thymic selection proceeds in a unique and highly organized thymic microenvironment resulting in the generation of a functional, self-tolerant T cell repertoire. In vitro models to study T lineage commitment and development have provided valuable insights into this process. However, these systems lack the complete three-dimensional thymic milieu necessary for T cell development and, therefore, are incomplete approximations of in vivo thymic selection. Some of the challenges related to modeling T cell development can be overcome by using in situ models that provide an intact thymic microenvironment that fully supports thymic selection of developing T cells. Thymic slice organotypic cultures complement existing in situ techniques. Thymic slices preserve the integrity of the thymic cortical and medullary regions and provide a platform to study development of overlaid thymocytes of a defined developmental stage or of endogenous T cells within a mature thymic microenvironment. Given the ability to generate ~20 slices per mouse, thymic slices present a unique advantage in terms of scalability for high throughput experiments. Further, the relative ease in generating thymic slices and potential to overlay different thymic subsets or other cell populations from diverse genetic backgrounds enhances the versatility of this method. Here we describe a protocol for the preparation of thymic slices, isolation and overlay of thymocytes, and dissociation of thymic slices for flow cytometric analysis. This system can also be adapted to study non-conventional T cell development as well as visualize thymocyte migration, thymocyte-stromal cell interactions, and TCR signals associated with thymic selection by two-photon microscopy.
Keywords: Immunology, Issue 114, Thymic slices, organ culture, thymus, positive selection, negative selection, T cell development.
Introduction
T cells differentiate through a series of developmental intermediates in the thymus during which time they encounter several checkpoints that ensure the generation of a functional, self-tolerant T cell repertoire1-3. Positive selection promotes the survival of thymocytes with T cell receptors (TCR) capable of recognizing, with low to moderate affinity, peptide presented by major histocompatibility complex molecules (MHC) on cortical thymic epithelial cells (cTEC)2,3. Negative selection and regulatory T (Treg) cell development contribute to the establishment of self-tolerance via the elimination or diversion of thymocytes that respond strongly to self-peptide presented by MHC2,4. Immature CD4+CD8+ double positive (DP) thymocytes expressing TCRs that pass the selection process differentiate into mature T cell subpopulations, the majority of which are MHC class I-restricted CD8+ cytotoxic or MHC class II-restricted CD4+ helper single positive (SP) T cells, before exiting the thymus to perform effector functions in the secondary lymphoid organs1-3.
Adding to the complexity of T cell development is the dynamic migration and cellular encounters of developing thymocytes throughout the stromal cell network5-9. These stromal cells play distinct roles in thymocyte development and are differentially distributed between the thymic cortical and medullary regions where positive and negative selection occur10. Although positive selection takes place primarily in the cortex, there is accumulating evidence that DP thymocytes migrate to the medulla and continue to require TCR signals before they differentiate into mature T cells suggesting that the medulla may provide additional signals necessary for completion of positive selection and lineage differentiation11,12. Further, despite the presence of specialized medullary thymic epithelial cells (mTEC) that express and present tissue-restricted antigens facilitating deletion of autoreactive thymocytes13,14, a large proportion of negative selection occurs in the cortex in response to ubiquitously expressed self-peptide presented by dendritic cells15,16. Thus, accurate models of T cell development must provide a highly organized thymic microenvironment, with intact cortical and medullary regions, that facilitates interaction between thymocytes and stromal cells, and supports thymocyte migration as these cells undergo positive and negative selection.
To complement ex vivo analyses of thymocytes as a means of studying positive and negative selection, a number of in vitro, in situ, and in vivo models of T cell development have been developed17-22. It has been notoriously difficult to recapitulate positive selection in vitro, but coculture of stem cell populations or T cell precursors with stromal cells expressing Notch ligand, notably OP9-DL1/4 cells, has the capability to support T lineage commitment and limited positive selection making it an invaluable in vitro model to study T cell development23-25. Limitations of this system, however, include the fact that these cells lack the unique peptide processing machinery found in thymic stromal cells and the three-dimensional thymic microenvironment.
Though more technically cumbersome, in situ and in vivo models of thymic selection can overcome some of the barriers related to in vitro systems. Reaggregate thymic organ cultures (RTOC) contain defined mixtures of thymocytes and thymic stromal cells18,26,27. These thymic epithelial cell reaggregates maintain MHC class I and II expression and can support development of both conventional T cell subsets, yet still lack defined cortical and medullary structures. Fetal thymic organ culture (FTOC) is a popular model of T cell development that can be seeded with thymocytes via hanging-drop culture of lymphodepleted thymic lobes or via injection of thymocytes into lymphoreplete thymic lobes and support efficient development of CD4+ and CD8+ T cells over time in culture18,28-31. At the initiation of culture of fetal thymic lobes there is a paucity of mTECs, but defined cortical and medullary structures may develop over time depending on conditions. An important consideration is that this model may preferentially support fetal versus adult T cell development. Finally, intrathymic injection of defined thymic precursors in adult mice is technically challenging but clearly provides an environment to supportT cell development in vivo. These in situ and in vivo models are excellent tools to study T cell development and their use should be considered on an experiment-by-experiment basis.
