Abstract
Cytohesins are a family of highly homologous guanine nucleotide exchange factors (GEFs) that act on ADP-ribosylation factors (ARFs). The small ARF-GEFs are involved in integrin signaling, actin cytoskeleton remodeling, and vesicle transport. Here, we selected and applied a specific inhibitor for ARF nucleotide-binding site opener (ARNO)/cytohesin-2, an RNA aptamer that clearly discriminates between cytohesin-1 and cytohesin-2. This reagent bound to an N-terminal segment of cytohesin-2 and did not inhibit ARF-GEF function in vitro. When transfected into HeLa cells, it persisted for at least 6 h without requiring stabilization. Its effect in vivo was to down-regulate gene expression mediated through the serum-response element and knockdown mitogen-activated protein kinase activation, indicating that cytohesin-2 acts by means of mitogen-activated protein kinase signaling. We conclude that the N-terminal coiled-coil and parts of the Sec7 domain of cytohesin-2 are required for serum-mediated transcriptional activation in nonimmune cells, whereas cytohesin-1 is not. Our results indicate that intramer technology can be used not only for assigning novel biological functions to proteins or protein domains but also to prove nonredundancy of highly homologous proteins.
The cytohesin family members are cytoplasmic signaling proteins that are thought to be involved in integrin signaling (1), actin cytoskeleton remodeling events (2, 3), and vesicle transport. They belong to a class of highly homologous guanine nucleotide exchange factors (GEFs), the small ADP-ribosylation factor (ARF)-GEFs, which catalyze the exchange of GDP for GTP on ARFs. (4). All cytohesin family members are modular proteins with an N-terminal coiled-coil, a central Sec7 domain (5) that harbors the GEF activity, a C-terminal pleckstrin homology (PH) domain that mediates membrane localization by means of interaction with specific polyphosphoinositides, and an adjacent C-terminal polybasic domain that cooperates with the PH domain to enhance membrane binding (6–8) (Fig. 1A).
Fig. 1.
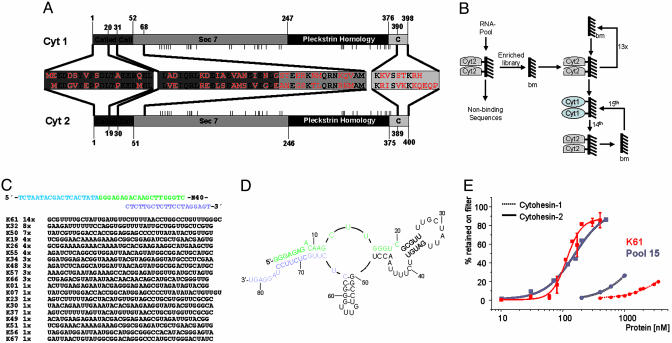
Aptamer selection and characterization. (A) Domain composition of cytohesin-1 and cytohesin-2 containing the N-terminal coiled-coil domain (amino acids 1–51), the Sec7 domain (amino acids 52–246), the PH domain (amino acids 247–375), and the C-terminal polybasic domain (amino acids 376–400) (9, 10). The expanded sequences highlight regions in which the two proteins indicate the highest diversity. Although these regions represent only 17% of the total length, they differ in 37 (red) from a total of 72 amino acids. Single amino acid changes outside of these regions are represented by short vertical dashes. (B) Selection scheme for cytohesin-2-specific aptamers. An RNA library with a starting diversity of 1014 was selected on Sepharose-immobilized cytohesin-2. Preselection against blank Sepharose (bm) in cycles 2–13 efficiently preenriched the pool with protein-binding RNAs. Cycles 14 and 15 included a counterselection against cytohesin-1, immobilized in the same way. (C) Sequencing of pool-15 RNAs revealed the sequences shown (random insert sequences; primer sequences are shown above. Blue, T7 promoter; green, 5′ primer binding site; purple, 3′ primer binding site), listed in order according to their abundance. In total, 19 different sequences were identified. (D) Secondary structure of the most abundant clone K61, generated by mfold (11, 12). Green, 5′ primer site; purple, 3′ primer site. (E) Comparison of binding affinities of the K61 aptamer (red) and pool-15 RNA (blue) for binding to cytohesin-1 (dotted curves) and cytohesin-2 (solid curves), determined by filter binding assays.
