Abstract
Analysis of promoter sequences of all known human cytotoxic endonucleases showed that endonuclease G (EndoG) is the only endonuclease that contains a CpG island, a segment of DNA with high G+C content and a site for methylation, in the promoter region. A comparison of three human prostate cancer cell lines showed that EndoG is highly expressed in 22Rv1 and LNCaP cells. In PC3 cells, EndoG was not expressed and the EndoG CpG island was hypermethylated. The expression of EndoG correlated positively with sensitivity to cisplatin and etoposide, and the silencing of EndoG by siRNA decreased the sensitivity of the cells to the chemotherapeutic agents in the two EndoG-expressing cell lines. 5-aza-2′-deoxycytidine caused hypomethylation of the EndoG promoter in PC3 cells, induced EndoG mRNA and protein expression, and made the cells sensitive to both cisplatin and etoposide. The acetylation of histones by trichostatin A, the histone deacetylase inhibitor, induced EndoG expression in 22Rv1 cells, while it had no such effect in PC3 cells. These data are the first indication that EndoG may be regulated by methylation of its gene promoter, and partially by histone acetylation, and that EndoG is essential for prostate cancer cell death in the used models.
Keywords: Endonuclease G, DNA methylation, Prostate cancer, Cell death, Cisplatin
1. Introduction
Prostate cancer is the most commonly diagnosed non-cutaneous malignancy. It is the second leading cause of cancer deaths among men in the United States and overseas [1–3]. Prostate cancer, as well as cancer in general, is recognized as both a genetic and epigenetic disease, because both genetic and epigenetic changes play crucial roles in the transformation of cells to malignancy and in the progression of cancer [4]. Epigenetic changes are believed to be the most common alteration at the DNA level in prostate cancer [5,6].
Two types of DNA epigenetic changes occur in prostate cancer: regional DNA hypermethylation and regional/global DNA hypomethylation. Hypermethylation of the promoter region that contains CpG island occurs in a large number of genes and is associated with gene silencing in the vast majority of prostate cancer cases [1,3,4]. Studies have shown that hypermethylation of this region holds promise as a tumor biomarker for early diagnosis and risk assessment of prostate cancer. Furthermore, the prevalence of epigenetic changes in prostate cancer and the potential reversibility of DNA methylation alterations by DNA methylation inhibitors suggest that these changes are a viable target for cancer chemotherapy and chemoprevention strategies [7–9].
One of the key properties of cancer cells that leads to chemotherapy failure is the cells’ ability to avoid cell death by deregulation of apoptosis [10,11]. The two best characterized and most broadly used chemotherapeutic drugs that inhibit DNA methylation are 5-aza-2′-deoxycytidine (AzaC), clinically referred to as decitabine [12,13], and another recently developed inhibitor of DNA methylation, zebularine [14]. One of the main goals of the application of these drugs is the restoration of apoptosis by normalizing DNA methylation [8]. Evidence has accumulated that a number of genes involved in cell death pathways are often silenced by DNA hypermethylation, contributing to inactivation of the apoptosis that is commonly observed in prostate cancer cells [15–18]. However the potential role of DNA methylation in modulating cell death endonucleases in cancer cells has not been studied.
Cytotoxic endonucleases, also called “cell death endonucleases,” are the recently recognized group of enzymes responsible for premortem and postmortem DNA fragmentation associated with cell death by apoptosis or necrosis [19,20]. Major representatives of this group of enzymes include: deoxyribonuclease I (DNase I) [21], deoxyribonuclease II (DNase II) [22], EndoG [23], caspase-activated DNase (CAD) [24], and DNase gamma [25]. Cell death endonucleases were found in all studied cells and tissues, including the prostate [26,27]. These enzymes differ in certain catalytic characteristics and DNA sequence specificity, and yet produce a similar type of DNA damage, including single- and double-strand DNA breaks. Endonuclease-generated breaks have been shown to strongly interfere with DNA synthesis in both normal and cancer cells [28]. While often considered downstream effectors of apoptotic cascades, endonucleases can cause DNA fragmentation and imminent, irreversible cell death when acting alone after overexpression or introduction into the cell [21,22,24]. Some cell death endonucleases seem to be dispensable in normal apoptosis [29–31]. However, recent studies from our group and others demonstrated that inactivation of endonucleases causes protection of normal and cancer cells against a variety of injuries in vitro and in vivo [32–35], suggesting that the endonucleases are essential for injury-related cell death. One of the endonucleases, EndoG, seems to be particularly important in cancer cells because it regulates their sensitivity to chemotherapeutic agents [32]. EndoG has been recently recognized as a key endonuclease in the caspase-independent apoptosis [36,37] and necrosis [38,39].
The molecular mechanism of EndoG regulation is complex and largely unknown. As described below, the sequence analysis of human EndoG gene has shown the presence of a CpG island in the promoter region, suggesting that this gene is regulated by DNA methylation. In view of these considerations, we undertook the present study to investigate the role of DNA methylation in the regulation of EndoG in human prostate cancer cells.
2. Materials and methods
2.1. Prostate cancer cells
Human prostate cancer cell lines, including well-differentiated, 22Rv1 (ATCC # CRL-2505) and LNCaP cells (ATCC # CRL-1740), and poorly differentiated PC-3 cells (ATCC #CRL-1435) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The cells were maintained in accordance with ATCC recommendations. All cells were seeded at density 0.5 × 106 cells per 100 mm plate, and the media was changed every other day for 6 days. Cisplatin (Bedford laboratories, Bedford, OH) or etoposide (Sigma-Aldrich) was added to serum-free media for 24 h. To evaluate cytotoxicity, the lactate dehydrogenase (LDH) release assay kit (Promega, Madison, WI) was used. Toxicity was expressed as the percentage of LDH released to the medium to that of the total LDH. Cell viability was measured using clonogenic assay as described by us previously [65].
2.2. EndoG siRNA silencing
EndoG silencing was performed according to an algorithm similar to one previously described for breast cancer cells [32]. Briefly, 22Rv1 cells were seeded in Permanox 8-well chambers and grown to 70–80% confluence. To knockdown EndoG mRNA, cells were transfected with designed DY547-siRNA duplexes labeled by 5′UTR of its sense strand (sense siRNA 5′-DY547-AUGCCUGGAACAACCUGGAdTdT-3′ antisense siRNA 3′-UCCAGGUUGUUCCAGGCAUdTdT-5′) or Control Non-Targeting siRNA #1 (Dharmacon, Lafayette, CO). Control siRNA was labeled in the same way as the targeted siRNA. The cells were transfected with complexes containing 50 μM siRNA mixed with TransIT-TKO transfection reagent (Mirus, Houston, TX) according to manufacturer recommendations in serum-free medium for 48 h. Then cells were washed and treated with 80 uM cisplatin for an additional 24 h, washed again and fixed with 4% paraformaldehyde, containing 0.012% saponine (w/v).
2.3. Immunohistochemistry and image analysis
The fixed cells were rehydrated and probed with polyclonal anti-EndoG antibody (Millipore, Billerica, MA) at 1:200 dilution at +4°C overnight. Secondary anti-rabbit-AlexaFluor 594 conjugate (Invitrogen, Carlsbad, CA) at 1:400 dilution was applied for 1 h at room temperature. After subsequent washing the cells were mounted under coverslips using the Prolong® Antifade kit with DAPI (Invitrogen). The cells were visualized using an Olympus IX-81 microscope (Olympus America Inc., Center Valley, PA); images and acquisitions were made with a digital camera HAMAMATSU ORCA-ER (Hamamatsu Photonics K.K., Hamamatsu City, Japan) and software Slidebook 4.1 (SciTech Pty Ltd., Australia). For quantification three experimental points were taken (10 fields of view in each). Cells were masked, and integral optical density (IOD) of each channel was measured using the automated option of the mentioned software. The data were presented as averages of IODx/IODDAPI for each channel.