Thymic slices, however, have recently emerged as a versatile, complementary model to study thymic selection in situ with the possibility to accommodate unique, complex, and generally higher throughput experiments. Thymic slices maintain the integrity of the cortical and medullary regions and provide a framework of stromal cells that supports thymocyte migration during development as well as efficient positive and negative selection11,32-39. Thymocyte subsets added atop thymic slices migrate into the tissue and to their appropriate microenvironmental niche34,37. The overlaid thymocytes can be distinguished from thymic slice endogenous cells via congenic markers or fluorescent labels and can be maintained in culture for several days. Thymic slice organotypic cultures can be used to study various aspects of T cell development including thymic selection, thymocyte behavior (migration and cellular interactions), and thymocyte localization, among others. Given the ability to generate ~20 thymic slices per mouse, the scalability of experiments is generally greater than other in situ models of thymic selection. Although the preparation of thymic slices requires specialized equipment, such as the vibratome, and the life time of thymic slices in culture is limited owing to loss of cells over time via cell death and the lack of an encapsulating membrane, thymic slices provide an excellent model for analysis of thymic selection of synchronized populations of thymocytes within a mature thymic microenvironment. Here we describe the preparation of thymic slices (including harvesting the thymus, agarose embedding of thymus lobes and vibratome sectioning of the embedded tissue), isolation and overlaying of thymocytes, and dissociation of thymic slices for flow cytometric analysis.
Protocol
Protocols for all animal studies were approved by the Animal Care Committee at the Centre de recherche - Hôpital Maisonneuve-Rosemont.
1. Harvesting Mouse Thymus for Preparation of Thymic Slices and Single Cell Suspensions
Euthanize the mouse with CO2 followed by cervical dislocation.
In a laminar flow hood, pin the mouse ventral side up to a dissection board. Spray the mouse with 70% ethanol. Remove any excess alcohol by dabbing with gauze to prevent ethanol from entering the thoracic cavity and damaging the tissue.
Lift the skin at the base of the sternum with a pair of forceps and make a cut through the skin. Extend the cut of the skin upwards to each forelimb.
Make an additional cut at the base of the sternum to separate the diaphragm from the rib cage. Then cut each side of the rib cage towards the clavicle. Flip the rib cage over towards the head exposing the thoracic cavity. The two lobes of the thymus lie on top of the heart.
Remove the connective tissue around the thymic lobes using forceps, micro-dissection scissors, and/or sharp curved scissors. Use a pair of fine tip curved forceps to lift the individual lobes from underneath or via remaining connective tissue. Note: Do not grasp the thymus directly.
Place the thymic lobes in a 15 ml conical tube containing PBS and set aside on ice until needed.
2. Agarose Embedding and Vibratome Slicing of Thymic Lobes
Prepare 4% agarose solution for embedding by dissolving 2 g low-melting point agarose in 50 ml sterile PBS and microwaving on a low setting until the agarose has completely dissolved. Cover flask with aluminum foil and place in a water bath at 55 °C until ready for use.
In a tissue culture hood, prepare complete RPMI-1640 media that contains 10% fetal bovine serum (FBS), 4 mM L glutamine, 1x penicillin/streptomycin, and 10 µM 2-mercaptoethanol.
Add 1.5 ml complete RPMI-1640 media per well to a 6-well cell culture plate and place a cell culture insert in each well. Set the plate aside in an incubator at 37 °C until needed.
Prepare a slushy ice water bath with water and ice in an ice bucket.
Carefully remove all remaining connective tissue surrounding each thymic lobe using fine tip forceps. Do this while the thymic lobe is submerged in PBS in a tissue culture dish, after transfer onto a tissue wipe soaked with PBS, or under a dissecting microscope.
Allow the agarose to cool below 40 °C, when the flask is just warm to the touch, to avoid overheating the tissue. Pour the cooled 4% agarose into a tissue mold to a height of ~1 cm.
Use a pair of forceps to carefully transfer the thymic lobe to a tissue wipe and roll it gently to dry it without damaging the tissue. Ensure that the tissue dries completely or it will slide out of the agarose during slicing.
Carefully insert the lobe into the agarose and position it either horizontally (to increase surface area of each slice) or vertically (to increase number of slices) at the bottom of the mold. Place the mold in ice water for 5-10 min to allow the agarose to solidify.
During this time, prepare the vibratome for slicing by mounting the buffer tray, assembling the specimen disc, and inserting the vibratome blade. Place a piece of laboratory tape on the specimen disc. Sterilize the clean, assembled workspace with 70% ethanol.
Once the agarose solidifies, invert the mold and press gently at its center to release the agarose embedded lobe.
Use a sharp blade to trim the excess agarose surrounding the lobe leaving ~2 mm of agarose on each side and ~0.5 cm at the bottom.
