Abstract
Previous biochemical evidence suggests that a cytochrome P450 specific to male antennae of the pale-brown chafer, Phyllopertha diversa, has evolved as a pheromone-degrading enzyme. By using a bioinformatics approach, we have now cloned three P450 cDNAs: CYP4AW1, CYP4AW2, and CYP6AT1. RT-PCR indicated that CYP4AW2 is expressed in all tissues examined, that CYP6AT1 is antennae-rich, and that CYP4AW1 is antennae-specific. Both tissue specificity and electrophysiological studies strongly support that CYP4AW1 in P. diversa is a pheromone-degrading enzyme involved in pheromone inactivation. Highly sensitive, pheromone-specific olfactory receptor neurons in male antennae were completely desensitized by direct application of metyrapone into the sensillar lymph. When tested in the same or different individuals, the metyrapone treatment had no effect on olfactory receptor neurons tuned to the plant volatile (Z)-3-hexenyl acetate, which might be inactivated by an esterase. Metyrapone treatment did not affect pheromone reception in the Japanese beetle, Popillia japonica, in the scarab beetle, Anomala octiescostata, or in the Oriental beetle, Exomala orientalis. Metyrapone-induced anosmia was restricted to the pheromone detectors in P. diversa, which became insensitive to physiological concentrations of pheromones for a few minutes. As opposed to previous trials, the specificity of the inhibitor and pheromone system led to unambiguous evidence for the role of pheromone-degrading enzymes in the fast inactivation of pheromones.
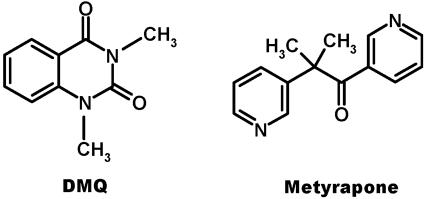
Insects rely heavily on their chemical communication skills to perform fundamental behaviors. Females, for example, advertise their readiness to mate and recruit males for reproduction by releasing sex pheromones. To avoid “chemical conspicuousness,” they release minute amounts of the species-specific chemical signals. To detect low levels of such unique signals in a noisy environment, males have evolved sensory systems with remarkable sensitivity that are highly tuned to these species-specific pheromones. Females of the pale-brown chaffer Phyllopertha diversa (Coleoptera or Scarabaeidae), for example, produce an alkaloid pheromone, 1,3-dimethyl-2,4-(1H,3H)-quinazolinedione (DMQ) (Fig. 1) (1), which is specifically detected by highly sensitive olfactory receptor neurons in male antennae (2, 3).
Fig. 1.
Structures of P. diversa sex pheromone DMQ (1) and metyrapone, a compound that inhibits pheromone degradation by a male antennae-specific cytochrome P450 (12).
While taking an odorant-oriented flight toward a pheromone-emitting female, a male encounters intermittent chemical signals comprising short bursts of high flux separated by periods during which the flux is zero (4, 5). Sustainable flight and orientation requires that the sensory system be reset on a millisecond timescale while navigating through the “clean” space between filaments. Two dichotomous hypotheses have been suggested for the fast inactivation of chemical signals. Based on the estimation that in the presence of an antennae-specific pheromone-degrading enzyme (PDE) (6) the pheromone has a half-life of 15 ms, it has been suggested that pheromone signals are deactivated by an enzymatic process (7). On the other hand, the enzymatic degradation has been considered too slow (on a scale of minutes) when tested in intact antennae (8). Furthermore, it has been suggested that the discrepancy between the in vivo and in vitro experiments is due to the involvement of pheromone-binding proteins that protect the pheromone from degradation (9). This controversy could be addressed by comparing the olfactory systems of “intact” insects to those having defective pheromone-degrading enzymes. Given that to date PDEs have never been isolated or cloned, a molecular knockout is not feasible. Previous attempts to impair pheromone-degrading enzymes with inhibitors (10, 11) were inconclusive, possibly because of interactions of the inhibitors with odorant receptors and/or pheromone-binding proteins.
The female-produced pheromone of P. diversa (1) is rapidly degraded in vitro by protein extracts from male antennae, but it is not metabolized by antennal extracts from 12 other species tested. We have previously demonstrated that the antennal enzyme involved in pheromone degradation is membrane-bound, requires NAD(P)H, and is sensitive to cytochrome P450 inhibitors (12). Given that several attempts to isolate the enzyme were (not surprisingly) unrewarding, we took a bioinformatics approach to obtain the cDNAs encoding antennae-specific cytochrome P450s. As reported here, we have cloned three cytochrome P450s, a ubiquitous, an antennae-rich, and an antennae-specific enzyme. In addition, we have carried out extensive electrophysiological experiments to test the effect of a cytochrome P450 inhibitor on the olfactory system of P. diversa. These in vivo treatments with metyrapone (2-methyl-1,2-di-3-pyridyl-1-propanone) induced temporary anosmia in the pheromone-detecting olfactory receptor neurons (ORNs), had no effect on detectors tuned to a plant-derived semiochemical, and showed no effect on the pheromone detectors of three other species of scarab beetles.