2.3. RNA extraction and real-time RT-PCR
The total RNA was extracted using RNeasy Mini kit from Qiagen (Valencia, CA) according to manufacturer recommendations. The quality of RNA was determined using 1.2% formaldehyde-agarose gel. Reverse transcription reaction was performed using the GeneAmp Gold RNA PCR core kit (Applied Biosystems, Foster City, CA) and Oligo d(T)16. In general, 1 μg of total RNA was reverse-transcribed in a 50-μl reaction followed by real-time RT-PCR in a 25-μl reaction using SmartCycler (Cepheid, Sunnyvale, CA). Reaction mix was prepared using Platinum SYBR Green qPCR Supermix-UDG (Invitrogen, Carlsbad, CA) according to manufacturer recommendations and primers: 5′-CTACCTGAGCAACGTCGCG-3′ and 5′-TCCAGGTTGTTCCAGGCATT-3′. 18s ribosomal subunit RNA was amplified in parallel reaction using primers 5′-TTCGAACGTCTGCCCTATCAA-3′ and 5′-ATGGTAGGCACGGCGACTA-3′. Two-temperature cycles with annealing/extension temperature at 62°C for EndoG and 64°C for 18s were used. The melting curve analyses were performed at the end of the reaction (after the 45th cycle) between 60°C and 95°C to assess the quality of the final PCR products. The C(t) values were calculated by using the basal fluorescence at 15 units. cDNA samples were diluted for real-time 1:5 and 1:200 for EndoG and 18s, respectively. Three replicate reactions were performed for each sample, and the average C(t) was calculated. The standard curve of the reaction effectiveness was performed using the serially diluted (5 points) mixture of all experimental cDNA samples for EndoG and 18s separately. Calculation of the relative RNA concentration was performed using Cepheid SmartCycle software (Version 2.0d). Data are presented as ratio of EndoG/18s mRNA.
2.5. Cell extracts
Cells (2–4 × 106) were rinsed twice with phosphate buffered saline (PBS). Cells were suspended in buffer containing 50 mM Tris-HCl pH 7.9, 0.25 M sucrose, and the Complete Mini Proteinase Inhibitor Cocktail (Roche Diagnostics, Mannheim, Germany) (1 tablet/10 ml). Cells were then homogenized using a minihomogenizer (Fisher Scientific, Houston, TX). The samples were sonicated in the Virsonic 475 (Virtis, Gardiner, NY) (5 × 20 sec) to decrease viscosity. DNA was removed by centrifugation at 195,000 × g for 2 h. The extracts were dialyzed against storage buffer (55% Glycerol, 10 mM Tris-HCl pH 7.6, 0.5 mM dithiothreitol) and kept at −20°C for up to 2 weeks without loss of endonuclease activity. The protein concentration was measured using the Bradford method [66].
2.6. Plasmid incision assay (PIA)
This assay was used for the measurement of endonuclease activity as previously described [67]. The activity was measured in 20 μl samples containing 1 μg plasmid pBR322 DNA (New England Biolabs, Beverly, MA), 10 mM Tris-HCl, pH 7.7, 25 μg/ml bovine serum albumin V fraction, 0.5 mM dithiothreitol, 5 mM MnCl2, and 2 μl of cell extract in serial dilutions 1:5. After 1 h incubation at 37°C, the reaction was stopped by the addition of 5 μl 1% SDS and 100 mM EDTA. Then digested DNA was subjected to 1% agarose gel electrophoresis at 7 V/cm for 1 h at room temperature. The gel was stained in 0.5 μg/ml ethidium bromide solution for 20 min and photographed under UV light. The EagleEye scanning densitometer (Stratagene, La Jolla, CA) was utilized to quantify the relative amount of endonuclease-treated plasmid DNA present in a covalently closed circular DNA (form 1), open circular DNA (form 2), or linear DNA (form 3). One DNase/endonuclease unit was the amount of the enzyme required to convert 1 μg DNA form 1 to DNA forms 2 and 3.
2.7. Methylation-sensitive McrBC-PCR assay
Methylation status of EndoG promoter was determined by McrBC-PCR assay as described previously [68,69]. Genomic DNA (1 μg) was digested with 20 units of McrBC endonuclease (New England Biolabs, Beverly, MA) for 12 h at 37°C. Undigested DNA served as the control. Following McrBC treatment, PCR was used to amplify the promoter sequence. The primers were designed to amplify CpG islands located within the 5′ region of the EndoG gene: sense 5′GCCGCGAGTCGTACGTGCTGTGCTA, reverse 5′GGGGCGCGACGTTGCTCAGGTAGA. Each PCR reaction contained 0.25 μg of undigested or McrBC-digested DNA and 50 pmol of each primer in 25 μl of 1X GC-RICH PCR System (Roche Diagnostic, Indianapolis, IN). The cycling conditions consisted of an initial denaturation at 95°C for 10 min, followed by 30 cycles of denaturation at 95°C for 30 s, primer annealing at 66°C for 60 s, and extension at 72°C for 60 s. The semi-quantitative aspect of the procedure was verified by a linear increase in PCR product recovery with increasing cycle number and DNA template concentration. The PCR products were separated on 2% agarose gels, stained with ethidium bromide, and photographed. The band intensity was measured by ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
2.8. Western blotting
Protein was separated in 11.5% gel according to the Laemmli procedure [70]. The total protein extract from cells (10–100 μg) was dissolved in 50 mM Tris-HCl, pH 6.8, 1% SDS, 2 mM EDTA, 1% 2-mercaptoethanol and 7.5% glycerol, and denatured by heating at 100°C for 10 min. Electrophoresis was performed at 100 V for 2 h. Proteins were transferred onto the nitrocellulose membrane in Novex transferring buffer (Invitrogen) at 40 V for 3 h, and stained with Ponseau S (Sigma) to control equal protein load as described elsewhere [71]. After soaking in the blocking solution overnight at 4°C, the membrane was incubated with polyclonal anti-EndoG antibody (Millipore) diluted 1:1000 and washed in Tris-buffered saline (TBS). Primary antibodies were detected with anti-rabbit IgG-horseradish peroxidase (HRP) using SuperSignal chemiluminiscent kit (Pierce Biotechnology, Rockford, IL).
2.9. Statistical analysis
The results were expressed as mean ± standard error of mean (SEM). Statistical analysis was performed using SPSS software (SPSS Inc., Chicago, IL USA). To evaluate the significance of differences between two the groups of experiments, the analysis of variance (ANOVA) and Student’s t test were used. Additionally to evaluate the significance of several time points in comparison to one control point, the Bonferroni adjustment of the t-test was used. A value of P < 0.05 was considered statistically significant.