Although the members of the small GEFs are strikingly homologous, they demonstrate significant differences in their biological functions (2, 4). For example, cytohesin-1 is a positive effector of leukocyte function-associated antigen-1 (LFA-1)-mediated cell adhesion, whereas this activity has not been shown for cytohesin-2, also known as ADP-ribosylation factor nucleotide-binding site opener (ARNO) (7), or other small GEFs (1). In addition, cytohesin-1 is involved in T cell activation mediated by intercellular adhesion molecule 2 and in stimulation of T cell proliferation by IL-2 (13).
The considerable sequence homology of cytohesin-1 and ARNO/cytohesin-2 (Fig. 1 A) set against the significant differences in their biological function demands further explanation and highlights the need to clearly differentiate between these related regulatory factors. We set out to obtain specific inhibitors of these proteins that can discriminate between cytohesin-1 and -2 in vitro and in living cells. One could then, for example, determine whether members of the cytohesin family of small ARF-GEFs are involved in regulating gene expression in nonimmune cells.
Methods
Protein Expression and Purification. Cytohesin-1 and cytohesin-2 (triglycine splice variants), cytohesin-2-Sec7, and cytohesin-2-Sec7PHC were heterologously expressed in Escherichia coli BL21(DE3). The proteins contained an N-terminal His6 tag and were purified by standard Ni2+-nitrilotriacetic acid affinity chromatography. Full-length proteins also were purified by ion exchange chromatography on a Bio-Scale Q column (Bio-Rad). For aptamer selection, cytohesin-1 and cytohesin-2 were immobilized on CNBr-activated Sepharose (Amersham Biosciences).
RNA Library and in Vitro Selection. The synthetic single-stranded DNA library 5′-TCT AAT ACG ACT CAC TAT AGG GAG AGA CAA GCT TGG GTC-N40-CTC TTG CTC TTC CTA GGA GT-3′ (N40, randomized part) was amplified by using the selection primers P20I, 5′-ACT CCT AGG AAG AGC AAG AG-3′, and P39F, 5′-TCT AAT ACG ACT CAC TAT AGG GAG AGA CAA GCT TGG GTC-3′. In vitro transcription with T7 RNA polymerase yielded the corresponding RNA library. For the first cycle, 10 nmol of this library was incubated with cytohesin-2-Sepharose in selection buffer (4.3 mM Na2HPO4/1.4 mM KH2PO4/2.7 mM KCl/147 mM NaCl/3.0 mM MgCl2, pH 7.4) with 0.8 unit/μl RNasin and 4.0 mM DTT for 30 min at 37°C. After washing with selection buffer, binding species were eluted with denaturing buffer (30 mM Tris·HCl/20% glycerol/2% SDS/1.0 M DTT, pH 6.8). Eluted RNA was amplified as described in ref. 14. The next 12 cycles included a preselection against nonderivatized Sepharose. Cycles 14 and 15 included a counterselection against cytohesin-1-Sepharose. After cycle 15, the pool was cloned into the vector pGEM4z (Promega) and sequenced.