Materials and Methods
Insects. Males of P. diversa, collected at the Doho Park in Tsukuba, Japan, were kept at 23°C, relative humidity 60-70%, under a 16:8-h light-dark photoregime for electrophysiological recordings or were immediately frozen in dry ice and kept at -80°C until use for molecular work. Anomala octiescostata, Exomala orientalis, and Popillia japonica were from lab colonies.
cDNA Cloning. Fifty male antennae, 10 hindlegs, 5 pronota, and 3 thorax-abdomen complexes of P. diversa were collected on ice-cold Petri dishes. Total RNA was extracted with TRIzol (Invitrogen). First-strand cDNA was synthesized from 1 μg of total RNA by using a SMART RACE cDNA synthesis kit (Clontech) and subsequently treated with RNase H (New England Biolabs). Partial cDNA fragments encoding CYP4AW1 and CYP4AW2 were amplified by using Cyp4-1 [5′-GA(C/T)AC(A/C/G/T)TT(C/T)ATGTT(C/T)(A/G)A(A/G)GG(A/C/G/T)CA(C/T)G-3′] and Cyp4-2 [5′-GC(A/G)A A(C/T)T T(C/T)TG(A/C/G/T)CC(A/G/T)AT(A/G)CA(A/G)TT-3′] primers. These degenerate primers were designed on the basis of conserved sequences of the helix I domain of CYP4 family and the heme-binding domain of cytochrome P450s (13). The following PCR program was used: 94°C for 1 min; 35 cycles of 94°C for 30 s, 50°C for 30 s, 72°C for 30 s; 72°C for 10 min. CYP6AT1 partial cDNA fragment was amplified by universal primer mix (Clontech) and Cyp6-1 degenerate primer [5′-GGICCI(A/C)GIAA(C/T)TG(C/T)ATIGG-3′; I, inosine] based on a conserved region (GPRSCVG) of the heme-binding domain of cytochrome P450s (14). Touch-down PCR was done at 94°C for 1 min; 3 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; 3 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min; 3 cycles of 94°C for 30 s, 45°C for 30 s, and 72°C for 1 min; 30 cycles of 94°C for 30 s, 40°C for 30 s, and 72°C for 1 min; 72°C for 5 min. Full-length sequences of CYP4AW1 and CYP6AT1 cDNAs were amplified by three and five gene-specific primers, respectively. To exclude PCR-derived artifact (mutation), sequences for the antennae-specific and antennae-rich cDNAs were obtained from at least 11 independent clones.
RT-PCR. For detection of actin transcript as internal control, 45 cycles of a stepwise program (94°C for 1 min, 45°C for 2 min, and 72°C for 3 min) were carried out by using Actin-1 [5′-AA(C/T)TGGGA(C/T)GA(C/T)ATGGA(A/G)AA-3′] and Actin-2 [5′-GCCAT(C/T)TC(C/T)TG(C/T)TC(A/G)AA(A/G)TC-3′] degenerate primers (15). For detection of CYP4AW1 and CYP4AW2 gene expression, the following two pairs of gene-specific primers were used: CYP4AW1f (5′-GACCGAAACGTTGATTCCTTT-3′) and CYP4AW1r (5′-TGCTTCACTTTTTAGCACAAC-3′); CYP4AW2f (5′-GTAGAATCCTCCGACTCTCTG-3′) and CYP4AW2r (5′-TCCATCATTCTTTAATGCAAT-3′). For CYP6AT1 transcript, the following two gene-specific primers were used: CYP6AT1f (5′-CGACAATGGATTGGA AGAT TCCAGGCACC-3′) and CYP6AT1r (5′-GCTGCATAAAAAGCTCTGGGTTTAACATC-3′). Each cDNA fragment was amplified with 20-40 cycles (94°C for 30 s, 40°C for 30 s, and 72°C for 1 min). Obtained PCR products were separated on 0.8% agarose gel, cropped by gel print 2000i (BioPhotonics, Ann Arbor, MI), and processed by photoshop 6.0 (Adobe Systems, San Jose, CA). Fragments were verified by DNA sequencing.
