Abstract
Digital histopathological images provide detailed spatial information of the tissue at micrometer resolution. Among the available contents in the pathology images, meso-scale information, such as the gland morphology, texture, and distribution, are useful diagnostic features. In this work, focusing on the colon-rectal cancer tissue samples, we propose a multi-scale learning based segmentation scheme for the glands in the colon-rectal digital pathology slides. The algorithm learns the gland and non-gland textures from a set of training images in various scales through a sparse dictionary representation. After the learning step, the dictionaries are used collectively to perform the classification and segmentation for the new image.
Keywords: digital pathology, gland segmentation, texture, dictionary learning
1. DESCRIPTION OF PURPOSE
Digital histopathological images provide detailed spatial information of the tissue at micrometer resolution.1 Among the available contents in the pathology images, meso-scale information, such as the gland morphology, texture, and distribution, are useful diagnostic features.
Researchers in2–6 design gland and nuclei segmentation algorithms that aid in determining the grading and/or sub-typing of the prostate, breast, and brain cancers. Gland morphology is studied in.7–9 In gland extraction, one common assumption is that the contours of the glands appear as conic curves in 2D imaging planes.10, 11 While this may be a satisfying assumption for normal tissue, for the malignant tissue, however, such a morphological prior does not hold.
In this work, focusing on colon-rectal cancer tissue samples, we propose a multi-scale learning based segmentation scheme for the glands in the colon-rectal digital pathology slides. The algorithm learns the gland and non-gland textures from a set of training images in various scales through a sparse dictionary representation, regardless of the tissue type (benign vs malignant). After the learning step, the dictionaries are used collectively to perform the classification and segmentation for the new image.
2. METHOD
In,12 a sparse dictionary approach is employed for extracting texture features from the foreground and background, for the purpose of interactive segmentation. In digital pathology images, it is noted that certain images features can be captured in a multi-scale fashion. As a result, we extend the sparse dictionary representation based segmentation into a multi-scale framework.
Specifically, denote the training images as
| (1) |
Their corresponding ground truth segmentations are
| (2) |
where 1 indicates the gland region. Then, M0 image patches of the size m × m are sampled, with replacement and overlapping, from the training images.13 A 3-channel RGB patch is denoted as
| (3) |
where gi is from gland region and ti is non-gland. Such a sample-with-replacement strategy successfully enlarges the sample set and reduces the estimation variance. All the patches are grouped into two categories: gland and non-gland. Denote the patch list of the gland tissue as
| (4) |
and the non-gland tissue has the list
| (5) |
Here, we slightly abuse the notation such that gi, ti ∈ ℝ3m2 are vectors.
With the two lists of patches (vectors), the K-SVD algorithm is used to learn the underlining structure of the texture space as in Algorithm 114
Algorithm 1.
K-SVD Dictionary Construction
| 1: | Initialize (T0 resp.) |
| 2: | repeat |
| 3: | Find sparse coefficients Γ (γi’s) using any pursuit algorithm. |
| 4: | for j = 1, …, d0, update fj, the j-th column of (T0 resp.), by the following process do |
| 5: | Find the group of vectors that use this atom: ζj:= {i : 1 ≤ i ≤ d0, γi(j) ≠ 0} |
| 6: | Compute where is the i-th row of Γ |
| 7: | Extract the i-th columns in Ej, where i ∈ ζj, to form |
| 8: | Apply SVD to get |
| 9: | fj is updated with the first column of U |
| 10: | The non-zeros elements in is updated with the first column of V × Δ(1, 1) |
| 11: | end for |
| 12: | until Convergence criteria is met |
The over-complete basis for G0 (T0 resp.) is denoted as
| (6) |
Such a dictionary construction is performed in the highest (native) resolution of the image. In order to better capture the texture information at various scales, wavelet decomposition is performed on the Ii’s, and the learning is performed at each decomposition level. More explicitly, let the original image be at level 0, then level i ≥ 1 is the 2i down-sampled version of the original image. In all the levels, the same patch size is used for learning. Equivalently, the same patch will cover bigger tissue region and therefore capture larger scale information of the image texture. The dictionaries (gland and non-gland) in the i-th level are denoted as and , with i ≥ 0.
After the gland and non-gland dictionaries at all levels have been constructed, we perform classification on the patches in the given image to be segmented. The classification is also performed in a multi-scale fashion, which is detailed as follows.
