Spliceosomal introns are prevalent in our genomes and also in our minds as unsolved evolutionary mysteries. Are introns primordial features of eukaryotic genes? Or have they been acquired during eukaryotic evolution? These questions are central to a still-simmering debate among biologists. To describe the phylogenetic pattern of introns across eukaryotes, two general models have emerged: the introns-late view claims that all introns have been gained into preformed genes, with their current-day distributions explained by processes of both gain and loss; whereas introns-early proponents posit that most introns can be explained by frequent loss from intron-rich ancestral genes that predate eukaryotic cells (1–4). But the key questions remain unanswered. Both views agree that intron loss does occur, but the main disagreement concerns what fraction of present-day introns have been gained, and how. Spliceosomal introns are dominant features of most eukaryotic genes and genomes, yet we have little knowledge about their mechanisms of acquisition (1). By using evolutionary comparisons between nematode genes, Coghlan and Wolfe (5), in this issue of PNAS, make major strides in understanding spliceosomal intron gain and provide us with a clearer picture of intron evolution in eukaryotic genomes. They not only demonstrate that 122 introns have been gained recently in Caenorhabditis genes, but also provide solid evidence that 28 of them are actually derived from “donor” introns present in the same genome. Indeed, a few of these new introns apparently derive from other introns in the same gene!
Getting a Hold on Intron Gain
Previous phylogenetic interpretations of introns indicate that many, if not most, introns have been gained once without subsequent loss (1, 6). These inferences are powerful when considering the pattern of the vast number of introns known to take residence in eukaryotic genes, but they have been impotent in illuminating the underlying molecular mechanism(s) of intron insertion. Scant few cases of intron gain have revealed anything about the mechanism by which the hordes of introns have apparently been inserted into eukaryotic genes (7, 8). If intron gain is so common, then why is it not well understood? Indeed, what do we actually know about spliceosomal intron gain?
The answer is, a fair bit. Introns tend not to insert randomly into genes but instead are preferentially gained at a constrained nucleotide sequence MAG↓ R, termed the “proto-splice site” (↓ represents the location where the intron inserts) (7, 9). Further, spliceosomal introns are nonrandomly distributed with respect to codons: about half of all introns are between amino acid codons (phase 0) as opposed to the two other possible positions within codons (phases 1 and 2). A recent analysis by Qiu et al. (10) dispels the idea that these sequences represent sites of intron loss but instead act as insertion “targets;” this work systematically extends more limited previous work to demonstrate that proto-splice sites (9) are ancestrally present at sites of later intron gain. More importantly, Qiu et al. (10) also show these unoccupied sites follow precisely the same pattern of phase bias as recent introns that have been gained at such sites.
Thus, there are some clues about the process of intron gain, but what mechanisms are responsible for creating new DNA sequence at a previously unoccupied site? Transposable elements are likely suspects, but other possibilities are gene conversion, tandem exon duplication, insertion by self-splicing (group II) introns, and reverse-splicing of existing introns (7, 8). These mechanisms are responsible for a few cases; however, there is no generally demonstrated model for the propagation of most new introns into genes. Indeed, all of these mechanisms could be responsible for at least some recently gained introns.
Herein lies the difficulty for understanding the mechanism(s) of intron gain. Only a few cases provide clues to intron origins, and those that do are so few in number that it seems unwise to generalize from them. The simple reason for the paucity of good cases is that spliceosomal introns diverge in sequence at about the rate of silent substitution. Thus, the actual nucleotide sequences of introns, although potentially clear indicators of their own evolutionary history, are ephemeral features of their existence. Only very recently gained introns (in which substitution has not erased all sequence similarities, at ≈100 million years of divergence) allow for the possibility of understanding the underlying process(es) of insertion (7).
Among complete genome sequences, a few possible comparisons between closely related genomes have the potential to reveal cases of recent intron gain and thus provide clues to the underlying process. Comparisons among humans, mice, and rats have come up virtually empty (11), showing exceedingly few intron differences between these genomes, most of which can be attributed to intron loss. Perhaps this result should not have come as a surprise, because vertebrate genes have apparently experienced evolutionary stasis in their intron content (7). Another approach is to look within particular genomes for evidence of homologous introns occupying unrelated genes. Recent attempts by Fedorov et al. (12) to do this failed to reveal even a single case for humans, Drosophila melanogaster, Arabidopsis thaliana, and Caenorhabditis elegans (although, curiously, the latter contrasts with the findings of ref. 5). This dearth of data led Fedorov et al. (12) to advise, “To understand the real mechanism of intron acquisition, we must find and analyze several examples of recently acquired introns. Such cases, which will involve the appearance of a novel sequence within a phylogenetic pattern, would shed light on the question of intron gain.” Two kinds of data are needed: (i) lots of cases of newly gained introns, and among those, (ii) some that are recent enough to discover their source(s).