Single Sensillum Recordings. Field-collected beetles were used within 2-3 days after capture, whereas lab-raised insects were 7-12 days old because they require several days to mature after emerging. The beetles were immobilized with dental wax inside 1.5-ml microfuge tubes, with their heads sticking out through a cut in the bottom of the tube. The antennae were immobilized in dental wax, and the three lamellae were separated and held immobile with thin tungsten pins. The indifferent electrode was a thin Ag-AgCl wire inserted in the abdomen. Receptor potential and nerve impulse activity were obtained simultaneously with glass electrodes (M1B150F-4, World Precision Instruments, Sarasota, FL) filled with sensillum-lymph Ringer solution (16). The tip of the capillary was sharpened (diameter, 2-3 μm) with PC-10 micropipette puller (Narishige, Tokyo). Extracellular recordings were established under a stereomicroscope (Olympus SZH-10) by inserting the electrode through the cuticle adjacent to a sensillum placodeum. The signal was amplified by using a Nihon Kohden MEZ-8300 amplifier. Data were stored by using axotape 2.0.2 (Axon Instruments, Foster City, CA) and visualized on a Nihon Kohden oscilloscope VC-11 and a Thermal Array Recorder (RTA-1100). Net spikes were calculated as the number of spikes generated during 1 s after the onset of stimulation minus the number of spikes during the same period before stimulation. The results were statistically processed (17) and represented as diagrams. The antennal preparation was continuously flushed by a charcoal-filtered and moistened air stream at 0.5 m/s through an 8-mm (i.d.) glass tube. The tube had a 2-mm hole (5 cm from the end) for insertion of a stimulus pipette and was placed 10 mm away from the preparation. A dichloromethane or hexane solution of each test compound was applied to a piece of filter paper (0.5 × 1 cm) that was dried for 2 min and inserted into a Pasteur pipette (14.5 cm long). Throughout the paper, the amounts of semiochemical loaded onto the filter papers (rather than the absolute amounts released from the pipettes) are referred to as the doses of stimuli. Air was blown for 300 ms through the pipette by a stimulus controller (CD-02/E; Syntech, Hilversum, The Netherlands). External application of metyrapone along with pheromone was done by loading each compound onto separate filter papers and loading them into the same pipette. Internal application was done by adding metyrapone (10-100 mM) to the sensillum-lymph Ringer solution (16). To facilitate diffusion of metyrapone, a slight pressure [14 mmHg (1 mmHg = 133 Pa)] was applied to the Ringer solution in the recording electrode.
Results and Discussion
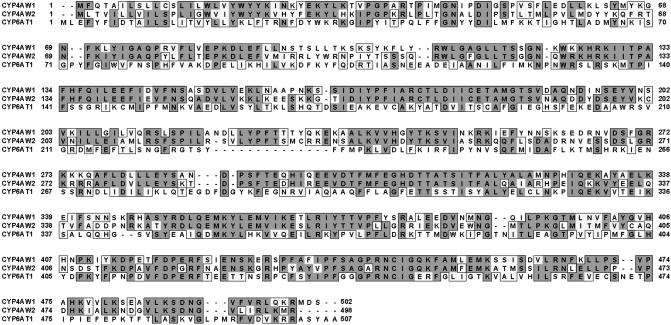
With degenerate primers designed on the basis of conserved sequences of cytochrome P450 heme-binding and helix I domains and antennal cDNA pool, we were able to clone three new cytochrome P450 cDNAs (Fig. 2). They were classified according to the P450 Gene Family Nomenclature Committee as CYP4AW1, CYP4AW2, and CYP6AT1 cDNAs, which encode predicted proteins with 502-, 498-, and 507-aa residues, respectively. The N-terminal sequences of the predicted mature proteins are mainly hydrophobic and present a cluster of prolines preceded by a cluster of basic residues, as observed for other eukaryotic P450s associated with microsomal membrane (18).
Fig. 2.
Sequence alignment of three cytochrome P450s cloned from P. diversa. CYP4AW1 and CYP4AW2 share 59% amino acid identity, whereas CYP6AT1 and the two other P450s have only 22% identical amino acids.
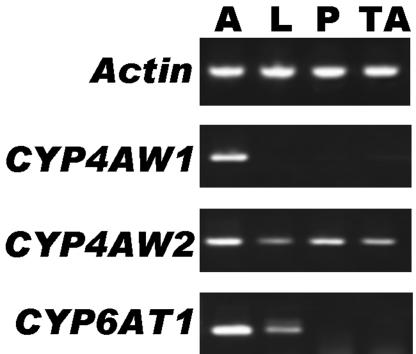
To examine transcription of the cytochrome P450 genes in various tissues, we carried out RT-PCR by using gene-specific primers along with primers for a control actin gene. Although 25 cycles of RT-PCR indicated that the three genes were expressed specifically in antennae, starting at 28 cycles it became clear that CYP6AT1 is predominantly expressed in the antennae, with low levels of expression in legs and no detection in pronota, or thorax-abdomen complexes (data not shown). Based on biochemical evidence that leg extracts do not degrade DMQ (12), CYP6AT1 is unlikely to be a PDE. Under the same conditions, CYP4AW2 was detected in all tissues (Fig. 3), whereas CYP4AW1 was specifically detected in antennae. No bands corresponding to a CYP4AW1 cDNA fragment were detected in nonolfactory tissues, even with 40 cycles of reaction (Fig. 3). Thus, we concluded that CYP4AW1 is an antennae-specific gene, and, because we have previously found evidence for antennae-specific cytochrome P450(s) in P. diversa that metabolize(s) the pheromone (12), we propose that CYP4AW1 is a pheromone-degrading enzyme (PdivPDE). To refine the data, we have sequenced CYP4AW1 and CYP6AT1 cDNAs from at least 11 independent clones. blast searches indicated that CYP4AW1 shares 40% amino acid identity with a CYP4C1 from a tropical cockroach (19), whereas the closest hit for CYP6AT1 was 40% amino acid identity with CYP6K1 from the German cockroach (20).