At the original resolution, given a new image patch g : ℝm×m → ℝ3, by slightly abuse of notation, we also consider g as a 3m2 dimensional vector. Then, we compute the sparse coefficient η of g, with respect to the dictionary .
| (7) |
where k is the sparsity constraint. The reconstruction error is defined as
| (8) |
This problem is solved by the Orthogonal Matching Pursuit (OMP) method.15 Similarly, the sparse coefficient ξ of g under the dictionary is also computed as:
| (9) |
The reconstruction error is defined as:
| (10) |
Moreover, we define
| (11) |
It is noted that the errors and indicate how well (or how bad) the new image patch can be reconstructed from information learned from the gland and non-gland region. Hence, the higher the e0 is, the more this patch is considered as the non-gland region and vice versa. Furthermore, for each pixel (x, y) ∈ Ω, a patch p(x, y) is extracted as the sub-region of I defined on [x, x + m] × [y, y + m]. Following the procedure above, we compute the image e0(x, y) accordingly. A probability map P0(x, y) for the gland region is defined as
| (12) |
Such procedure is performed for all the scales and a series of probability maps Pi(x, y) with i = 0, 1, …, L are constructed. Note that each Pi(x, y) is reconstructed to the original scale (with all detail coefficients set to 0) and is therefore of the same size as P0. Finally, define
| (13) |
as the maximum response along the scale dimension. The final probability is threshold at 0.5 for the final mask of gland region.
3. EXPERIMENTS AND RESULTS
80 images are extracted from The Cancer Genome Atlas (TCGA) colon-rectal digital pathology diagnosis images. The resolution is 0.5μm/pixel. The image sizes are all 1024×1024 pixels.
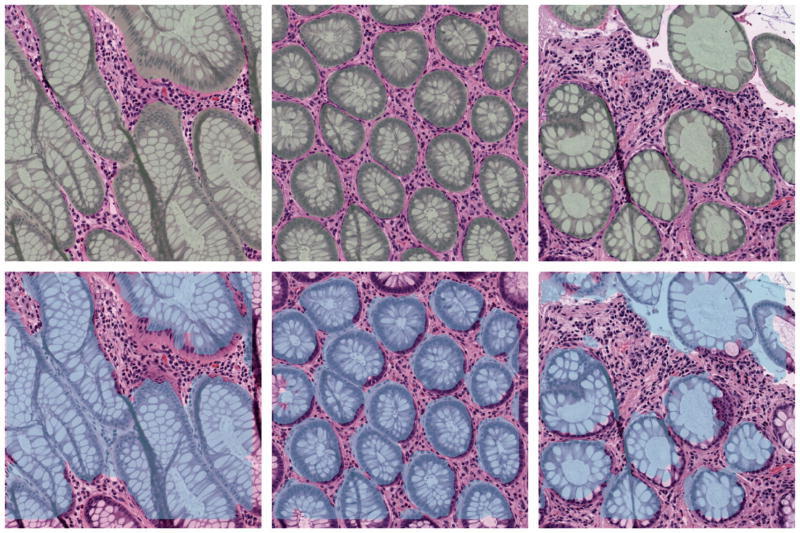
Three randomly picked results are shown in Figure 1. The top row shows the image with the shading indicating the manual segmentation. The corresponding panel below shows the proposed automatic segmentation results.
Figure 1.
Segmentation on three benign tissues patches.
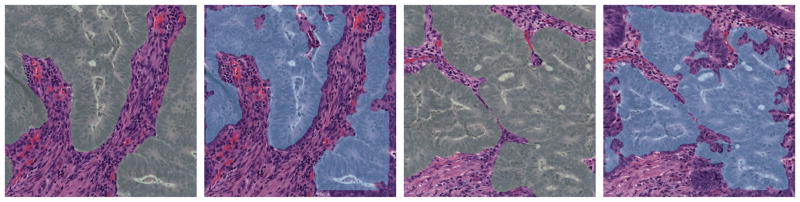
In addition to the benign tissue, the algorithm also performs consistently on malignant tissue, as shown in Figure 2. Here two cases are shown from left to the right: The first/third are the manual segmentation and the second/forth are the algorithm outputs.
Figure 2.
Segmentation on two malignant tissues patches.
Quantitatively, a leave-one-out scheme is performed for testing. Specifically, the 79 images and their manual segmentations are used for learning. The learned model is then applied on the left-out image. The Dice coefficient is measured for all the 80 cases and a mean of 0.81 accuracy (0.056 standard deviation) is achieved. Moreover, the Hausdorff distance is measured and a mean of 70μm (10.0μm standard deviation) is achieved.
4. CONCLUSION AND FUTURE RESEARCH DIRECTIONS
In this work, we propose a multi-scale learning based segmentation scheme for the glands in the colon-rectal digital pathology slides. The algorithm learns the gland and non-gland textures from a set of training images in various scales through a sparse dictionary representation. After the learning, the dictionaries are used collectively to perform the classification and segmentation for the new image.
More recently, the deep neural network (DNN) framework is becoming more and more widely adopted, including for the task of gland segmentation.16, 17 The DNN framework has the advantage that minimal manually curated features are needed for the classification process. The proposed framework utilizes the sparse dictionary for the learning and feature extraction. However, the signal sparse reconstruction based on dictionary is a linear combination. DNN harnesses the hierarchical nonlinear transformation and may result in better performance. The ongoing research include combining the manually designed features with the DNN based features, for a better overall performance.