Secure each agarose block with a drop of tissue glue to the piece of tape on the specimen disc. Multiple blocks can be glued on the tape.
Align the vibratome blade with the top of the agarose block. Fill the buffer tray with sterile PBS until the blade and agarose block(s) are fully immersed.
Section the agarose embedded tissue to obtain slices with a thickness of 400-500 µm. Set the vibratome to 0.225 mm/sec speed, 100 Hz frequency, and 5° angle.
Use a bent spatula to collect the thymic slices into a tissue culture plate containing sterile PBS as they are cut. Discard the first and last slice.
Examine the slices under a light microscope at 4X magnification. Choose the slices with intact thymic tissue and surrounding agarose (Figure 1A).
Transfer the slices to the plate prepared in step 2.3 by using a pipette tip to gently slide the thymic slice from the bent spatula onto the cell culture insert. Each insert can accommodate ~3 slices. Make sure the slices do not touch each other or the cell culture insert walls.
Maintain the plate at 37 °C until needed.
3. Preparation of Thymocytes for Overlaying
Isolate thymic lobes as described in section 1. Dissociate the lobes manually to make a single cell suspension using a sterilized 15 ml tissue grinder filled with 5 ml of sterile PBS containing 2% FBS. Transfer the cells to a 15 ml conical tube.
Centrifuge the cells at 545 x g for 5 min at 4 °C. Discard the supernatant and resuspend the cells in 1 ml of 1x ACK lysis buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM Na2EDTA) at room temperature for 3 min to lyse red blood cells.
Fill the tube to 15 ml with PBS containing 2% FBS. Centrifuge the cells at 545 x g for 5 min at 4 °C.
Resuspend the cells in 10 ml of PBS containing 2% FBS and pass cells through a 255 µm mesh filter.
Count cells using a hemocytometer. Centrifuge the cells at 545 x g for 5 min at 4 °C, and resuspend the cell pellet in PBS containing 2% FBS at a concentration of 1 x 107 cells per ml.
Label the cells with cellular dyes such as carboxyfluorescein succinimidyl ester (CFSE) according to manufacturer protocols if not using thymocytes from mice expressing congenic markers or genetically encoded fluorescent reporters to distinguish from slice endogenous cells.
After the final wash of the labeled cells, resuspend the cell pellet in complete RPMI-1640 media at 1-3 x 106 cells per 15 µl.
4. Overlaying Thymocytes onto the Thymic Slices
Use a pipette to aspirate any liquid surrounding the thymic slices on the insert. Note: Take care to avoid damaging the agarose. If the agarose is damaged, the overlaid thymocytes will not attach to the slice surface due to loss of surface tension.
Without touching the slice with the pipette tip, overlay 15 µl of thymocytes prepared in section 3 onto each thymic slice.
Incubate the plate at 37 °C for 2 hr to allow the cells to migrate into the thymic slices.
After 2 hr, rinse the thymic slices 3 times with 1 ml PBS to remove excess overlaid thymocytes that have not yet migrated into the tissue.
Incubate the plate at 37 °C until the thymic slices are ready to be harvested, typically 1-72 hr depending on the experiment.
5. Dissociation of Thymic Slices for Flow Cytometry
Prepare a microcentrifuge tube for each slice by adding 150 µl of PBS containing 2% FBS.
Add 1 ml of PBS containing 2% FBS to the tissue culture insert with the thymic slices and gently stir with a bent spatula to detach the slices from the surface of the cell culture insert.
Transfer the slice from the insert to the microcentrifuge tube using a bent spatula.
Manually disrupt the slice using a microcentrifuge tube sample pestle. Add 150 µl of PBS containing 2% FBS to a final volume 300 µl.
Filter the dissociated tissue through a 40 µm filter into a new microcentrifuge tube. The filtered cells are ready to be stained for flow cytometric analysis40.
Filter cells a second time before passing them on a flow cytometer to ensure that all the agarose is removed and does not clog the flow cytometer.
Acquire the cells on a flow cytometer and analyze data as has been previously described40.
Representative Results
Thymic slices support analysis of different aspects of T cell development such as positive and negative selection. For successful experiments, the quality of the thymic slice is paramount.Thus, thymic slices should be examined to ensure the integrity of the thymic tissue and that the agarose surrounding the thymic slice is intact (Figure 1A). Surface tension can be compromised when the agarose is damaged causing a significant decrease in the number of thymocytes that migrate into the tissue. Thus, thymic slices with indications of tissue damage or nicks/tears in the agarose should be discarded (Figure 1B).