Cell Culture and Luciferase Assays. Cells (6.5 × 104 per well) were distributed in a 24-well plate 24 h before transfection, washed with PBS, and provided with serum-free DMEM (GIBCO). After 1 h of incubation, the cells were cotransfected with 800 ng of SRE-Luc plasmid (15) and 200 ng of pEGFP-N1 plasmid (Clontech) per well, along with the appropriate nucleic acids {K61, M69, small interfering RNAs (siRNAs), pRK-wtARNO, N1-ARNO[E156K]} by lipofection by using Metafectene (Biontex Laboratories, Munich). Synthetic siRNAs [anti-ARNO siRNA366 (sense), 5′-CCU GGC AGU GCU CCA UGC UdTdT-3′; anti-ARNO siRNA753 (sense), 5′-UGA CCU GAC CCA CAC CUU CdTdT-3′; anti-Cyt1siRNA754 (sense), 5′-UGA CCU CAC UCA CAC UUU CdTdT-3′] were prepared according to the manufacturer's protocol (Dharmacon, Lafayette, CO). Over-expression of WT ARNO/cytohesin-2 was mediated by plasmid pRK-wtARNO, that of the mutant ARNO[E156K] by plasmid N1-ARNO[E156K], both under the control of the cytomegalovirus promoter. After 12 h of incubation cells were stimulated with DMEM and 10% FCS (GIBCO) and harvested after an additional 4–6 h of incubation. Cells were lysed in reporter lysis buffer (Promega) by multiple freezing and thawing. Total protein concentrations were determined by the Bradford assay. Enhanced green fluorescent protein (EGFP) fluorescence intensities (excitation 485 nm, emission 520 nm) of cell lysates were measured, and luminescence intensities were determined by using the luciferase assay system (Promega). The luminescence signals were normalized to protein concentrations and to the corresponding EGFP fluorescence intensities.
Real-Time PCR. Total RNA was extracted from cells after transfection with 15 pmol of K61 or pool RNA and stimulation with FCS by using the RNA/DNA isolation kit (Qiagen, Hilden, Germany). After reverse transcription, real-time PCRs were performed by using the selection primers P20I and P39F and the iQ SYBR Green Supermix kit (Bio-Rad), according to the manufacturer's protocol. Synthetic K61 DNA or the double-stranded DNA library was used as standard. Measurements were made by using an iCycler real-time PCR machine (Bio-Rad).
MAPK Activity Assay. After transfection with the appropriate nucleic acids (K61, M69, and siRNA366, -754, and -753), cells were stimulated with 10% FCS in DMEM for 30 min. MAPK activities of Erk1 and Erk2 were determined by using the p44/p42 MAPK assay kit (Cell Signaling Technology, Beverly, MA), according to the manufacturer's protocol. The Erk1/2 kinases were immunoprecipitated and then incubated with a truncated mutant of their physiological substrate Elk-1. The fraction of phosphorylated Elk-1 was determined by Western blotting using a specific phospho-Elk-1 (Ser-383) Ab. Quantification of the bands was performed by densitometry.
Results and Discussion
RNA Aptamers Bind Cytohesin-2 and Discriminate Cytohesin-1. To generate inhibitors capable of discriminating between cytohesin-1 and cytohesin-2, we used combinatorial in vitro selection (16–19) to screen a library containing ≈1014 RNA species for anti-cytohesin-2 aptamers. We applied the selection scheme shown in Fig. 1B, employing a series of selections for cytohesin-2 binding and counterselections against cytohesin-1 binding, to set the stringency high enough for isolating discriminatory, high-affinity sequences. After 15 rounds of iterative selection and amplification, the pool was cloned and sequenced. Fig. 1C shows the sequences of 62 clones, listed in order of their abundance. Although individual clones differ significantly, they can be grouped into families with some sequence relation (Fig. 1C). The secondary structure of the most abundant sequence, clone, K61, is shown in Fig. 1D. Filter binding assays to measure the binding affinity revealed that the K61 monoclone or selected pool-15 RNA bound to cytohesin-2 with a Kd of 115 nM or 154 nM, respectively. Significantly, K61 appears to bind cytohesin-2 with high specificity because the dissociation constant with the related cytohesin-1 indicates ≈35-fold less efficient binding (Fig. 1E and Table 1). At present, we cannot fully exclude the possibility that K61 exhibits some binding affinity to other members of the cytohesin family, namely cytohesin-3 and -4. However, the fact that these cytohesins have even lower homology to cytohesin-2 than cytohesin-1 renders this possibility unlikely.