3. Results
3.1. CpG islands in promoter regions of the genes of cell death endonucleases
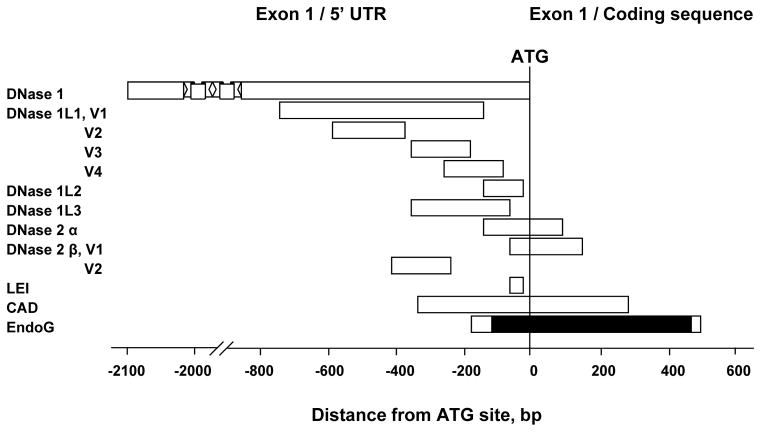
The mammalian genome depends on patterns of methylated cytosines for normal function, but until recently the structural organization of the methylation landscape of the human genome was unclear [40]. It has been reported that the human genome consists of short (<4 kb) unmethylated domains enriched in promoters, CpG islands, and first exons, embedded in a matrix of long methylated domains [40]. A large number of studies have shown that methylation of promoter CpG islands plays an important role in gene silencing [41,42]. The broadly accepted definition of a CpG island as a 200-bp fragment of DNA with G + C content greater than 50% and observed CpG/expected CpG ratio higher than 0.6 failed to exclude many sequences (such as Alu repeats and unknown sequences) that are not associated with regulatory regions of genes [43]. Recent studies indicate that the usage of a modified algorithm to search for CpG islands using a more stringent definition (G + C content higher than 55% and a length greater than 500 bp with observed CpG/expected CpG ratio 0.65) resulted in the exclusion of the majority of Alu repetitive and unknown sequences associated with the 5′ region of genes [43]. In view of these considerations, we applied this algorithm to the analysis of endonuclease genes, which could be regulated by DNA methylation. All known human cell death endonucleases and their sequence variants were analyzed using the CpG Island Searcher program (available at http://www.cpgislands.com [44]): DNase 1, DNase 1L1 variants 1, 2, 3 and 4; DNase 1L2, DNase 1L3 (DNase gamma), DNase 2α, DNase 2β variants 1 and 2, L-DNase II (LEI), caspase-activated DNase (CAD) and EndoG. This analysis showed that EndoG is the only gene that satisfied the criteria of containing a long CpG island in the promoter and exon 1 of the gene (Fig. 1).
Fig. 1.
Identification of CpG island only in promoter and exon 1 regions of EndoG gene among all known human cell death endonucleases including sequence variants. The criteria for CpG island identification were: length > 500bp, GC > 55%, observed/expected ratio > 0.65). DNase 1 (GenBank accession # NM 005223.3), DNase 1L1 variants 1 (NM 006730.2), 2 (NM 001009932.1), 3 (NM 001009933.1) and 4 (NM 001009934.1), DNase 1L2 (NM 001374.2), DNase 1L3 (DNase γ) (NM 004944.2), DNase 2α (NM 001375.2), DNase 2β variants 1 (NM 021233.2) and 2 (NM 058248.1), L-DNase II (LEI) (NM 030666.2), caspase-activated DNase (CAD) (NM 004402.2) and EndoG (NM 004435.2). CpG island in EndoG promoter/exon 1 is shown in solid black color.
3.2. Methylation of EndoG promoter in human prostate cancer cells
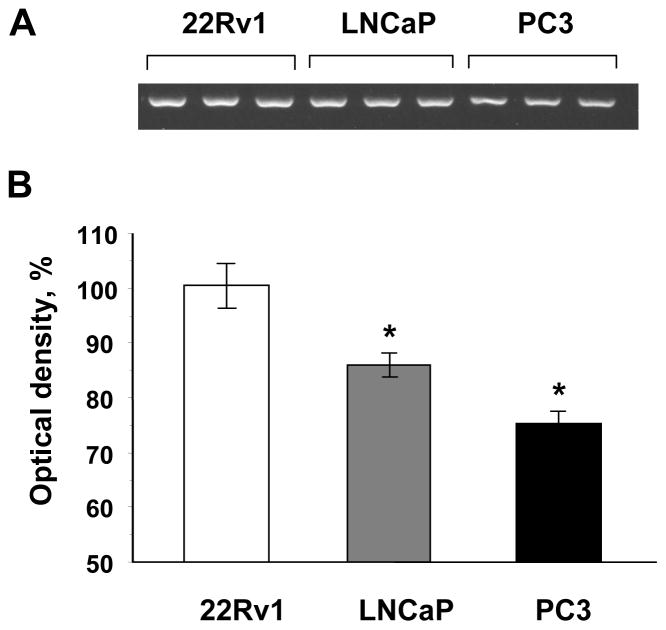
The methylation status of the EndoG promoter/exon 1 in prostate cancer cells was determined by using the methylation-sensitive McrBC-PCR method. McrBC is an endonuclease, that does not act on unmethylated DNA, but cleaves DNA containing 5-methylcytosine in one or both strands and thus nullifies PCR amplification [45]. This experiment showed that in the three prostate cancer cell lines, EndoG promoter methylation was lowest in 22Rv1 cells and highest in PC3 cells (Fig. 2).
Fig. 2.
Methylation of EndoG promoter/exon1 region in human prostate cancer cell lines. (A) EndoG promoter/exon1 DNA methylation measured by the McrBC-PCR screening method. (B) Quantification by densitometry. Optical density of 22Rv1 cells was considered 100%. n = 3 per group, * p <0.05 compared to 22Rv1 cells.
3.3. EndoG expression
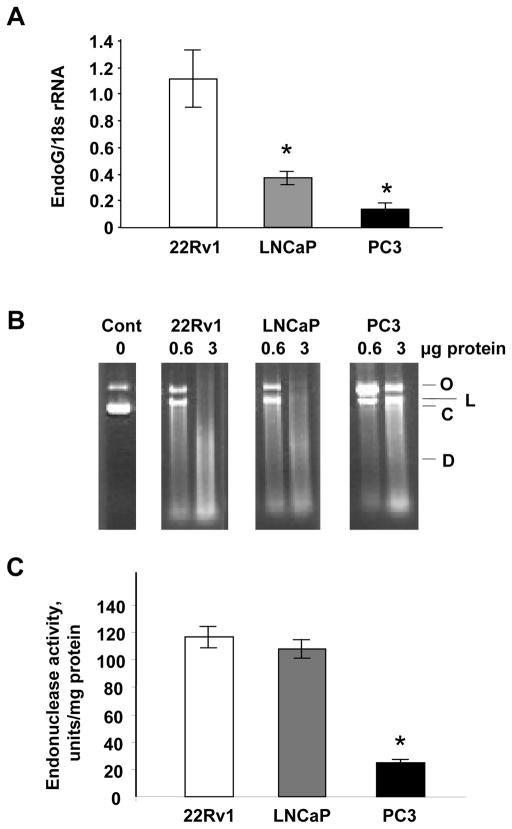
We next determined whether and how EndoG expression correlates with the methylation of the EndoG gene promoter. The expression of EndoG determined by real-time RT-PCR showed that the level of EndoG inversely correlates with the level of the methylation in the EndoG promoter (Fig. 3A). Plasmid incision assay in the presence of Mn2+ ion was used to measure activity of EndoG, the only known Mn-dependent endonuclease, as described by us previously [46]. As expected, EndoG activity was higher in 22Rv1 and LNCaP cells than in PC3 cells (Fig. 3B,C). The Western blotting data described below also confirmed these observations.
Fig. 3.
EndoG expression in human prostate cancer cell lines. (A) Expression of EndoG measured by using real-time RT-PCR. n = 4 per group, *p <0.001 compared to 22Rv1 and <0.05 compared to LNCaP cells. (B) Mn-dependent endonuclease (EndoG) activity in total protein extracts from the cells as measured using plasmid incision assay. O, open circular DNA; L, linear DNA; C, covalently closed supercoiled DNA, D, digested (fragmented) DNA. Each cell extract was tested using two concentrations of protein, 0.6 or 3 μg per sample. (C) Quantification of the plasmid incision assay. n = 4 per group, * p<0.001 compared to 22Rv1 or LNCaP cells.