TCR transgenic (tg) mice are often employed to study thymic selection due to the expression of a defined, functional TCR on the surface of every thymocyte that increases the frequency of positive selection over polyclonal populations. In order to capture each stage of positive selection, pre-selection TCR tg thymocytes can be overlaid atop thymic slices, and the development of mature, CD4+ or CD8+ SP T cells followed over time by flow cytometry40. Thymocytes isolated from MHC class I-restricted OT-I TCR tg Rag1-/- β2m-/- mice are arrested at the DP stage prior to thymic selection due to the necessity of β2m to associate with and stabilize the MHC class I heavy chain41,42. When overlaid onto β2m-/- thymic slices, the pre-selection OT-I TCR tg thymocytes remain at the DP stage and do not generate significant CD8+ SP T cells (Figure 2A, left). In contrast, when overlaid on WT thymic slices containing endogenous selecting ligands, the ability of this model to support positive selection is evidenced by CD8+ T cell development at 72 hr (Figure 2A, right). Quantification of CD8+ T cell development as a read-out of positive selection is shown in Figure 2B.
Thymic slices can also be used to study negative selection. A large proportion of negative selection occurs in response to ubiquitous antigen16. To model negative selection to ubiquitous antigen, total thymocytes from OT-I TCR tg Rag1-/- and wild-type (WT) mice were labelled with CFSE and CTV, respectively, then mixed in a 1:1 ratio and overlaid onto WT slices (Figure 3A). After washing off excess thymocytes, the thymic slices were placed in complete RPMI-1640 media with or without 1 nM of the cognate, agonist peptide of the OT-I TCR, SIINFEKL (OVA peptide). All MHC class I expressing cells will present the antigen, modeling the presentation of ubiquitously expressed antigen in the thymus. After 24 hr of incubation at 37 °C, the proportion of OT-I TCR tg cells (% live CFSE+) to WT control cells (% live CTV+) was compared by flow cytometry (Figure 3B). A decrease in the relative proportion of OT-I TCR tg cells represents deletion of OTI TCR tg cells in response to OVA peptide.
It is important to note that due to heterogeneity in the size of thymic slices as well as cell entry into thymic slices, one should include appropriate analysis and internal controls to normalize for these variables. Here, WT cells mixed with OT-I TCR tg cells and overlaid atop slices were used as an internal control as this population should not be significantly affected by the presence or absence of OVA peptide. The extent of negative selection of OTI TCR tg cells was quantified based on proportions rather than absolute numbers. First, the ratio between OT-I TCR tg (% live CFSE+) to WT (% live CTV+) cells on each WT thymic slice in the presence or absence of OVA peptide was calculated. Each of these ratios was then divided by the mean of the ratio between OT-I TCR tg (% live CFSE+) to WT (% live CTV+) cells on WT slices in the absence of OVA peptide. On average, 80% of the OT-I TCR tg cells were depleted in response to ubiquitous antigen as depicted in Figure 3C.
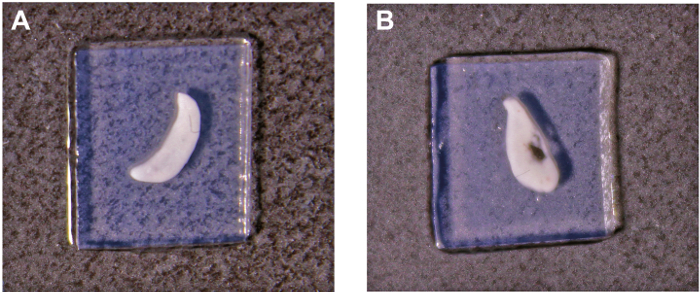 Figure 1: Representative images of thymic slices prepared from a vertically embedded thymic lobe. (A) Good quality slice with the embedded tissue and surrounding agarose intact. (B) Poor quality slice with damage due to tearing of the embedded tissue during slicing. Please click here to view a larger version of this figure.
Figure 1: Representative images of thymic slices prepared from a vertically embedded thymic lobe. (A) Good quality slice with the embedded tissue and surrounding agarose intact. (B) Poor quality slice with damage due to tearing of the embedded tissue during slicing. Please click here to view a larger version of this figure.
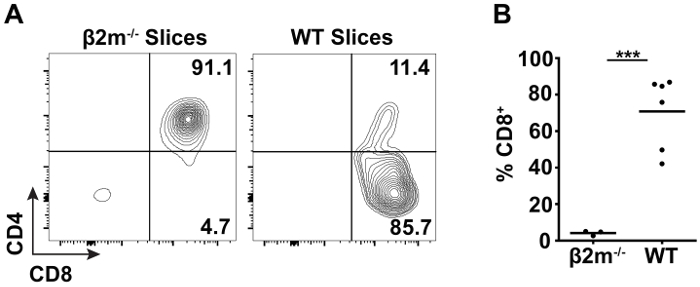 Figure 2:Flow cytometric analysis of positive selection in thymic slices. OT-I TCR tg Rag1-/- β2m-/- thymocytes were labelled with CFSE and overlaid onto non-selecting β2m-/- or selecting WT slices. After 72 hr of incubation at 37 °C, CD8+ T cell development was analyzed by flow cytometry. (A) Representative data of cell surface CD4 and CD8 expression on live CFSE+ TCRβhigh thymocytes from β2m-/- and WT slices. (B) Quantification of live CFSE+ TCRβhigh CD8+ T cell development on WT slices as compared to non-selecting controls. Each dot represents an individual thymic slice and the line represents the mean. (***p < 0.001, unpaired t test) Please click here to view a larger version of this figure.