Table 1. Affinities of K61 aptamer and pool-15 RNAs to cytohesin-1 and cytohesin-2.
|
Kd, nM
|
|||
|---|---|---|---|
| RNA | Cytohesin-2 | Cytohesin-1 | Discrimination factor |
| K61 | 115 ± 3 | ≈4,000 | ≈35 |
| Pool-15 | 154 ± 6 | ≈2,000 | ≈13 |
Dissociation constants (Kd) were determined by incubating 1.0 nM 32P-labeled RNA with increasing concentrations of the proteins in selection buffer. RNA/protein complexes were retained on nitrocellulose filters (Protran, 0.45 μm) and washed with 200 μl of selection buffer. Retained RNA was quantified by Phosphorlmager.
The domain specificity of K61 was investigated by using three deletion constructs of cytohesin-2: one that comprised the coiled-coil/Sec7-domains (amino acids 1–246), one that consists of the Sec7 domain only (amino acids 52–246), and one lacking just the N-terminal coiled-coil domain (amino acids 52–400; Sec7/PH/C). We found that K61 binds to the isolated Sec7 domain with a Kd of 800 nM (Table 2). The affinity does not change significantly when the PH and C domains are added. In contrast, addition of the coiled-coil to the Sec7 domain improves the affinity of K61 by 2-fold (CC/Sec7). Taken together, these results support the notion that the major epitope of cytohesin-2 recognized by K61 is within the coiled-coil/Sec7 domains. The fact that the N termini of cytohesins 1 and 2 are indeed highly variable (Fig. 1 A) provides a rationale for the high discriminatory activity of the aptamer.
Table 2. Domain specificities of K61 aptamer to cytohesin-2 constructs.
|
Kd of cytohesin-2 construct, nM
|
|||
|---|---|---|---|
| RNA | CC/Sec7 | Sec7 | Sec7/PH/C |
| K61 | 400 ± 20 | 800 ± 10 | 1,000 ± 25 |
Dissociation constants (Kd) were determined by incubating 1.0 nM 32P-labeled RNA with increasing concentrations of the deletion derivatives in selection buffer. RNA/protein complexes were retained on nitrocellulose filters (Protran, 0.45 μm) and washed with 200 μl of selection buffer. Retained RNA was quantified by Phosphorlmager. CC/Sec7, N-terminal coiled-coil/Sec7 domain; Sec7/PH/C, C-terminal Sec7/pleckstrin homology/polybasic C-domain.
K61 Does Not Affect GEF Activity of the Cytohesin-2 Sec7 Domain. Among the selected clones, besides being the most abundant, K61 appeared to possess the highest discriminatory activity. In this respect, it differs from an aptamer sequence, M69, isolated in a previous cytohesin-1 binding study (3), because M69 inhibited ARF-GEF activity of cytohesin-1 and cytohesin-2 and exclusively bound the Sec7 domains of both proteins (3). In the case of K61, the presence of the coiled-coil domain is required to achieve tight binding, although the Sec7 domain presumably participates in K61 binding because the Sec7/K61 complex shows a Kd of 800 nM (Table 2). We therefore investigated whether K61 is also able to modulate ARF-GEF activity of the Sec7 domain of cytohesin-2 by performing an in vitro GTP exchange assay in the presence and absence of K61. As a negative control, we used the unselected RNA pool from cycle 0 (pool-0) that had no detectable affinity for cytohesin-2 but contained the same constant sequence regions (5′ and 3′ primer sites) (Fig. 1D) as K61. Notably, K61 did not modulate the GTP exchange activity of cytohesin-2 on ARF-1 (Fig. 2 A and B), indicating that its binding to the N-terminal domain leaves the catalytic activity of the Sec7 domain unaltered.
Fig. 2.