Intron Insertions in Worm Genes
The analysis of Coghlan and Wolfe (5) is the first systematic study to both identify large numbers of clear intron gains and pinpoint in some cases their evolutionary source: other introns from the same or different genes. The approaches used to identify new introns and discern their origins are depicted in Fig. 1. This study represents the largest number of recently gained introns identified at one time. It should be emphasized that this set of 122 new introns represents very conservatively identified cases. Previous studies that indicated ≈6,500 intron differences between C. elegans and Caenorhabditis briggsae genes (13) suggest that many more introns have been gained (and lost; see below) in worm genes. Being mindful of their methodological focus to identify only clear cases of intron gains in a sea of many good candidates, Coghlan and Wolfe (5) wisely do not make any general claims from their data about overall rates of intron gain in these worms, because only bare minimum estimates (of unclear relevance) would be possible.
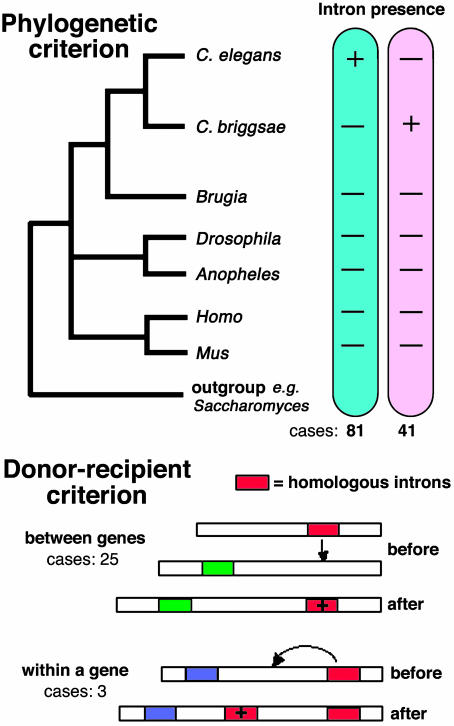
Fig. 1.
Diagnostic criteria used in ref. 5 to identify recently gained introns and, in some cases, possible donors. A set of recently gained introns, each having a pattern like one depicted (Upper), was determined by a rigorous phylogenetic scheme: (i) protein sequences from complete C. elegans and C. briggsae genomes were compared to detect homologous genes, (ii) gene sequence comparisons between these homologs identified introns whose positions were uniquely present in either C. elegans or C. briggsae and not found in any of the other species compared (including a distantly related worm, Brugia malayi, two insects and two mammals), (iii) sequence alignments of these intron-containing genes were quality-checked to verify that the introns were unambiguously positioned in a highly conserved region, and (iv) formal phylogenetic analyses (using an appropriate outgroup) established orthology of the genes. The resulting novel introns found in either C. elegans or C. briggsae were inferred as being gained since the divergence of these two species. All of the recently gained introns were then further scrutinized to determine whether their source(s) could be identified from among other introns in the same species. Two outcomes of this analysis are shown (Lower): of those new introns for which homologous introns could be identified, these sources were from either introns in other (unrelated) genes or different introns in the same gene. Inferred ancestral states are labeled “before” and present states, “after.”
Beyond the numbers of new introns found, the more surprising result of ref. 5 is the discovery of likely molecular donors. By their criteria, all of the new introns are no older than the divergence time between C. elegans and C. briggsae, estimated at ≈100 million years ago, the approximate evolutionary distance at which homologous introns become indiscernible. Considering introns of this vintage, one does not fully expect to find all donors. Of those donors identified, the evidentiary “gunsmoke” of sequence similarity may be sufficiently diffuse to not prove the cases beyond a reasonable doubt. Of 32 novel introns that had significant similarity to other introns in the same genome, ref. 5 rejected four, because they matched large numbers of other introns. But even with their stringent statistically validated comparison methods, there is room for some remaining doubt: (i) many of the putatively homologous introns share repetitive DNA sequences, and (ii) most of the new introns had matches to multiple additional introns in the genome. It seems unlikely that all of these donor recipients are false-positives matches, and prima facie evidence for this diagnosis comes from the unlikely possibility that three new introns hail from the same gene in which their older donors reside (Fig. 1 Lower). To more clearly identify donors will require additional cases but, more importantly, comparisons among more closely related species, as suggested by ref. 5 in their concluding remarks.