Figure 2:Flow cytometric analysis of positive selection in thymic slices. OT-I TCR tg Rag1-/- β2m-/- thymocytes were labelled with CFSE and overlaid onto non-selecting β2m-/- or selecting WT slices. After 72 hr of incubation at 37 °C, CD8+ T cell development was analyzed by flow cytometry. (A) Representative data of cell surface CD4 and CD8 expression on live CFSE+ TCRβhigh thymocytes from β2m-/- and WT slices. (B) Quantification of live CFSE+ TCRβhigh CD8+ T cell development on WT slices as compared to non-selecting controls. Each dot represents an individual thymic slice and the line represents the mean. (***p < 0.001, unpaired t test) Please click here to view a larger version of this figure.
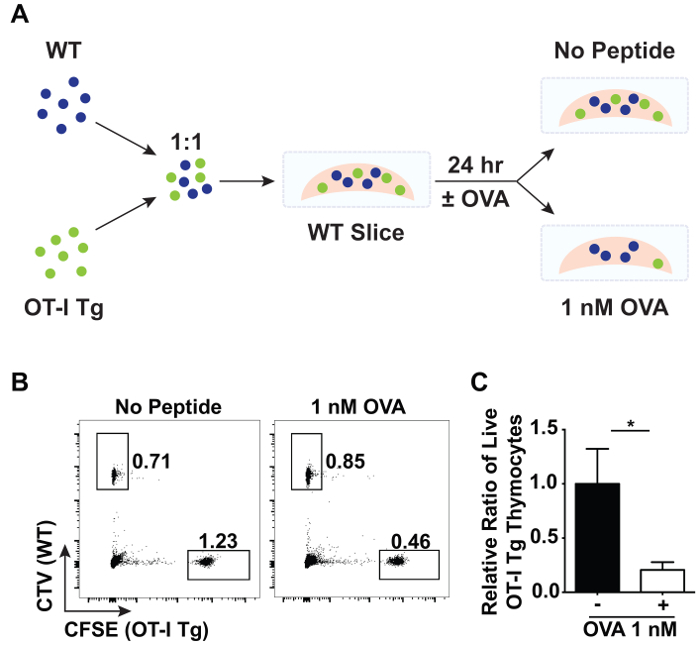 Figure 3:Flow cytometric analysis of negative selection in thymic slices. A 1:1 ratio of total CFSE-labeled OT-I TCR tg Rag1-/- and CTV-labeled WT thymocytes were overlaid on WT slices in the presence or absence of the OVA peptide, SIINFEKL. After 24 hr of incubation at 37 °C, the relative proportions of overlaid cells were analyzed by flow cytometry. (A) A schematic of the experimental set-up showing an overlay of CTV-labeled WT cells (blue) mixed 1:1 with CFSE-labeled OT-I TCR tg cells (green) onto a WT thymic slice in the presence or absence of OVA peptide. (B) Representative data depicting the proportion of live CFSE+ OT-I TCR tg cells and CTV+ WT cells from slices that were incubated with or without OVA peptide. (C) Data was normalized between slices, and the relative ratio of live CFSE+ OT-I TCR tg cells to live CTV+ WT cells is shown. Error bars indicate standard deviation. N = 3 for each condition. (*p < 0.05, unpaired t test). Please click here to view a larger version of this figure.
Figure 3:Flow cytometric analysis of negative selection in thymic slices. A 1:1 ratio of total CFSE-labeled OT-I TCR tg Rag1-/- and CTV-labeled WT thymocytes were overlaid on WT slices in the presence or absence of the OVA peptide, SIINFEKL. After 24 hr of incubation at 37 °C, the relative proportions of overlaid cells were analyzed by flow cytometry. (A) A schematic of the experimental set-up showing an overlay of CTV-labeled WT cells (blue) mixed 1:1 with CFSE-labeled OT-I TCR tg cells (green) onto a WT thymic slice in the presence or absence of OVA peptide. (B) Representative data depicting the proportion of live CFSE+ OT-I TCR tg cells and CTV+ WT cells from slices that were incubated with or without OVA peptide. (C) Data was normalized between slices, and the relative ratio of live CFSE+ OT-I TCR tg cells to live CTV+ WT cells is shown. Error bars indicate standard deviation. N = 3 for each condition. (*p < 0.05, unpaired t test). Please click here to view a larger version of this figure.