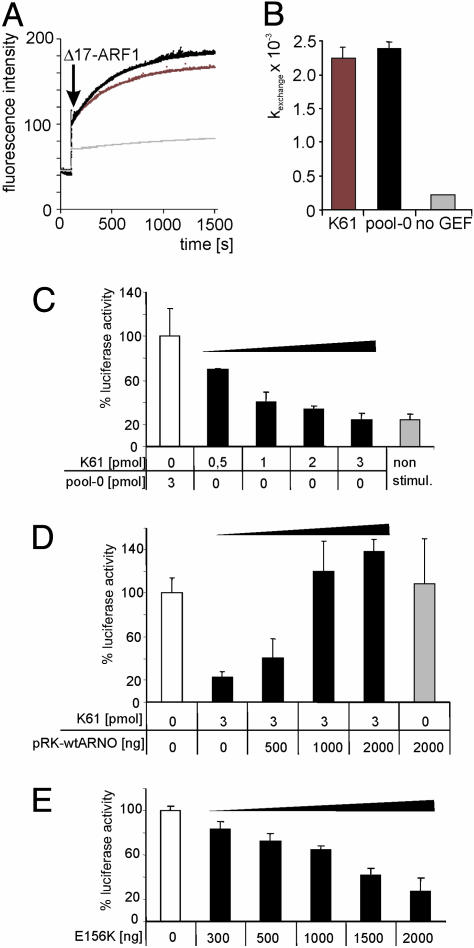
Activity of aptamer K61 in GDP/GTP exchange on ARF1 and inhibition of SRE-mediated transcription. (A) Binding of aptamer K61 to ARNO/cytohesin-2 does not affect GDP/GTP exchange on ARF1. The exchange rate on [Δ17]ARF1-GDP was determined by measuring changes in tryptophan fluorescence (20, 21). A mix of cytohesin-2 and 100 μM guanosine 5′-[γ-thio]triphosphate was supplemented with a 5-fold molar excess of K61 (red) or pool-0 RNA (black). The reaction was started by adding 500 nM [Δ17]ARF1-GDP (arrow) and shows a rapid increase in fluorescence resulting from the GEF activity of cytohesin-2. In the absence of cytohesin-2, only basal levels of GDP/guanosine 5′-[γ-thio]triphosphate exchange were obtained (gray). (B) Exchange rate constants (kexch) in the presence of K61 (red) or pool-0 (black) RNA or in the absence of exchange factor (gray). The binding of aptamer K61 to its target protein does not affect cytohesin-2-mediated GEF activity on [Δ17]ARF1 because the assay profile is comparable to adding the nonbinding pool-0 RNA. (C) Aptamer K61 inhibits SRE-mediated transcription. Cotransfection of K61 with the luciferase reporter plasmid and the pEGFP-N1 plasmid as an internal standard led to concentration-dependent inhibition of SRE-mediated transcription (black bars). Three picomoles of K61 was sufficient to reduce the luciferase level to nonstimulated basal levels (gray bar). Values of luminescence were adjusted to the transfection efficiency resulting from the EGFP standard as well as the total amount of protein after cell lysis. Each experiment was carried out three times with triplicate measurements. Shown here are the results of one representative experiment. As a negative control, 3 pmol of pool-0 RNA was used and normalized to 100% (white bar). (D) The complete inhibition of serum-mediated transcriptional activation by K61 can be fully rescued by cotransfection of the expression vector pRK-wtARNO in a concentration-dependent manner (black bars). Full rescue is seen at 1,000 ng of pRK-wtARNO. Luciferase activity in the absence of K61 and wtARNO (white bar) was used as the 100% control. The specificity of this genetic rescue is emphasized by the fact that transfection of 2,000 ng of wtARNO plasmid in the absence of K61 has no effect on serum-mediated transcriptional activation (gray bar). (E) Overexpression of the cytohesin-2 GEF-deficient mutant (E156K) (22) results in inhibition of luciferase activity in a concentration-dependent manner (black bars). Transfection of 2,000 ng of the expression plasmid ARNO[E156K] reduced luciferase activity to approximately the same levels as 3 pmol of K61. All values were normalized to the luciferase activity in the absence of ARNO[E156K] (white bar).