3.4. Sensitivity of prostate cancer cells to cisplatin and etoposide
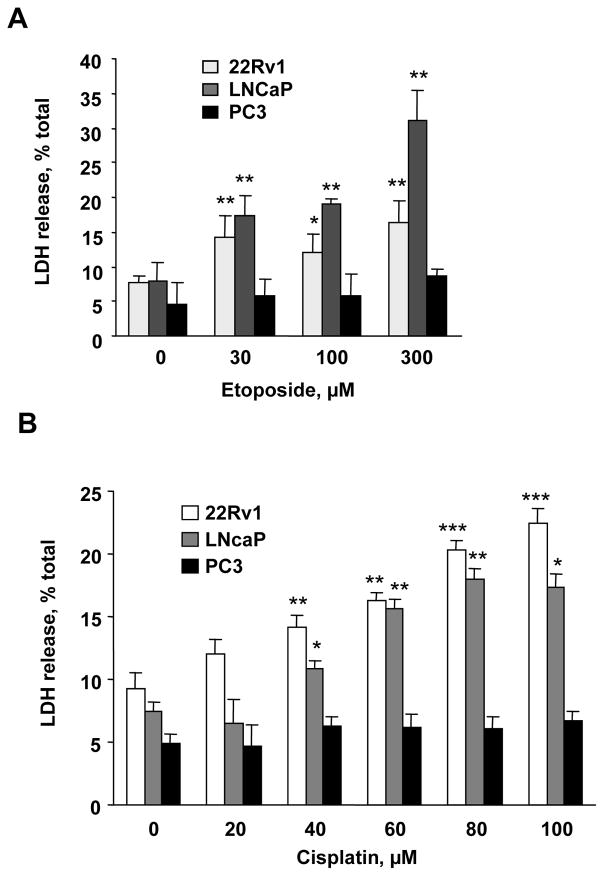
To determine whether the level of EndoG expression affects the sensitivity of prostate cancer cells to chemotherapeutic drugs, we exposed the three cell lines to two anticancer agents, cisplatin (0–100 μM) and etoposide (0–300 μM), which are known to induce cell death in vitro [16,47]. As expected, the two cell lines that expressed EndoG, 22Rv1 and LNCaP, were highly sensitive to both chemotherapeutic agents (Fig. 4). EndoG-deficient PC3 cells, in contrast, were insensitive to these drugs in the range of concentrations used.
Fig. 4.
Sensitivity of prostate cancer cells to cisplatin and etoposide. (A) 22Rv1, LNCaP and PC3 cells were treated with cisplatin (0–100 μM) or (B) etoposide (0–300 μM) for 24 h. Cell death was measured using LDH release assay. n = 4 per group, *p <0.05, **p <0.01, ***p < 0.001 compared to zero point for the same cell line.
3.5. Cisplatin-induced death of prostate cancer cells can be prevented by EndoG silencing
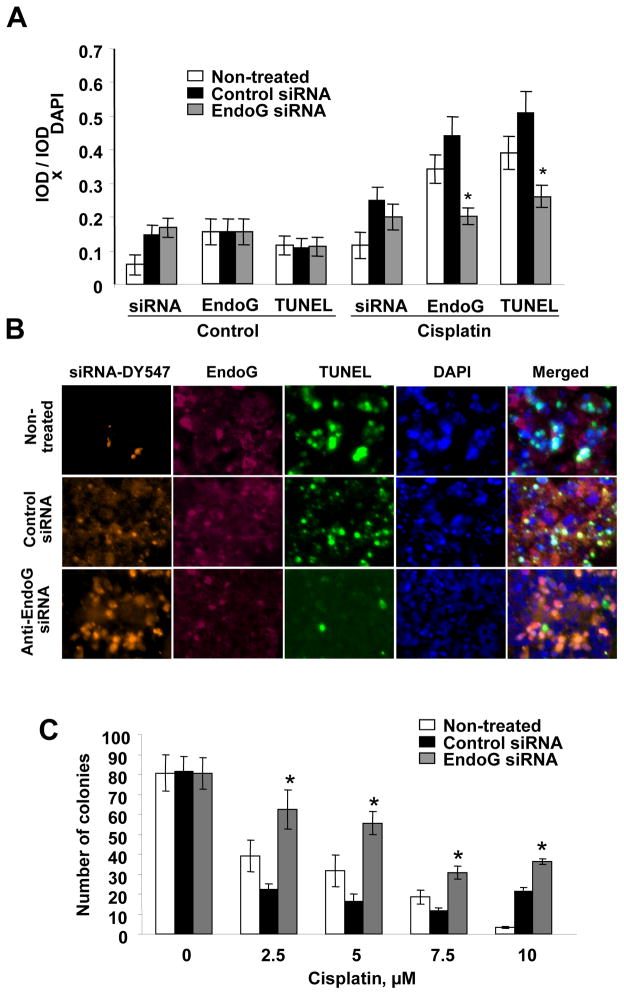
Although EndoG is known to participate in cell death, we needed to determine whether the role of EndoG was the same in prostate cancer cells subjected to injury by cytotoxic agents as has been described in other cells. To determine a causal relationship, we silenced EndoG in 22Rv1 cells by applying siRNA. To show that siRNA was delivered to the cells, we used DY547-labeled siRNA. After DY547-siRNA transfection, 22Rv1 cells were exposed to 80 μM cisplatin, a concentration that had induced significant cell death in the experiments described above. Next, we conducted TUNEL assay to measure DNA fragmentation, and observed that it was decreased, showing that silencing of EndoG leads to significant decrease of EndoG expression and protects cells from DNA fragmentation (Fig. 5A, B). As expected, EndoG silencing resulted in the increased viability of cisplatin-treated 22Rv1 cells as measured using clonogenic assay (Fig 5C). Our results suggest that EndoG is responsible for cisplatin-induced death in prostate cancer cells.
Fig 5.
Cisplatin-induced death of prostate cancer cells can be prevented by silencing EndoG. (A) EndoG-positive 22Rv1 cells treated with DY547-labeled anti-EndoG siRNA had EndoG expression and DNA fragmentation (TUNEL) significantly decreased compared to the cells treated with transfection reagent only (“non-treated”) or with non-specific control siRNA, and were protected from cell death induced by cisplatin (80 μM) exposure for 24 h. n=4 per group, *p<0.05 compared to non-treated or cells treated with control siRNA. (B) Representative images of the cells after treatment with cisplatin. Bar, 10 μm. (C) Silencing of EndoG in 22Rv1 cells by anti-EndoG siRNA caused increased viability of the cells during exposure with cisplatin (2.5–10 μM) (n=4 per group, *p<0.01 to non-treated or cells treated with control siRNA).