Discussion
Here we describe a protocol for the preparation of thymic slices and representative results of efficient positive and negative selection of overlaid pre-selection MHC class I-restricted TCR transgenic thymocytes by flow cytometry. This system has been used with similar success to support positive selection of MHC class II-restricted CD4+ T cells from pre-selection DP thymocytes32, and, in the presence of agonist antigen, negative selection and thymic Treg development11,12,36,38,39,43,44. This protocol can be modified to study thymic selection in the presence or absence of inhibitors, defined peptides, as well as of genetically modified thymocytes or stromal cell populations. Thymic slices are also amenable to the overlay of cell populations other than thymocyte subsets. For example, it was recently shown that dendritic cells overlaid on thymic slices efficiently migrate into the tissue and can support negative selection and Treg development39,43. Studies to date have used pre-selection DP or total thymocytes to study positive and negative selection. Although T cell development can proceed for at least 4 days in thymic slices, if interested in initiating experiments with an earlier progenitor population, it should be taken into consideration that a limitation of this system is that the quality of the thymic slices begins to decline after 1-2 days in culture due to cell death and lack of an encapsulating membrane.
The protocol described herein can also be used for generating slices from human thymic tissue with subtle modifications. Before embedding in agarose, human thymic tissue should be cut into fragments, roughly the size of an adult mouse thymic lobe. Notably, human thymic samples are rich in connective tissue, and, relative to murine thymus, it is more difficult to prepare good quality slices. In our experience, fetal rather than neonatal human thymic tissue is more conducive for slice preparation. It has been shown that human thymocyte subsets localize correctly on both human and mouse thymic slices37. Thymic selection of overlaid human thymocyte subsets, however, has not yet been reported in the thymic slice model. The efficiency of positive selection from a polyclonal population is low, and due to the low proportion (~0.5-2%) of overlaid thymocytes in the thymic slice, it may be difficult to assess such small numbers of positively selected cells. It may be feasible to introduce a human TCR transgene in human T cell progenitor cells prior to overlay on human thymic slices to increase positive selection efficiency. This also does not preclude the possibility of monitoring changes in endogenous T cell populations in the presence or absence of exogenous peptides or other cell populations and inhibitors of TCR signals, among others.
This protocol can also be adapted for visualization of thymocyte migration, TCR signals, and cellular interactions12,32-38. Until recently, most studies of thymocyte behavior in situ have been limited to the cortex as the medulla is centrally located in the thymus, just beyond the limit of detection by two-photon microscopy. In contrast, thymic slices provide the unique advantage in that the slices have intact thymic cortical and medullary regions and the slice surface allows direct access to the medulla by two-photon imaging. In addition, thymic slices overlaid with cells labelled with calcium indicator dyes such as Indol can be used to monitor TCR signals using cytosolic calcium levels as an indicator of TCR signals associated with positive and negative selection by two-photon microscopy11,32,36,38,39,44. To prepare samples for microscopy, the slices can be used directly after step 4.5 of the protocol. In this case, care should be taken to choose appropriate fluorescent markers suitable for imaging.
Thymic slice organ cultures are an excellent tool to study various aspects of mouse and human T cell development in situ. However, successful application of this technique requires special attention to critical steps in the protocol. To maximize the number of thymic slices obtained for high throughput experiments, the age of the mouse used must be taken into consideration as the size of the thymus changes throughout the life time of the mouse. We typically use 4-8 week old mice that generate ~20 thymic slices per mouse. However, fetal, neonatal or older mice thymi can theoretically be used to generate a limited number of slices depending on the experimental needs of the user. For obtaining good quality slices, it is paramount to remove all connective tissue surrounding the thymic lobes prior to embedding in agarose. Remaining connective tissue is not efficiently cut by the vibratome blade during slicing and can damage the thymic lobes and resulting slices. The bulk of connective tissue should be removed during thymus harvest (step 1.5) and allows the lobes to be removed from the thoracic cavity without significant manipulation of the tissue itself. After isolating the lobes, carefully remove any residual connective tissue as outlined in step 2.5. In addition, maintaining the integrity of the agarose surrounding the embedded tissue is imperative for overlaying thymocytes effectively on the slice surface. Thus, it is critical to inspect both the thymus slice and the surrounding agarose when choosing the thymic slices for use in the experiment. The vibratome is typically located outside of tissue culture hood, increasing the potential risk of contamination. Cleaning the vibratome thoroughly in between experiments and spraying the work area and vibratome with 70% ethanol prior to slicing can limit the potential for contamination. It is also important to use sterile PBS for steps 2.13 and 2.15 as mentioned in the protocol. Finally, it is also crucial to distinguish overlaid thymocytes that migrate into the tissue from the cells that are endogenous to the thymus. This can be achieved by labelling the overlaid cells with fluorescent dyes as mentioned in step 3.7. Alternatively, thymocytes from mice expressing congenic markers or genetically encoded fluorescent reporters can be used45,46. In general, however, thymic slices are easy to manipulate and can be adapted to the specific experimental needs of the user, supporting broad applicability that has only begun to be explored.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
We would like to thank Marilaine Fournier for her comments on the manuscript and Josée Tessier for technical assistance. C57BL/6-Tg (OT-I)-RAG1<tmMom> #4175 were obtained through the NIAID Exchange Program, NIH. Support for this research is provided by a grant from the SickKids Foundation and CIHR-IHDCYN (NI15-002), an operating grant from the CIHR-III (MOP-142254), and start-up funds from the FRQS (Établissement de jeunes chercheurs) and Hôpital Maisonneuve-Rosemont Foundation to HJM. HJM is a junior 1 scholar of the FRQS, a CIHR New Investigator (MSH-141967), and a Cole Foundation Early Career Transition award recipient.