Cytohesin-2 Is a Positive Effector of Gene Expression by Means of the SRE. The cytohesin class of small ARF-GEFs is implicated in the control of cytoskeleton dynamics (23). Likewise, activation of the small GTPase Rac is required for cytohesin-2-induced cell motility, and GTPases of the Rho and ARF families are thought to work together to regulate membrane traffic and cytoskeletal remodeling (24). The Rho family of small GTPases also is involved in the stimulation of serum response factor transcriptional activity (c-Jun/c-Fos) induced by serum (25). We therefore hypothesized that cytohesins also may play a role in transcriptional activation by means of the serum-response element (SRE) and investigated whether K61 affects the transcription of a luciferase reporter gene under the control of the SRE promoter in HeLa cells stimulated by serum growth factors. Different concentrations of K61 RNA were transfected in the presence of Metafectene. As a negative control, we used the nonselected pool-0 RNA. As shown in Fig. 2C, K61 was able to down-regulate SRE-controlled luciferase expression to basal, nonstimulated levels in a concentration-dependent manner, with maximal inhibitory activity at 3 pmol of transfected RNA, whereas 3 pmol of unselected pool RNA had no effect. Down-regulation of SRE-controlled luciferase expression was indeed because of cytohesin-2 inhibition, specifically mediated by binding of the discriminatory aptamer, because the inhibitory activity of 3 pmol of K61 was fully reversed, i.e., squelched, by addition of WT ARNO/cytohesin-2. One thousand nanograms of transfected WT ARNO expression vector was sufficient to completely restore activity; at 500 ng of WT ARNO expression vector, partial restoration was achieved (Fig. 2D).
Domain Specificity of Cytohesin-2 in SRE Transcription. To further analyze an involvement of cytohesin-2 in SRE-dependent transcriptional regulation and to test whether the catalytic activity of the Sec7 domain is involved, we investigated an effect of overexpressing a dominant negative cytohesin-2 mutant on SRE-dependent reporter gene expression. We transfected HeLa cells with a vector that encodes the GEF-deficient ARNO/cytohesin-2 E156K point mutant (26). Fig. 2E shows that luciferase expression was reciprocally proportional to the amount of expressed ARNO E156K and resulted in down-regulation of SRE-promoter activity to basal levels.
It is possible that GEF activity of the cytohesin-2 Sec7 domain is necessary for activation of the SRE gene promoter. However, targeting the N terminus of cytohesin-2 with K61, which leaves the GEF activity intact, is just as effective in reducing serum-mediated transcriptional activation (compare Fig. 2 C and E). This finding indicates that the N terminus of cytohesin-2 participates in transcriptional activation, at least to some extent. Because the E156K mutant is dominant negative, it may affect transcription through other more indirect means, such as by preventing correct localization for the SRE response or preventing interaction of cytohesin-2 with a protein necessary for activation. In addition, these data indicate that the effect obtained by direct inhibition of cytohesin-2 by the K61 intramer can mimic almost exactly a genetic approach, providing an intriguing example of how chemical genetics can lead to novel biological insights as well as alternative ways to manipulate protein function.
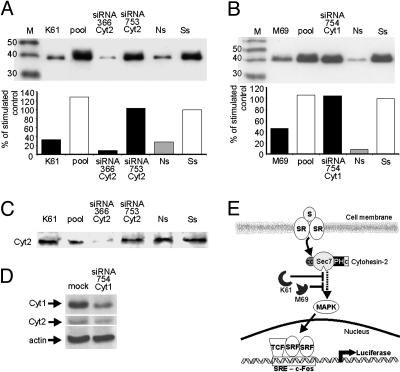
Cytohesin-2 Is a Positive Effector of MAPK Activation. To further pinpoint the effects of K61 on serum-stimulated transcriptional activation, we investigated the effect of this intramer on MAPK activation. We compared the effect of K61 with that of the unselected pool and two cytohesin-2 siRNA duplexes, siRNA366 and siRNA753. As shown in Fig. 3C, siRNA366 specifically knocks down cytohesin-2 expression, whereas siRNA753 has no detectable effect on ARNO/cytohesin-2 levels. The same cell lysates in Fig. 3C were used in an Erk1/2 kinase assay in which MAPK activation was visualized by a specific Ab that detects the phosphorylated substrate Elk-1. Densitometric quantification of the bands corresponding to phosphorylated Elk-1 revealed that pool-0 did not reduce serum-mediated MAPK activity, whereas the inhibitor K61 led to significant down-regulation to 33% (Fig. 3A). The cytohesin-2-specific siRNA366 resulted in a reduction of MAPK activity to 8% of the level observed with the unselected RNA pool, whereas siRNA753, which is not able to reduce cytohesin-2 expression (Fig. 3C), left Elk-1 phosphorylation unchanged. These data independently provide additional evidence for the involvement of the cytohesin-2 N-terminal region in serum-mediated transcriptional activation via the Erk1/2 components of the MAPK signaling pathway.