The work has not been submitted for publication or presentation elsewhere.
References
- 1.Gurcan MN, Boucheron LE, Can A, Madabhushi A, Rajpoot NM, Yener B. Histopathological image analysis: A review. Biomedical Engineering, IEEE Reviews in. 2009;2:147–171. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Naik S, Doyle S, Agner S, Madabhushi A, Feldman M, Tomaszewski J. Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro; IEEE; 2008. pp. 284–287. [Google Scholar]
- 3.Basavanhally AN, Ganesan S, Agner S, Monaco JP, Feldman MD, Tomaszewski JE, Bhanot G, Madabhushi A. Computerized image-based detection and grading of lymphocytic infiltration in her2+ breast cancer histopathology. Biomedical Engineering, IEEE Transactions on. 2010;57(3):642–653. doi: 10.1109/TBME.2009.2035305. [DOI] [PubMed] [Google Scholar]
- 4.Kong H, Gurcan M, Belkacem-Boussaid K. Partitioning histopathological images: an integrated framework for supervised color-texture segmentation and cell splitting. Medical Imaging, IEEE Transactions on. 2011;30(9):1661–1677. doi: 10.1109/TMI.2011.2141674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper L, Gutman DA, Long Q, Johnson BA, Cholleti SR, Kurc T, Saltz JH, Brat DJ, Moreno CS. The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PloS one. 2010;5(9):e12548. doi: 10.1371/journal.pone.0012548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong J, Cooper LA, Wang F, Gao J, Teodoro G, Scarpace L, Mikkelsen T, Schniederjan MJ, Moreno CS, Saltz JH, Brat DJ. Machine-based morphologic analysis of glioblastoma using whole-slide pathology images uncovers clinically relevant molecular correlates. PLoS ONE. 2013;8(11) doi: 10.1371/journal.pone.0081049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopez CM, Agaian S, Sanchez I, Almuntashri A, Zinalabdin O, Rikabi A, Thompson I. Exploration of efficacy of gland morphology and architectural features in prostate cancer gleason grading. Systems, Man, and Cybernetics (SMC), 2012 IEEE International Conference on; IEEE; 2012. pp. 2849–2854. [Google Scholar]
- 8.Cohen A, Rivlin E, Shimshoni I, Sabo E. Memory based active contour algorithm using pixel-level classified images for colon crypt segmentation. Computerized Medical Imaging and Graphics. 2015 doi: 10.1016/j.compmedimag.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Janowczyk A, Chandran S, Madabhushi A. SPIE Medical Imaging. International Society for Optics and Photonics; 2010. A weighted mean shift, normalized cuts initialized color gradient based geodesic active contour model: applications to histopathology image segmentation; p. 76230Y–76230Y. [Google Scholar]
- 10.Fu H, Qiu G, Shu J, Ilyas M. A novel polar space random field model for the detection of glandular structures. Medical Imaging, IEEE Transactions on. 2014;33(3):764–776. doi: 10.1109/TMI.2013.2296572. [DOI] [PubMed] [Google Scholar]
- 11.Gunduz-Demir C, Kandemir M, Tosun AB, Sokmensuer C. Automatic segmentation of colon glands using object-graphs. Medical image analysis. 2010;14(1):1–12. doi: 10.1016/j.media.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Gao Y, Bouix S, Shenton M, Tannenbaum A. Sparse texture active contour. Image Processing, IEEE Transactions on. 2013;22(10):3866–3878. doi: 10.1109/TIP.2013.2263147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Efron B, Tibshirani R, Tibshirani R. An introduction to the bootstrap. Chapman & Hall/CRC; 1993. [Google Scholar]
- 14.Aharon M, Elad M, Bruckstein A. K-SVD: An algorithm for designing overcomplete dictionaries for sparse representation. Signal Processing, IEEE Transactions on. 2006;54(11):4311–4322. [Google Scholar]
- 15.Pati YC, Rezaiifar R, Krishnaprasad P. Orthogonal matching pursuit: Recursive function approximation with applications to wavelet decomposition. Signals, Systems and Computers, 1993. 1993 Conference Record of The Twenty-Seventh Asilomar Conference on; IEEE; 1993. pp. 40–44. [Google Scholar]
- 16.Kainz P, Pfeiffer M, Urschler M. Semantic segmentation of colon glands with deep convolutional neural networks and total variation segmentation. 2015 doi: 10.7717/peerj.3874. arXiv preprint arXiv:1511.06919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Qi X, Yu L, Heng P-A. DCAN: Deep contour-aware network for accurate gland segmentation. GlaS challenge in Medical Image Computing and Computer-Assisted Interventation; 2015; Springer; 2015. [Google Scholar]