Fig. 3.
RT-PCR data. cDNA fragments were amplified with templates from male antennae (A), hindlegs (L), pronota (P), and thorax-abdomen complexes (TA) and primers for actin (control) or gene-specific primers. To confirm their identity, P450 bands were sequenced.
PdivPDE is, therefore, an ideal molecular target to test the hypothesis that inactivation of pheromones is an enzymatic process. PdivPDE is specific to antennae (see above). Pheromone degradation by antennal enzyme(s) is inhibited by metyrapone (12). Electrophysiological studies (2, 3, 21-25) led to mapping the pheromone-detecting sensilla placodea and those tuned to other semiochemicals in P. diversa and other scarab beetle species.
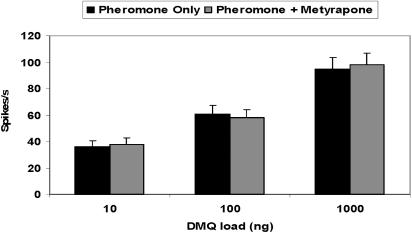
Initial electrophysiological studies of the effect of the cytochrome P450 inhibitor on the pheromone reception were done by conventional puffing of test stimulus and/or metyrapone on antennal preparations. There was no difference in response of pheromone detectors in P. diversa antennae to pheromone alone or in 1:1 combinations of pheromone and metyrapone (Fig. 4). In addition, we did not observe any significant influence on the receptor potential, nor were spikes generated by direct applications of metyrapone even at very high doses (10,000 ng, data not shown). By contrast, it has been observed that application of an esterase inhibitor, decyl-thio-trifluoropropanone, to pheromone detectors in moth species accelerated the repolarization of receptor potentials, generated spikes, and elicited delayed, transient, irregularly fluctuating decrease of transepithelial potential (10, 11). These findings led to the conclusion that inhibition by decyl-thio-trifluoropropanone was due to an antagonistic action at the pheromone-binding sites either at pheromone-binding proteins or pheromone receptors (10). Our data suggest that metyrapone did not enter the sensillar lymph, probably because of the lack of affinity of P. diversa odorant-binding proteins (26) to this ligand. If metyrapone did reach the sensillar lymph, it caused no interference with the reception of pheromone.
Fig. 4.
Dose-response relationships recorded from ORNs in P. diversa male antennae (6 beetles and 21 cells) tuned to the female sex pheromone. Responses to pheromone alone were indistinguishable from those obtained by simultaneous applications of pheromone and metyrapone.
Next, we tested the direct application of metyrapone to the sensillum lymph by diffusion through the recording glass electrode (hereafter referred to as topical application). Although it was not possible to measure/estimate accurately the amount of metyrapone that diffused into the sensillum lymph, we aimed at obtaining very low in situ concentrations of the inhibitor so as to avoid complete inhibition of the cytochrome P450s. Thus, initial trials were done with 10 mM metyrapone in the recording electrode. If the treatment had no effect, subsequent trials were done with 50 and 100 mM. The rationale for the low in situ concentration approach was to minimize possible side effects, i.e., interactions of metyrapone with other olfactory proteins (pheromone-binding proteins, odorant receptors, etc) rather than a PDE.
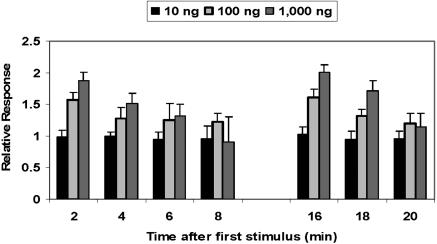
To simplify the analysis and interpretation of the data, we first tested the effect of pheromone doses on adaptation of the olfactory system. Although the pheromone detectors responded initially on a dose-dependent manner in the range of 10-1,000 ng of DMQ (Fig. 5), adaptation increased as the ORNs were subjected to repeated stimulus of high doses of the pheromone so that after four applications the response to 100-fold pheromone (1,000 ng) was almost identical to that elicited by 10 ng. When flushed with clean air, the olfactory system recovered in <10 min. As there was no adaptation to repeated applications of low doses of pheromone, this dose (10 ng) was used in all DMQ-repeated puffs after the prestimulus (which was applied at 10-, 100-, or 1,000-ng doses). Even when high doses (100 and 1,000 ng) of DMQ were applied as prestimulus, adaptation was minimal (Fig. 6A). When observed, the effect of the prestimulus was small and limited to the first 2 min after the prestimulus.
Fig. 5.
Dose-response relationships recorded from pheromone ORNs (eight beetles and 36 cells) by repeated applications of DMQ. At high doses (e.g., 1,000 ng), adaptation increases as the stimulus is repeated. After four applications, the response is ≈100 times lower. No adaptation was observed at 10 ng, i.e., 10 times the threshold of the pheromone detectors.