Inhibition of DNA methylation induces EndoG and increases sensitivity of PC3 cells to cisplatin and etoposide
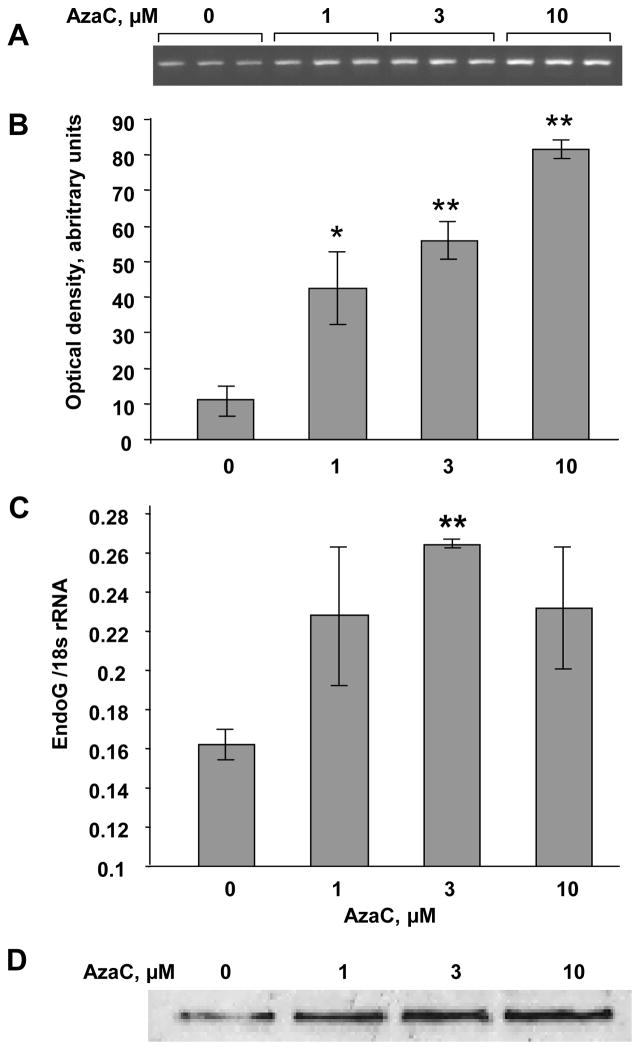
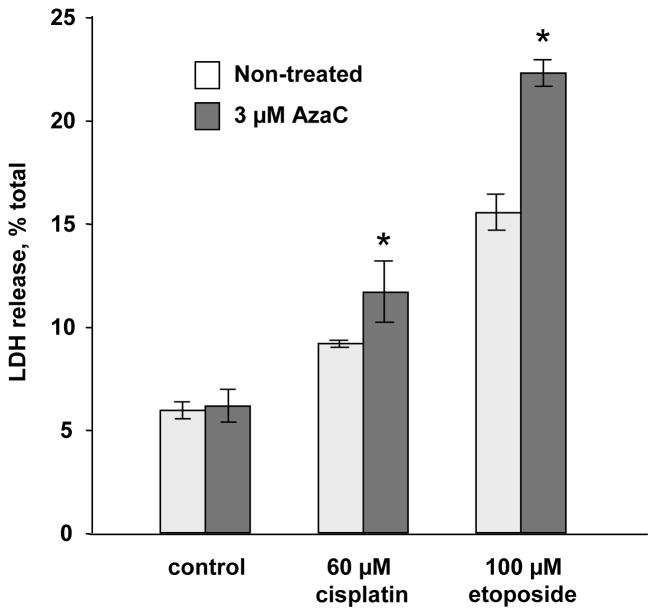
To determine whether EndoG expression is regulated by DNA methylation, we treated PC3 cells with 5′-aza-2-deoxycytidine (decitabine), a DNA methylation inhibitor. Using McrBC-PCR method, we determined that the treatment of PC3 cells with decitabine inhibited methylation of the CpG island in the EndoG gene (Fig. 6A,B). The same concentration of decitabine also increased EndoG expression as determined by real-time RT-PCR and Western blotting (Fig. 6C,D). These data suggest that EndoG expression is regulated by DNA methylation. Importantly, the induction of EndoG by demethylation caused a significant increase in sensitivity to cisplatin and etoposide (Fig. 7).
Fig. 6.
Inhibition of DNA methylation induces EndoG expression in PC3 cells. The cells were exposed with AzaC (0–10 μM) for 24 h. (A, B) EndoG promoter methylation was measured using the McrBC-PCR screening method. n = 3 per group, n = 3 per group, * p <0.05, ** p <0.01 compared to zero point. (C) EndoG expression assessed by real-time RT-PCR. n = 4 per group, * p <0.01, (D) EndoG protein expression determined by Western blotting. The membrane was stained with Ponseau S to control equal protein load (75 μg/well).
Fig. 7.
Inhibition of DNA methylation increases cisplatin and etoposide sensitivity of PC3 cells. The cells were exposed to AzaC (3 μM) for 24 h. EndoG mRNA induction was confirmed by real time RT-PCR (in Figure 6C above). The cells were then exposed to cisplatin (60 μM, 24 h) or etoposide (100 μM, 24 h). Cell death was measured by the LDH release. n=4 per group, *p<0.05 compared to corresponding non-treated cells.
3.6. Role of histone acetylation in regulation of EndoG expression in prostate cancer cells
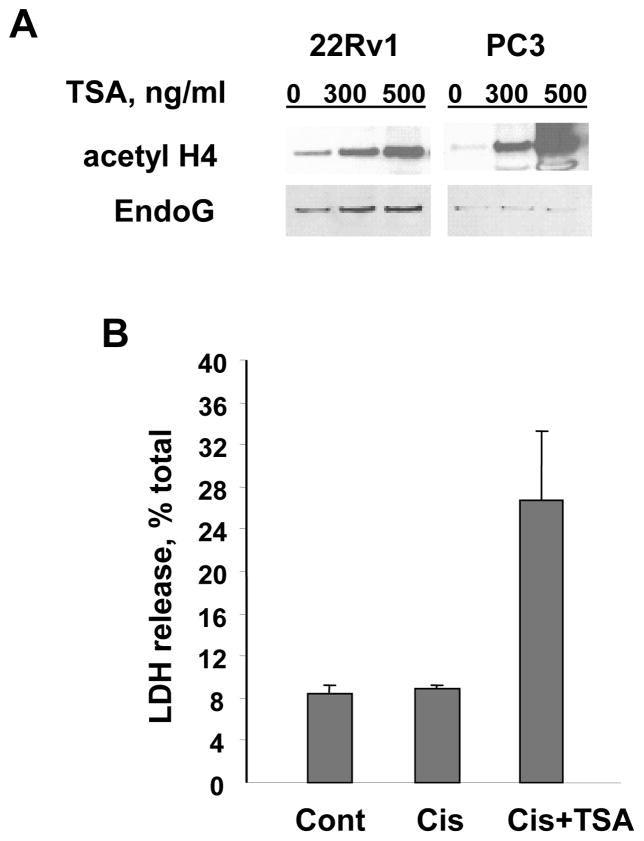
Histone modification, in particular histone acetylation, is another epigenetic mechanism that is important in regulation of genes in prostate cancer [7,8,48]. To determine whether and how histone acetylation regulates EndoG expression, two prostate cancer cell lines were treated with the histone deacetylase inhibitor trichostatin A (TSA), and EndoG protein expression was studied using Western blotting. The exposure of the cells to TSA induced high levels of EndoG expression in EndoG-positive 22Rv1 cells, whereas in EndoG-deficient PC3 cells, EndoG was not induced (Fig. 8A). As expected, EndoG induction by TSA caused increased sensitivity to cisplatin (Fig. 8B). These data demonstrate that chromatin acetylation is important for EndoG expression. Taken together with the above experiments, these data also indicate that DNA methylation plays a primary role in EndoG regulation as compared to histone acetylation. In other words, the CpG island of the EndoG gene has to be hypomethylated in order to allow regulation of EndoG expression by histone acetylation.
Fig. 8.
Histone acetylation induced by TSA causes EndoG induction and increased sensitivity to cisplatin. (A) Histone acetylation induced by TSA caused EndoG induction only in EndoG-expressing 22Rv1 cells but not in EndoG-negative PC3 cells as determined by Western blotting. The membranes were stained with Ponseau S to control equal protein load (30 μg/well). Histone H4 acetylation was assessed using polyclonal acetyl-histone H4 antibody (Upstate, Lake Placid, NY). (B) Induction of EndoG by TSA (100ng/ml) in 22Rv1 cells was associated with the increased sensitivity to cisplatin (Cis, 25 μM).