Lessons Learned and Open Questions
Coghlan and Wolfe (5) break fertile new ground for intron evolution studies in revealing that introns do insert into genes at relatively recent time scales and that, in so doing, these introns can yield clues about their origins and the mechanism(s) of gain. Where do new introns come from? The answer is, apparently, other introns. If so, this would be most consistent with a reverse splicing model. Additional data consistent with this model are marshaled, given its requirement for germline expression of donors and recipients (7). The authors' limited data sample for donors does not indicate a germline bias, but the recipients are clearly germline-expressed. Another hint lies in the strong disparity of new introns present in genes involved in RNA splicing; whether this bias indicates a direct connection to the splicing machinery or just to this class of abundantly transcribed messages is unclear. Taken together, this is considerable evidence in favor of the reverse-splicing mechanism for intron gain, albeit a model clearly in need of additional testing.
Among other information gained from ref. 5 is that the intron phase bias appears largely, if not wholly, determined by insertion biases. The phase distribution of their novel introns is statistically indistinguishable from (older) introns present in the rest of the genome, results congruent with the phylogenetically based analyses of Qiu et al. (10). Finally, there is the strong implication by ref. 5 that their new introns inserted at proto-splice sites. Although they do not directly demonstrate the presence of insertion sites in the intron-lacking ancestor, they do provide compelling alternative evidence: the sequence flanking new introns is more constrained (to the predicted target site) than an analogous sequence flanking old introns. This suggests these sequences are crucial for precise intron insertion, but once the intron is present, they are more relaxed to substitution. These are obvious clues for designing experiments to evaluate mechanistic models of intron gain.
What about intron loss? What fraction of the thousands of intron differences between C. elegans and C. briggsae are due to lost introns? Although Coghlan and Wolfe (5) do not address intron loss, the comparative methods they have developed, along with additional species to compare, will certainly provide an answer to this question, along with estimates of rates of gain. However, the ostensibly high frequency of intron gains implied by Coghlan and Wolfe's data would seem to stand in contrast to another recent analysis (14) that inferred intron loss was the dominant process responsible for high rates of intron turnover in Caenorhabditis. Yet this massive loss interpretation was based on an apparently erroneous inference of numerous introns in Caenorhabditis ancestors. Perhaps, then, intron evolution in Caenorhabditis is largely dominated by intron gain. In any case, there is certainly enough smoke to sentence worms to lengthy sentences of hard labor in the laboratories of intron enthusiasts.
Acknowledgments
I thank Jeff Palmer, Dawn Simon, Ken Wolfe, and Arlin Stoltzfus for informative discussions and helpful comments. Apologies are given to those whose work could not be cited due to space restrictions.
See companion article on page 11362.
References
- 1.Logsdon, J. M., Jr. (1998) Curr. Opin. Genet. Dev. 8, 637–648. [DOI] [PubMed] [Google Scholar]
- 2.de Souza, S. J., Long, M., Klein, R. J., Roy, S., Lin, S. & Gilbert, W. (1998) Proc. Natl. Acad. Sci. USA 95, 5094–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch, M. & Richardson, A. O. (2002) Curr. Opin. Genet. Dev. 12, 701–710. [DOI] [PubMed] [Google Scholar]
- 4.Roy, S. W. (2003) Genetica 118, 251–266. [PubMed] [Google Scholar]
- 5.Coghlan, A. & Wolfe, K. H. (2004) Proc. Natl. Acad. Sci. USA 101, 11362–11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer, J. D. & Logsdon, J. M., Jr. (1991) Curr. Opin. Genet. Dev. 1, 470–477. [DOI] [PubMed] [Google Scholar]
- 7.Logsdon, J. M., Jr., Stoltzfus, A. & Doolittle, W. F. (1998) Curr. Biol. 8, R560–R563. [DOI] [PubMed] [Google Scholar]
- 8.Stoltzfus, A. (2004) Curr. Biol. 14, R351–R352. [DOI] [PubMed] [Google Scholar]
- 9.Dibb, N. J. & Newman, A. J. (1989) EMBO J. 8, 2015–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu, W. G., Schisler, N. & Stoltzfus, A. (2004) Mol. Biol. Evol. 21, 1252–1263. [DOI] [PubMed] [Google Scholar]
- 11.Roy, S. W., Fedorov, A. & Gilbert, W. (2003) Proc. Natl. Acad. Sci. USA 100, 7158–7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedorov, A., Roy, S., Fedorova, L. & Gilbert, W. (2003) Genome Res. 13, 2236–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein, L. D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M. R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., et al. (2003) PLoS Biol. 1, E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiontke, K., Gavin, N. P., Raynes, Y., Roehrig, C., Piano, F. & Fitch, D. H. (2004) Proc. Natl. Acad. Sci. USA 101, 9003–9008. [DOI] [PMC free article] [PubMed] [Google Scholar]