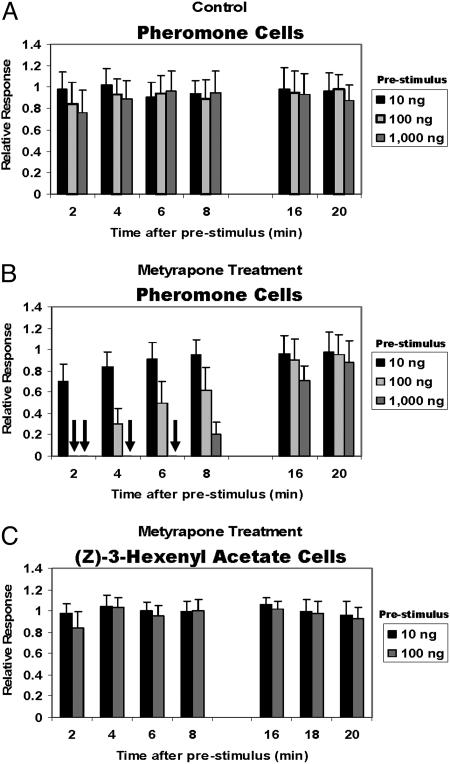
Fig. 6.
Responses of ORNs tuned to the female sex pheromone (A and B) and specific to a plant volatile (C). (A) In control experiments, prestimulus had little effect on the repeated applications of 10 ng of the pheromone, except for some adaptation observed at 2 min after high doses (1,000 ng) applied before repeated (10 ng) stimulus. (B) Application of metyrapone into the pheromone sensilla caused anosmia. The higher the concentration of the prestimulus, the longer the desensitization lasted. Arrows indicate the complete lack of response. (C) The neurons tuned to a plant volatile were unaffected by a similar treatment. Recordings were obtained from 7 beetles and 23 cells (A); 25 beetles and 122 cells (B); and 9 beetles and 24 cells (C).
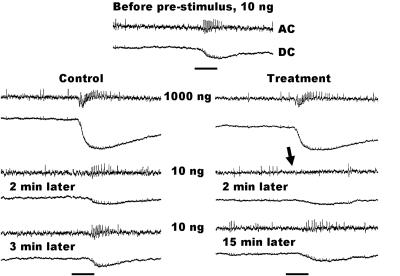
Metyrapone-treated ORNs were completely knocked out when subjected to a prestimulus of 1,000 ng of DMQ. Two minutes after the prestimulus, the pheromone-detecting ORNs were completely insensitive to 10 ng of DMQ. There was neither receptor potential nor spikes generated by 10 ng of DMQ (Figs. 6B and 7). In control experiments, the 2-min response is very similar to the response observed before the prestimulus, and 3 min after prestimulus, the system is almost completely recovered (from adaptation) (Fig. 7). By contrast, the metyrapone-treated ORNs remain completely silent for 6 min after each prestimulus; if flushed with clean air, they recovered after 15 min. These experiments were repeated with at least 25 different individuals with contacts made in 3-10 pheromone ORNs for each antennal preparation in a total of 122 cells (Fig. 6B). The most striking responses were observed when 1,000 ng of DMQ was used as a prestimulus, which completely incapacitated the pheromone detectors for at least 6 min. With 100 ng of prestimulus, there was no response to DMQ (10 ng) after 2 min, but after 4 and 6 min, the pheromone detectors recovered to about 30% and 50%, respectively. Even at the lowest dose (10 ng), there was a decrease in response. The small (30%) decrease in response was statistically different from adaptation. The concentrations of the inhibitor in the sensillar lymph may have decreased, but it did not completely deplete the enzymatic activity or the molecular process of pheromone inactivation. When subjected to high doses of the prestimulus, the inhibitor-decreased enzymatic activity may not be sufficient to inactivate (deactivate) pheromone molecules at high, but physiologically relevant, doses (Fig. 4).
Fig. 7.
Comparison of single sensillum recordings obtained from pheromone detectors (AC, nervous activity; DC, receptor potential) stimulated with DMQ without (control) and with metyrapone treatment. Note the complete lack of nervous activity (arrow) 2 min after prestimulus. In the control, a small decrease in response is due to adaptation. With no treatment, the neurons are almost entirely recovered (from adaptation) after 3 min, whereas the treated neurons remain completely insensitive for 6 min. After 15 min, treated neurons start responding to the same level untreated neurons did 2 min after prestimulus. (Scale bars representing stimulus duration = 300 ms.)