4. Discussion
Existing studies of endonucleases in either normal prostate tissue or prostate cancer cells are limited [49–55]. The degradation of genomic DNA into nucleosome-sized fragments was shown to be an early event in castration-induced androgen withdrawal that involves the death of the androgen-dependent epithelial cells, following an increase in endonuclease activity [49–51]. Anticancer drugs were shown to induce death of cancer cells through endonuclease-mediated DNA fragmentation [52,53], and inhibition of endonuclease activity had a protective effect [52]. Although endonuclease-mediated DNA fragmentation is commonly used as a marker of apoptosis in prostate cancer [54,55], there are virtually no studies performed on the mechanisms of DNA fragmentation by endonucleases or the regulation of these enzymes in prostate cancer.
In general, there is no difference between apoptosis and necrosis in terms of endonuclease-mediated DNA fragmentation. In both processes, the fragmentation of DNA is produced by the same group of cytotoxic endonucleases. Our recent studies showed that if the impact is weak or the used model is strictly apoptotic, for example, in etoposide-induced cell injury [32], EndoG and other endonucleases work as apoptotic enzymes. Strong in vitro injuries of cells or in vivo injuries usually force endonucleases to act as executioners of necrosis [33,35]. In the present study, as before [32,33,46], we use LDH release as a universal method to assess cell death.
DNA methylation of cell death-related genes was shown to be involved in pathogenesis and to correlate with the prognosis of prostate cancer [56]. Demethylation is known to regulate DNA damage-and death-related genes. For example, Pulukuri & Rao [57] demonstrated that demethylation of p53 and p21Waf1/Cip1 pathway in LNCaP cells led to the inhibition of cell proliferation as well as the induction of apoptosis. Our data are similar to those in a recent report by Das et al. [48] wherein it was observed that the loss of TMS1/ASC gene expression associated with complete methylation of the promoter region in LNCaP cells was restored by AzaC, while TSA had no effect. The role of DNA methylation in regulation of cytotoxic endonucleases has not been studied.
To study the potential regulation of EndoG by methylation, we first compared the promoter and exon 1 regions of all known human endonucleases involved in cell death. This comparison showed that EndoG promoter/exon 1 contains the longest CpG island of all cell death endonuclease sequences, and thus it is likely to be methylated. We found that two human prostate cancer cell lines, 22Rv1 and LNCaP, in which EndoG is highly expressed, contain unmethylated EndoG CpG islands, while in PC3 cells, EndoG is silenced and the EndoG CpG island is methylated. By using specific siRNA, EndoG was shown to be responsible for cisplatin or etoposide-induced cell death in 22RV1 and LNCaP cells. Unlike the two other prostate cancer cell lines, the EndoG-deficient PC3 cells were insensitive to cisplatin or etoposide-induced cell death. Inhibition of methylation by AzaC increased EndoG expression in PC3 cells and made them sensitive to the cytotoxic chemotherapeutic drugs.
The induction of EndoG by promoter hypomethylation may explain the synergy between AzaC and cisplatin during treatment of prostate cancer DU145 as observed by others [47]. The observation that EndoG is important in inducing the death of prostate cancer cells may explain the resistance of prostate cancer to cisplatin and camptothecin reported in some studies [58,59]. Our data provide further evidence that EndoG is the endonuclease that is important in cell death induced by cell injury.
It is interesting that PC3 is not only chemoresistant but is also an invasive cell line, while the two other prostate cancer cell lines are not invasive. This finding is in agreement with our previous study in which we determined that endonuclease activity in the differentiated non-invasive human breast cancer cell lines was higher than that in the poorly differentiated invasive cells [32]. In these cells, the expression of EndoG was strongly correlated with the degree of estrogen receptor expression and showed an inverse correlation with vimentin and matrix metalloproteinase-13. The EndoG-positive, well-differentiated, non-invasive cells were more sensitive to etoposide- or camptothecin-induced cell death than the EndoG-negative, poorly differentiated, invasive cells. These findings suggest that the presence of EndoG in non-invasive cancer cells determines their sensitivity to cytotoxic agents. Downregulation of endonucleases was also observed in diethylnitrosamine (DEN)-induced hepatomas in rats, compared to normal liver tissue [60]. The decrease was proportional to the degree of dedifferentiation, and endonucleases activity was lowest in poorly differentiated tumors. Immortalization of rat fibroblasts with the S1A segment of SA7 adenovirus also led to a significant decrease of endonuclease activity [61]. However, the particular endonuclease that was decreased in tumor cells was not identified in these studies.
Future studies may be necessary to determine the role of other epigenetic mechanisms in regulation of EndoG and their role in chemoresistance to prostate cancer and cancers of other organs. Chemotherapy is currently one of the frequently used therapeutic strategies for prostate cancer [62–64], and measurement of EndoG may be a potentially useful approach to evaluate chemosensitivity of cancer cells to determine optimal conditions for chemotherapy. Manipulating with sensitivity to cytotoxic agents to alter cancer progression has been suggested as a therapeutic approach for prostate cancer in several studies [10,11]. If in vivo studies confirm our observation that EndoG is a potential key mediator of prostate cancer cell death regulated by the methylation of EndoG gene promoter, future epigenetic therapeutics will need to be targeted to EndoG.
Acknowledgments
This research was supported in part by VA Merit Review grants (S.V.S., A.G.B.), and Tobacco Settlement Award, and National Institutes of Health grants 5R03CA114729-02 and 1R01DK078908-01 (A.G.B.).