References
- Carpenter AC, Bosselut R. Decision checkpoints in the thymus. Nat Immunol. 2010;11:666–673. doi: 10.1038/ni.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- Vrisekoop N, Monteiro JP, Mandl JN, Germain RN. Revisiting thymic positive selection and the mature T cell repertoire for antigen. Immunity. 2014;41:181–190. doi: 10.1016/j.immuni.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritesky GL, Jameson SC, Hogquist KA. Selection of self-reactive T cells in the thymus. Annu Rev Immunol. 2012;30:95–114. doi: 10.1146/annurev-immunol-020711-075035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousso P, Bhakta NR, Lewis RS, Robey E. Dynamics of thymocyte-stromal cell interactions visualized by two-photon microscopy. Science. 2002;296:1876–1880. doi: 10.1126/science.1070945. [DOI] [PubMed] [Google Scholar]
- Takahama Y. Journey through the thymus: stromal guides for T-cell development and selection. Nat Rev Immunol. 2006;6:127–135. doi: 10.1038/nri1781. [DOI] [PubMed] [Google Scholar]
- Halkias J, Melichar HJ, Taylor KT, Robey EA. Tracking migration during human T cell development. Cell Mol Life Sci. 2014;71:3101–3117. doi: 10.1007/s00018-014-1607-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Chtanova T, Ladi E, Robey EA. Thymocyte motility: mutants, movies and migration patterns. Curr Opin Immunol. 2006;18:191–197. doi: 10.1016/j.coi.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Ladi E, Yin X, Chtanova T, Robey EA. Thymic microenvironments for T cell differentiation and selection. Nat Immunol. 2006;7:338–343. doi: 10.1038/ni1323. [DOI] [PubMed] [Google Scholar]
- Klein L, Kyewski B, Allen PM, Hogquist KA. Positive and negative selection of the T cell repertoire: what thymocytes see (and don't see) Nat Rev Immunol. 2014;14:377–391. doi: 10.1038/nri3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JO, et al. Distinct phases in the positive selection of CD8+ T cells distinguished by intrathymic migration and T-cell receptor signaling patterns. Proc Natl Acad Sci U S A. 2014;111:E2550–E2558. doi: 10.1073/pnas.1408482111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Lancaster JN, Sasiponganan C, Ehrlich LI. CCR4 promotes medullary entry and thymocyte-dendritic cell interactions required for central tolerance. J Exp Med. 2015;212:1947–1965. doi: 10.1084/jem.20150178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- Takaba H, et al. Fezf2 Orchestrates a Thymic Program of Self-Antigen Expression for Immune Tolerance. Cell. 2015;163:975–987. doi: 10.1016/j.cell.2015.10.013. [DOI] [PubMed] [Google Scholar]
- McCaughtry TM, Baldwin TA, Wilken MS, Hogquist KA. Clonal deletion of thymocytes can occur in the cortex with no involvement of the medulla. J Exp Med. 2008;205:2575–2584. doi: 10.1084/jem.20080866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stritesky GL, et al. Murine thymic selection quantified using a unique method to capture deleted T cells. Proc Natl Acad Sci U S A. 2013;110:4679–4684. doi: 10.1073/pnas.1217532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G, Jenkinson EJ. Review article: thymus organ cultures and T-cell receptor repertoire development. Immunology. 2000;100:405–410. doi: 10.1046/j.1365-2567.2000.00067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare KJ, Jenkinson EJ, Anderson G. In vitro models of T cell development. Semin Immunol. 1999;11:3–12. doi: 10.1006/smim.1998.0151. [DOI] [PubMed] [Google Scholar]
- de Pooter R, Zuniga-Pflucker JC. T-cell potential and development in vitro: the OP9-DL1 approach. Curr Opin Immunol. 2007;19:163–168. doi: 10.1016/j.coi.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Lian Z, et al. Intrathymically injected hemopoietic stem cells can differentiate into all lineage cells in the thymus: differences between c-kit+ cells and c-kit < low cells. Stem Cells. 1997;15:430–436. doi: 10.1002/stem.150430. [DOI] [PubMed] [Google Scholar]
- Manna S, Bhandoola A. Intrathymic Injection. Methods Mol Biol. 2016;1323:203–209. doi: 10.1007/978-1-4939-2809-5_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschneider I, Komschlies KL, Greiner DL. Studies of thymocytopoiesis in rats and mice. I. Kinetics of appearance of thymocytes using a direct intrathymic adoptive transfer assay for thymocyte precursors. J Exp Med. 1986;163:1–17. doi: 10.1084/jem.163.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- de Pooter RF, Schmitt TM, Zuniga-Pflucker JC. In vitro generation of T lymphocytes from embryonic stem cells. Methods Mol Biol. 2006;330:113–121. doi: 10.1385/1-59745-036-7:113. [DOI] [PubMed] [Google Scholar]