We tested the effect of metyrapone treatment on nonpheromone detectors, particularly ORNs tuned to a green leaf volatile compound, (Z)-3-hexenyl acetate. Because these ORNs are extremely sensitive to this semiochemical (2, 24), we used lower doses (10 and 100 ng) as prestimulus and presented repeated 0.1-ng puffs so as to avoid overexcitation of the (Z)-3-hexenyl acetate ORNs. In other words, the stimulus was 10 times the threshold of the detector for (Z)-3-hexenyl acetate, in line with that used for testing the pheromone cells. As preliminary experiments showed no effect of metyrapone treatment (data not shown), we increased to 100 mM the concentration of the inhibitor in the pipette. We tested the effect of the metyrapone treatment on ORNs tuned to (Z)-3-hexenyl acetate and those specific to the sex pheromone with the same antennal preparations. The pheromone detectors in P. diversa were always influenced by the cytochrome P450 inhibitor, but the nonpheromone cells responded to (Z)-3-hexenyl acetate with profiles of receptor potentials and spikes indistinguishable from those of control experiments (Fig. 6C). Given the chemical nature of this semiochemical, it is expected that an esterase, not a cytochrome P450, would work as odorant-degrading enzyme in the (Z)-3-hexenyl acetate-detecting cells.
In addition to the tests on nonpheromone cells in P. diversa antennae, the effect of metyrapone treatment was tested in three other scarab beetle species. We applied metyrapone to the pheromone-detecting cells (3, 21, 27) in the antennae of the Japanese beetle, P. japonica, by prestimulus and stimulus with the female-produced sex pheromone, (R)-japonilure (28). The Japanese beetle antennae possess esterase(s) that degrade the lactone pheromone (29). Likewise, we tested the effect of metyrapone on the pheromone-detecting ORNs (25) in A. octiescostata, whose pheromone system is composed of two lactones (30). Only the major constituent, (R,Z)-5-(-)-(oct-1-enyl)oxacyclopentan-2-one (31), was used. In addition, the metyrapone treatment was tested in the pheromone-detecting ORNs in the antennae of the Oriental beetle, E. orientalis. The enzymatic process for metabolism of the pheromone of this species, (Z)- and (E)-tetradec-7-en-2-one (32, 33), has never been investigated. Despite several attempts (over 50 contacts established with pheromone detectors for each of the three species), no effect of metyrapone (100 mM) was observed even when high doses (1,000 ng) of the species-specific pheromones were applied as prestimuli (data not shown). As a positive control, these experiments were intercalated with metyrapone treatments on the pheromone detectors in P. diversa. In conclusion, the metyrapone treatment induced anosmia specifically on the pheromone detectors in P. diversa antennae, having no effect on the pheromone cells of three species of scarab beetles, namely P. japonica, A. octiescostata, and E. orientalis, nor on ORNs tuned to green leaf volatile in P. diversa.
The following lines of evidence strongly suggest that the metyrapone-induced temporary anosmia in P. diversa pheromone detectors is due to the specific action of the inhibitor on a pheromone-degrading enzyme. First, DMQ is specifically degraded by antennal extracts from P. diversa, and the enzymatic activity is due to cytochrome P450(s), which is inhibited in vitro by metyrapone (12). Second, P. diversa expresses at least one antennae-specific cytochrome P450 (this paper). Third, metyrapone is unlikely to interact with pheromone receptor(s) given that it does not influence receptor potential or generate spikes in the pheromone detectors of the four species tested. This finding is unambiguous evidence for the role of pheromone-degrading enzymes in the fast inactivation of pheromones.
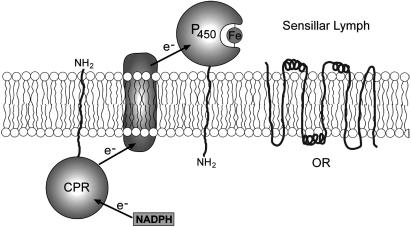
A priori, the classic location of cytochrome P450 may seem incompatible with the function of odorant-degrading enzymes. Typically, these enzymes are located in the endoplasmic reticulum and in the mitochondria of the mammalian and insect cells, whereas pheromone inactivation is supposed to take place in the extracellular environment. However, a number of different forms of cytochrome P450s have been detected (by using immunocytochemistry, electron microcopy, confocal microscopy, flow cytometry, etc.) in the plasma membrane of mammals, including P450 from 1A, 2B, 2C, 2D, 2E1, and 3A families in the plasma membrane of rat and human hepatocytes, rabbit pulmonary cells, and mammalian transfected cells (34-38). On the other hand, there is evidence in the literature that some NADPH cytochrome P450 reductases associated with cytochrome P450s that are localized in PM are not exposed to the outer surface, but rather cytosolically oriented and having a possible transmembrane electron transfer system (36, 39). We, therefore, hypothesized that the antennae-specific cytochrome P450 in P. diversa functioning as a pheromone-degrading enzyme is exposed to the sensillar lymph and linked to a cytosolic cytochrome P450 reductase (Fig. 8).
Fig. 8.