We thank Yulia Dadugina and Anna Stewart for excellent technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li LC, Carroll PR, Dahiya R. Epigenetic changes in prostate cancer: implication for diagnosis and treatment. J Natl Cancer Inst. 2005;97:103–115. doi: 10.1093/jnci/dji010. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA Cancer J Clin. 2002;52:23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 3.Perry AS, Foley R, Woodson K, Lawler M. The emerging roles of DNA methylation in the clinical management of prostate cancer. Endocr Relat Cancer. 2006;13:357–377. doi: 10.1677/erc.1.01184. [DOI] [PubMed] [Google Scholar]
- 4.Rennie PS, Nelson CC. Epigenetic mechanisms for progression of prostate cancer. Cancer Metastasis Rev. 1998;17:401–409. doi: 10.1023/a:1006121219097. [DOI] [PubMed] [Google Scholar]
- 5.Schulz WA, Hatina J. Epigenetics of prostate cancer: beyond DNA methylation. J Cell Mol Med. 2006;10:100–125. doi: 10.1111/j.1582-4934.2006.tb00293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walton TJ, Li G, Seth R, McArdle SE, Bishop MC, Rees RC. DNA demethylation and histone deacetylation inhibition co-operate to re-express estrogen receptor beta and induce apoptosis in prostate cancer cell-lines. Prostate. 2008;68:210–222. doi: 10.1002/pros.20673. [DOI] [PubMed] [Google Scholar]
- 7.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 8.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 9.Kopelovich L, Crowell JA, Fay JR. The epigenome as a target for cancer chemoprevention. J Natl Cancer Inst. 2003;95:1747–1757. doi: 10.1093/jnci/dig109. [DOI] [PubMed] [Google Scholar]
- 10.Watson RW, Fitzpatrick JM. Targeting apoptosis in prostate cancer: focus on caspases and inhibitors of apoptosis proteins. BJU Int. 2005;96(Suppl 2):30–34. doi: 10.1111/j.1464-410X.2005.05944.x. [DOI] [PubMed] [Google Scholar]
- 11.McKenzie S, Kyprianou N. Apoptosis evasion: the role of survival pathways in prostate cancer progression and therapeutic resistance. J Cell Biochem. 2006;97:18–32. doi: 10.1002/jcb.20634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Momparler RL. Epigenetic therapy of cancer with 5-aza-2′-deoxycytidine (decitabine) Semin Oncol. 2005;32:443–451. doi: 10.1053/j.seminoncol.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2(Suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 14.Cheng JC, Matsen CB, Gonzales FA, Ye W, Greer S, Marquez VE, Jones PA, Selker EU. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst. 2003;95:399–409. doi: 10.1093/jnci/95.5.399. [DOI] [PubMed] [Google Scholar]
- 15.Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, McCombie R, Herman JG, Gerald WL, Lazebnik YA, Cordon-Cardo C, Lowe SW. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 16.Lee MG, Huh JS, Chung SK, Lee JH, Byun DS, Ryu BK, Kang MJ, Chae KS, Lee SJ, Lee CH, Kim JI, Chang SG, Chi SG. Promoter CpG hypermethylation and downregulation of XAF1 expression in human urogenital malignancies: implication for attenuated p53 response to apoptotic stresses. Oncogene. 2006;25:5807–5822. doi: 10.1038/sj.onc.1209867. [DOI] [PubMed] [Google Scholar]
- 17.Qin HR, Iliopoulos D, Semba S, Fabbri M, Druck T, Volinia S, Croce CM, Morrison CD, Klein RD, Huebner K. A role for the WWOX gene in prostate cancer. Cancer Res. 2006;66:6477–6481. doi: 10.1158/0008-5472.CAN-06-0956. [DOI] [PubMed] [Google Scholar]
- 18.O’Kane HF, Watson CJ, Johnston SR, Petak I, Watson RW, Williamson KE. Targeting death receptors in bladder, prostate and renal cancer. J Urol. 2006;175:432–438. doi: 10.1016/S0022-5347(05)00160-6. [DOI] [PubMed] [Google Scholar]
- 19.Hengartner MO. Apoptosis. DNA destroyers. Nature. 2001;412:27, 29. doi: 10.1038/35083663. [DOI] [PubMed] [Google Scholar]
- 20.Samejima K, Earnshaw WC. Trashing the genome: the role of nucleases during apoptosis. Nat Rev Mol Cell Biol. 2005;6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- 21.Polzar B, Peitsch MC, Loos R, Tschopp J, Mannherz HG. Overexpression of deoxyribonuclease I (DNase I) transfected into COS-cells: its distribution during apoptotic cell death. Eur J Cell Biol. 1993;62:397–405. [PubMed] [Google Scholar]
- 22.Krieser RJ, Eastman A. The cloning and expression of human deoxyribonuclease II. A possible role in apoptosis. J Biol Chem. 1998;273:30909–30914. doi: 10.1074/jbc.273.47.30909. [DOI] [PubMed] [Google Scholar]
- 23.Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 24.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 25.Shiokawa D, Ohyama H, Yamada T, Tanuma S. Purification and properties of DNase gamma from apoptotic rat thymocytes. Biochem J. 1997;326(Pt 3):675–681. doi: 10.1042/bj3260675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Napirei M, Ricken A, Eulitz D, Knoop H, Mannherz HG. Expression pattern of the deoxyribonuclease 1 gene: lessons from the Dnase1 knockout mouse. Biochem J. 2004;380:929–937. doi: 10.1042/BJ20040046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koizumi T. Deoxyribonuclease II (DNase II) activity in mouse tissues and body fluids. Exp Anim. 1995;44:169–171. doi: 10.1538/expanim.44.169. [DOI] [PubMed] [Google Scholar]
- 28.Nagata S. Apoptotic DNA fragmentation. Exp Cell Res. 2000;256:12–18. doi: 10.1006/excr.2000.4834. [DOI] [PubMed] [Google Scholar]
- 29.Napirei M, Karsunky H, Zevnik B, Stephan H, Mannherz HG, Moroy T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 30.Irvine RA, Adachi N, Shibata DK, Cassell GD, Yu K, Karanjawala ZE, Hsieh CL, Lieber MR. Generation and characterization of endonuclease G null mice. Mol Cell Biol. 2005;25:294–302. doi: 10.1128/MCB.25.1.294-302.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David KK, Sasaki M, Yu SW, Dawson TM, Dawson VL. EndoG is dispensable in embryogenesis and apoptosis. Cell Death Differ. 2006;13:1147–1155. doi: 10.1038/sj.cdd.4401787. [DOI] [PubMed] [Google Scholar]
- 32.Basnakian AG, Apostolov EO, Yin X, Abiri SO, Stewart AG, Singh AB, Shah SV. Endonuclease G promotes cell death of non-invasive human breast cancer cells. Exp Cell Res. 2006;312:4139–4149. doi: 10.1016/j.yexcr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basnakian AG, Apostolov EO, Yin X, Napirei M, Mannherz HG, Shah SV. Cisplatin nephrotoxicity is mediated by deoxyribonuclease I. J Am Soc Nephrol. 2005;16:697–702. doi: 10.1681/ASN.2004060494. [DOI] [PubMed] [Google Scholar]
- 34.Yin X, Apostolov EO, Shah SV, Wang X, Bogdanov KV, Buzder T, Stewart AG, Basnakian AG. Induction of Renal Endonuclease G by Cisplatin Is Reduced in DNase I-Deficient Mice. J Am Soc Nephrol. 2007;18:2544–2553. doi: 10.1681/ASN.2006080896. [DOI] [PubMed] [Google Scholar]
- 35.Napirei M, Basnakian AG, Apostolov EO, Mannherz HG. Deoxyribonuclease 1 aggravates acetaminophen-induced liver necrosis in male CD-1 mice. Hepatology. 2006;43:297–305. doi: 10.1002/hep.21034. [DOI] [PubMed] [Google Scholar]
- 36.Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausio J. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J Biol Chem. 2001;276:41945–41949. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- 37.Bahi N, Zhang J, Llovera M, Ballester M, Comella JX, Sanchis D. Switch from caspase-dependent to caspase-independent death during heart development: essential role of endonuclease G in ischemia-induced DNA processing of differentiated cardiomyocytes. J Biol Chem. 2006;281:22943–22952. doi: 10.1074/jbc.M601025200. [DOI] [PubMed] [Google Scholar]