- Dervovic DD, Ciofani M, Kianizad K, Zuniga-Pflucker JC. Comparative and functional evaluation of in vitro generated to ex vivo CD8 T cells. J Immunol. 2012;189:3411–3420. doi: 10.4049/jimmunol.1200979. [DOI] [PubMed] [Google Scholar]
- White A, Jenkinson E, Anderson G. Reaggregate thymus cultures. J Vis Exp. 2008. [DOI] [PMC free article] [PubMed]
- Anderson G, Owen JJ, Moore NC, Jenkinson EJ. Thymic epithelial cells provide unique signals for positive selection of CD4+CD8+ thymocytes in vitro. J Exp Med. 1994;179:2027–2031. doi: 10.1084/jem.179.6.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G, Jenkinson EJ. Fetal thymus organ culture. CSH Protoc. 2007. [DOI] [PubMed]
- Mazda O, Watanabe Y, Gyotoku J, Katsura Y. Requirement of dendritic cells and B cells in the clonal deletion of Mls-reactive T cells in the thymus. J Exp Med. 1991;173:539–547. doi: 10.1084/jem.173.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceredig R, Jenkinson EJ, MacDonald HR, Owen JJ. Development of cytolytic T lymphocyte precursors in organ-cultured mouse embryonic thymus rudiments. J Exp Med. 1982;155:617–622. doi: 10.1084/jem.155.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairchild PJ, Austyn JM. Developmental changes predispose the fetal thymus to positive selection of CD4+CD8 T cells. Immunology. 1995;85:292–298. [PMC free article] [PubMed] [Google Scholar]
- Bhakta NR, Oh DY, Lewis RS. Calcium oscillations regulate thymocyte motility during positive selection in the three-dimensional thymic environment. Nat Immunol. 2005;6:143–151. doi: 10.1038/ni1161. [DOI] [PubMed] [Google Scholar]
- Le Borgne M, et al. The impact of negative selection on thymocyte migration in the medulla. Nat Immunol. 2009;10:823–830. doi: 10.1038/ni.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich LI, Oh DY, Weissman IL, Lewis RS. Differential contribution of chemotaxis and substrate restriction to segregation of immature and mature thymocytes. Immunity. 2009;31:986–998. doi: 10.1016/j.immuni.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda Y, et al. Mst1 regulates integrin-dependent thymocyte trafficking and antigen recognition in the thymus. Nat Commun. 2012;3:1098. doi: 10.1038/ncomms2105. [DOI] [PubMed] [Google Scholar]
- Dzhagalov IL, Chen KG, Herzmark P, Robey EA. Elimination of self-reactive T cells in the thymus: a timeline for negative selection. PLoS Biol. 2013;11:e1001566. doi: 10.1371/journal.pbio.1001566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkias J, et al. Opposing chemokine gradients control human thymocyte migration in situ. J Clin Invest. 2013;123:2131–2142. doi: 10.1172/JCI67175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au-Yeung BB, et al. Quantitative and temporal requirements revealed for Zap70 catalytic activity during T cell development. Nat Immunol. 2014;15:687–694. doi: 10.1038/ni.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar HJ, Ross JO, Herzmark P, Hogquist KA, Robey EA. Distinct temporal patterns of T cell receptor signaling during positive versus negative selection in situ. Sci Signal. 2013;6 doi: 10.1126/scisignal.2004400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, Nicol SA, Suen AY, Baldwin TA. Examination of thymic positive and negative selection by flow cytometry. J Vis Exp. 2012. p. e4269. [DOI] [PMC free article] [PubMed]
- Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- Hogquist KA, et al. T cell receptor antagonist peptides induce positive selection. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Weist BM, Kurd N, Boussier J, Chan SW, Robey EA. Thymic regulatory T cell niche size is dictated by limiting IL-2 from antigen-bearing dendritic cells and feedback competition. Nat Immunol. 2015;16:635–641. doi: 10.1038/ni.3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar HJ, Ross JO, Taylor KT, Robey EA. Stable interactions and sustained TCR signaling characterize thymocyte-thymocyte interactions that support negative selection. J Immunol. 2015;194:1057–1061. doi: 10.4049/jimmunol.1400169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis AK, Macmaster S, Nagy A. Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol. 2002;2 doi: 10.1186/1472-6750-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer BC, Schaefer ML, Kappler JW, Marrack P, Kedl RM. Observation of antigen-dependent CD8+ T-cell/ dendritic cell interactions in vivo. Cell Immunol. 2001;214:110–122. doi: 10.1006/cimm.2001.1895. [DOI] [PubMed] [Google Scholar]