Proposed orientation of the antennae-specific cytochrome P450 on the dendritic membranes of ORNs tuned to DMQ. Because the electrophysiology data strongly suggest that the pheromone-degrading enzyme is involved in the fast inactivation of pheromone, a cytoplasm orientation is highly unlikely in CYP4AW1. Pheromone penetration through the dendritic membrane before inactivation must be a slow process. The P450 orientation (facing the sensillar lymph) is based on growing evidence for the occurrence of P450 localized in the plasma membrane and linked to cytosolic cytochrome P450 reductase (CPR) (34-38).
Cytochrome P450 and NADPH-cytochrome P450 oxidoreductase have been previously cloned from Drosophila melanogaster (40, 41). Because they are preferentially expressed in the antennae, a possible function in odor clearance has been suggested (40). More recently, two cytochrome P450s, CYP4S4 and CYP4L4, have been cloned from Mamestra brassicae (42). Northern blot and in situ hybridization analyses demonstrated that CYP4L4 was predominantly expressed in the antennae, whereas expression of CYP4S4 was restricted to odorant-detecting sensilla trichodea in the moth antennae (42). The diversity of odorant-degrading enzymes (esterases, aldehyde oxidases, aldehyde dehydrogenases, epoxide hydrolases, glutathione S-transferases, and cytochrome P450s) (43) is related to the plethora of chemicals essential in a bug's life. Given the unique chemistry of P. diversa sex pheromone (1), it is not entirely surprising that evolution has recruited a cytochrome P450 to cope with the need of a fast process of signal inactivation.
Acknowledgments
We thank Craig Wheelock for critical review of the manuscript; Hong-Chao Men, Mitsuo Chiba, and Mikio Ono for field collections; Marie-Christine François for technical assistance; and Tania Morgan for preparation of the illustration in Fig. 8. This work was supported by National Science Foundation Grant 0234769, U.S. Department of Agriculture National Research Initiative Competitive Grants Program Grant 2003-35302, and National Institutes of Health-National Institute of Allergy and Infectious Diseases Grant 1U01AI058267-01.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PDE, pheromone-degrading enzyme; ORN, olfactory receptor neuron; DMQ, 1,3-dimethyl-2,4-(1H,3H)-quinazolinedione.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. CYP4AW1, AY605086, CYP4AW2, AY605987, CYP6AT1, and AY605088).
References
- 1.Leal, W. S., Zarbin, P. H. G., Wojtasek, H., Kuwahara, S., Hasegawa, M. & Ueda, Y. (1997) Nature 385, 213. [DOI] [PubMed] [Google Scholar]
- 2.Hansson, B. S., Larsson, M. C. & Leal, W. S. (1999) Physiol. Entomol. 24, 121-126. [Google Scholar]
- 3.Nikonov, A. A. & Leal, W. S. (2002) J. Chem. Ecol. 28, 1075-1089. [DOI] [PubMed] [Google Scholar]
- 4.Murlis, J. (1997) in Insect Pheromone Research: New Directions, eds. Cardé, R. T. & Minks, A. K. (Chapman & Hall, New York), pp. 221-247.
- 5.Murlis, J., Willis, M. A. & Carde, R. T. (2000) Physiol. Entomol. 25, 211-222. [Google Scholar]
- 6.Vogt, R. G. & Riddiford, L. M. (1981) Nature 293, 161-163. [DOI] [PubMed] [Google Scholar]
- 7.Vogt, R. G., Riddiford, L. M. & Prestwich, G. D. (1985) Proc. Natl. Acad. Sci. USA 82, 8827-8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kassang, G., von Proff, L. & Nicholls, M. (1988) Z. Naturforsch. 43, 275-284. [Google Scholar]
- 9.Kaissling, K.-E. (2001) Chem. Senses 26, 125-150. [DOI] [PubMed] [Google Scholar]
- 10.Pophof, B. (1998) J. Comp. Physiol. A 183, 153-164. [Google Scholar]
- 11.Pophof, B., Gebauer, T. & Ziegelberger, G. (2000) J. Comp. Physiol. A 186, 315-323. [DOI] [PubMed] [Google Scholar]
- 12.Wojtasek, H. & Leal, W. S. (1999) FEBS Lett. 458, 333-336. [DOI] [PubMed] [Google Scholar]
- 13.Snyder, M. J., Stevens, J. L., Andersen, J. F. & Feyereisen, R. (1995) Arch. Biochem. Biophys. 321, 13-20. [DOI] [PubMed] [Google Scholar]
- 14.Danielson, P. B. & Fogleman, J. C. (1997) Insect Biochem. Mol. Biol. 27, 595-604. [DOI] [PubMed] [Google Scholar]
- 15.Ishida, Y. & Leal, W. S. (2002) Insect. Biochem. Mol. Biol. 32, 1775-1780. [DOI] [PubMed] [Google Scholar]
- 16.Kaissling, K.-E. (1995) in Experimental Cell Biology of Taste and Olfaction: Current Techniques and Protocols, eds. Spielman, A. I. & Brand, J. G. (CRC, Boca Raton, FL), pp. 361-377.
- 17.Zacks, S. (1971) The Theory of Statistical Interference (Wiley, New York).