- 38.Jiang H, Sha SH, Forge A, Schacht J. Caspase-independent pathways of hair cell death induced by kanamycin in vivo. Cell Death Differ. 2006;13:20–30. doi: 10.1038/sj.cdd.4401706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Apostolov EO, Shah SV, Ok E, Basnakian AG. Carbamylated Low-Density Lipoprotein Induces Monocyte Adhesion to Endothelial Cells Through Intercellular Adhesion Molecule-1 and Vascular Cell Adhesion Molecule-1. Arterioscler Thromb Vasc Biol. 2007;27:826–832. doi: 10.1161/01.ATV.0000258795.75121.8a. [DOI] [PubMed] [Google Scholar]
- 40.Rollins RA, Haghighi F, Edwards JR, Das R, Zhang MQ, Ju J, Bestor TH. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruchusatsawat K, Wongpiyabovorn J, Shuangshoti S, Hirankarn N, Mutirangura A. SHP-1 promoter 2 methylation in normal epithelial tissues and demethylation in psoriasis. J Mol Med. 2006;84:175–182. doi: 10.1007/s00109-005-0020-6. [DOI] [PubMed] [Google Scholar]
- 42.Taghavi P, van Lohuizen M. Developmental biology: two paths to silence merge. Nature. 2006;439:794–795. doi: 10.1038/439794a. [DOI] [PubMed] [Google Scholar]
- 43.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- 45.Nakayama M, Gonzalgo ML, Yegnasubramanian S, Lin X, De Marzo AM, Nelson WG. GSTP1 CpG island hypermethylation as a molecular biomarker for prostate cancer. J Cell Biochem. 2004;91:540–552. doi: 10.1002/jcb.10740. [DOI] [PubMed] [Google Scholar]
- 46.Basnakian AG, Ueda N, Hong X, Galitovsky VE, Yin X, Shah SV. Ceramide synthase is essential for endonuclease-mediated death of renal tubular epithelial cells induced by hypoxia-reoxygenation. Am J Physiol Renal Physiol. 2005;288:F308–F314. doi: 10.1152/ajprenal.00204.2004. [DOI] [PubMed] [Google Scholar]
- 47.Fang X, Zheng C, Liu Z, Ekman P, Xu D. Enhanced sensitivity of prostate cancer DU145 cells to cisplatinum by 5-aza-2′-deoxycytidine. Oncol Rep. 2004;12:523–526. [PubMed] [Google Scholar]
- 48.Das PM, Ramachandran K, Vanwert J, Ferdinand L, Gopisetty G, Reis IM, Singal R. Methylation mediated silencing of TMS1/ASC gene in prostate cancer. Mol Cancer. 2006;5:28–32. doi: 10.1186/1476-4598-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brandstrom A, Westin P, Bergh A, Cajander S, Damber JE. Castration induces apoptosis in the ventral prostate but not in an androgen-sensitive prostatic adenocarcinoma in the rat. Cancer Res. 1994;54:3594–3601. [PubMed] [Google Scholar]
- 50.Banerjee S, Banerjee PP, Brown TR. Castration-induced apoptotic cell death in the Brown Norway rat prostate decreases as a function of age. Endocrinology. 2000;141:821–832. doi: 10.1210/endo.141.2.7339. [DOI] [PubMed] [Google Scholar]
- 51.Kyprianou N, English HF, Isaacs JT. Activation of a Ca2+-Mg2+-dependent endonuclease as an early event in castration-induced prostatic cell death. Prostate. 1988;13:103–117. doi: 10.1002/pros.2990130203. [DOI] [PubMed] [Google Scholar]
- 52.Shrivastava P, Sodhi A, Ranjan P. Anticancer drug-induced apoptosis in human monocytic leukemic cell line U937 requires activation of endonuclease(s) Anticancer Drugs. 2000;11:39–48. doi: 10.1097/00001813-200001000-00007. [DOI] [PubMed] [Google Scholar]
- 53.Ploski JE, Aplan PD. Characterization of DNA fragmentation events caused by genotoxic and non-genotoxic agents. Mutat Res. 2001;473:169–180. doi: 10.1016/s0027-5107(00)00147-0. [DOI] [PubMed] [Google Scholar]
- 54.Miocinovic R, McCabe NP, Keck RW, Jankun J, Hampton JA, Selman SH. In vivo and in vitro effect of baicalein on human prostate cancer cells. Int J Oncol. 2005;26:241–246. [PubMed] [Google Scholar]
- 55.Vijayababu MR, Kanagaraj PP, Arunkumar AA, Arunakaran JJ. Effects of quercetin on insulin-like growth factors (IGFs) and their binding protein-3 (IGFBP-3) secretion and induction of apoptosis in human prostate cancer cells. J Carcinog. 2006;5:10–20. doi: 10.1186/1477-3163-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzuki M, Shigematsu H, Shivapurkar N, Reddy J, Miyajima K, Takahashi T, Gazdar AF, Frenkel EP. Methylation of apoptosis related genes in the pathogenesis and prognosis of prostate cancer. Cancer Lett. 2006;242:222–230. doi: 10.1016/j.canlet.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 57.Pulukuri SM, Rao JS. Activation of p53/p21Waf1/Cip1 pathway by 5-aza-2′-deoxycytidine inhibits cell proliferation, induces pro-apoptotic genes and mitogen-activated protein kinases in human prostate cancer cells. Int J Oncol. 2005;26:863–871. [PubMed] [Google Scholar]
- 58.Nomura T, Yamasaki M, Nomura Y, Mimata H. Expression of the inhibitors of apoptosis proteins in cisplatin-resistant prostate cancer cells. Oncol Rep. 2005;14:993–997. [PubMed] [Google Scholar]
- 59.Chatterjee D, Wyche JH, Pantazis P. Induction of apoptosis in malignant and camptothecin-resistant human cells. Ann N Y Acad Sci. 1996;803:143–156. doi: 10.1111/j.1749-6632.1996.tb26383.x. [DOI] [PubMed] [Google Scholar]
- 60.Basnakian AG, Boubnov NV, Kirsanova ID. Votrin, II, Nuclear topoisomerase I and DNase activities in rat diethylnitrosamine-induced hepatoma, in regenerating and fetal liver. Biochem Int. 1991;24:429–437. [PubMed] [Google Scholar]
- 61.Basnakian AG, Topol LZ, Kirsanova ID, Votrin, Kiselev FL. Activity of topoisomerase I and endonucleases in cells transfected by a ras oncogene. Mol Biol (Mosk) 1989;23:750–757. [PubMed] [Google Scholar]
- 62.Dyrstad SW, Shah P, Rao K. Chemotherapy for prostate cancer. Curr Pharm Des. 2006;12:819–837. doi: 10.2174/138161206776056100. [DOI] [PubMed] [Google Scholar]
- 63.Kaku H, Saika T, Tsushima T, Nagai A, Yokoyama T, Abarzua F, Ebara S, Manabe D, Nasu Y, Kumon H. Combination chemotherapy with estramustine phosphate, ifosfamide and cisplatin for hormone-refractory prostate cancer. Acta Med Okayama. 2006;60:43–49. doi: 10.18926/AMO/30759. [DOI] [PubMed] [Google Scholar]
- 64.Nakabayashi M, Oh WK. Chemotherapy for high-risk localized prostate cancer. BJU Int. 2006;97:679–683. doi: 10.1111/j.1464-410X.2006.06092.x. [DOI] [PubMed] [Google Scholar]
- 65.Fischer TW, Zbytek B, Sayre RM, Apostolov EO, Basnakian AG, Sweatman TW, Wortsman J, Elsner P, Slominski A. Melatonin increases survival of HaCaT keratinocytes by suppressing UV-induced apoptosis. J Pineal Res. 2006;40:18–26. doi: 10.1111/j.1600-079X.2005.00273.x. [DOI] [PubMed] [Google Scholar]
- 66.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 67.Basnakian AG, James SJ. Quantification of 3′OH DNA breaks by random oligonucleotide-primed synthesis (ROPS) assay. DNA Cell Biol. 1996;15:255–262. doi: 10.1089/dna.1996.15.255. [DOI] [PubMed] [Google Scholar]
- 68.Rabinowicz PD, Palmer LE, May BP, Hemann MT, Lowe SW, McCombie WR, Martienssen RA. Genes and transposons are differentially methylated in plants, but not in mammals. Genome Res. 2003;13:2658–2664. doi: 10.1101/gr.1784803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martens JH, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. Embo J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 71.Hofnagel O, Luechtenborg B, Stolle K, Lorkowski S, Eschert H, Plenz G, Robenek H. Proinflammatory cytokines regulate LOX-1 expression in vascular smooth muscle cells. Arterioscler Thromb Vasc Biol. 2004;24:1789–1795. doi: 10.1161/01.ATV.0000140061.89096.2b. [DOI] [PubMed] [Google Scholar]