- 18.Werck-Reichhart, D. & Feyereisen, D. (2000) Genome Biol. 1, 3003.1-3003.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradfield, J. Y., Lee, Y.-H. & Keeley, L. L. (1991) Proc. Natl. Acad. Sci. USA 88, 4558-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen, Z. & Scott, J. G. (2001) Insect Mol. Biol. 10, 131-137. [DOI] [PubMed] [Google Scholar]
- 21.Wojtasek, H., Hansson, B. S. & Leal, W. S. (1998) Biochem. Biophys. Res. Commun. 250, 217-222. [DOI] [PubMed] [Google Scholar]
- 22.Larsson, M. C., Leal, W. S. & Hansson, B. S. (1999) J. Comp. Physiol. A 184, 353-359. [Google Scholar]
- 23.Larsson, M. C., Leal, W. S. & Hansson, B. S. (2001) J. Insect Physiol. 47, 1065-1076. [DOI] [PubMed] [Google Scholar]
- 24.Nikonov, A. A., Valiyaveettil, J. T. & Leal, W. S. (2001) Chem. Senses 26, 49-54. [DOI] [PubMed] [Google Scholar]
- 25.Nikonov, A. A., Peng, G., Tsurupa, G. & Leal, W. S. (2002) Chem. Senses 27, 495-504. [DOI] [PubMed] [Google Scholar]
- 26.Wojtasek, H., Picimbon, J.-F. & Leal, W. S. (1999) Biochem. Biophys. Res. Commun. 263, 832-837. [DOI] [PubMed] [Google Scholar]
- 27.Kim, J.-Y. & Leal, W. S. (2000) Arthropod. Struct. Dev. 29, 121-128. [DOI] [PubMed] [Google Scholar]
- 28.Tumlinson, J. H., Klein, M. G., Doolittle, R. E., Ladd, T. L. & Proveaux, A. T. (1977) Science 197, 789-792. [DOI] [PubMed] [Google Scholar]
- 29.Leal, W. S. (2004) in Semiochemicals in Pest Management and Alternative Agriculture, eds. Petroski, R. J., Tellez, M. R. & Behle, R. W., in press.
- 30.Leal, W. S., Hasegawa, M., Sawada, M., Ono, M. & Ueda, Y. (1994) J. Chem. Ecol. 20, 1643-1655. [DOI] [PubMed] [Google Scholar]
- 31.Leal, W. S. (1991) Naturwissenschaften 78, 521-523. [Google Scholar]
- 32.Leal, W. S. (1993) Naturwissenschaften 80, 86-87. [Google Scholar]
- 33.Leal, W. S., Hasegawa, M., Sawada, M. & Ono, M. (1994) J. Chem. Ecol. 20, 1705-1718. [DOI] [PubMed] [Google Scholar]
- 34.Loeper, J., Descatoire, V., Maurice, M., Beaune, P., Feldmann, G., Larrey, D. & Pessayre, D. (1990) Hepatology 11, 850-858. [DOI] [PubMed] [Google Scholar]
- 35.Loeper, J., Descatoire, V., Maurice, M., Beaune, P., Belghiti, J., Houssin, D., Ballet, F., Feldmann, G., Guengerich, F. P. & Pessayre, D. (1993) Gastroenterology 104, 203-216. [DOI] [PubMed] [Google Scholar]
- 36.Loeper, J., Louerat-Oriou, B., Duport, C. & Pompon, D. (1998) Mol. Pharmacol. 54, 8-13. [DOI] [PubMed] [Google Scholar]
- 37.Loeper, J., Le Berre, A. & Pompon, D. (1998) Mol. Pharmacol. 53, 408-414. [DOI] [PubMed] [Google Scholar]
- 38.Robin, M. A., Maratrat, M., Loeper, J., Durandschneider, A. M., Tinel, M., Ballet, F., Beaune, P., Feldmann, G. & Pessayre, D. (1995) Gastroenterology 108, 1110-1123. [DOI] [PubMed] [Google Scholar]
- 39.Neve, E. P. A. & Ingelman-Sundberg, M. (2000) J. Biol. Chem. 275, 17130-17135. [DOI] [PubMed] [Google Scholar]
- 40.Hovemann, B. T., Sehlmeyer, F. & Malz, J. (1997) Gene 189, 213-219. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Q., Hasan, G. & Pikielny, C. W. (1999) J. Biol. Chem. 274, 10309-10315. [DOI] [PubMed] [Google Scholar]
- 42.Maïbèche-Coisne, M., Jacquin-Joly, E., Francois, M. C. & Nagnan-Le Meillour, P. (2002) Insect. Mol. Biol. 11, 273-281. [DOI] [PubMed] [Google Scholar]
- 43.Vogt, R. G. (2003) in Insect Pheromone Biochemistry and Molecular Biology: The Biosynthesis and Detection of Pheromones and Plant Volatiles, eds. Bomquist, G. J. & Vogt, R. G. (Elsevier Academic, London), pp. 391-445.