Fig. 3.
Down-regulation of serum-mediated MAPK activation by the two aptamer (intramer) inhibitors K61 and M69 compared with siRNAs. (A) HeLa cells were transfected with different effectors to investigate their influence on the intrinsic kinase activity of endogenous Erk1 and -2. Erk1 and -2 were immunoprecipitated, and their activity was determined by the MAPK assay described in Methods, followed by visualizing the phosphorylation status of purified recombinant Elk-1 substrate by Western blotting using a specific anti-pElk1(Ser-383) Ab. Quantification of bands was by densitometry. Lane 1, size marker. Lane 2, kinase activity in the presence of 3 pmol of K61 is 33% compared with serum-stimulated (Ss) cells (lane 7, 100%). Lane 3, kinase activity in the presence of 3 pmol of pool-0 RNA. Lane 4, the cytohesin-2-specific siRNA366 reduced phosphorylation activity to 8%. Lane 5, siRNA 753 has a minimal effect on Erk1/2 activity. Lane 6, Erk1/2 phosphorylation activity from nonstimulated (Ns) cells. Lane 7, Erk1/2 phosphorylation activity from serum-stimulated cells (100%). (B) Transfection of the Sec7-specific aptamer M69 reduced Erk1/2 activity to 47% (lane 2) serum-stimulated cells (lane 6, 100%), whereas the anti-cytohesin-1 siRNA 754 had no effect (lane 4). Lane 3, kinase activity in the presence of 3 pmol of pool-0 RNA. Lanes 5 and 6 are the same as lanes 6 and 7 in A.(C) Cytohesin-2 expression in the same total lysates used in A detected with a cytohesin-2-specific Ab Cyt2–21 (Sigma). Treatment with siRNAs 366 and 753 resulted in marked differences in the expression of cytohesin-2, which correlated well with Erk1/2 activity from cells transfected with these siRNAs. (D) The anti-cytohesin-1 siRNA 754 knocks down the expression of cytohesin-1, as detected by mAb 7H2, but not cytohesin-2, as detected by mAb Cyt2–21. Detection was with Abs specific for cytohesin-1 (Top), cytohesin-2 (Middle), and actin (Bottom). (E) Model for the role of cytohesin-2 as an effector of serum-mediated transcriptional activation via the MAPK pathway. The domain specificity of K61 and M69 suggest participation of both the N-terminal and the Sec7 domains of cytohesin-2. The failure of siRNA 754 to reduce MAPK activation, despite knockdown of cytohesin-1 expression (D), distinguishes cytohesin-1 from cytohesin-2 as an effector in this signaling pathway. S, growth factors in serum that act on MAPK-activating receptors; SR, MAPK-activating receptors; CC, coiled-coil domain; TCF, ternary complex factor; SRF, serum response factor.
Does direct inhibition of the GEF activity of cytohesin-2 also result in down-regulation of MAPK activation? To investigate this question, we used aptamer M69 in the same assay. This aptamer was previously shown to discriminate between Sec7 domains of the large GEF family members, such as Gea2 from yeast, and those of the small GEF family, such as cytohesins 1 and 2. In vitro, M69 inhibited GDP/GTP exchange on ARF-1, independently of whether cytohesin-1 or cytohesin-2 was used as the exchange catalyst (3). As shown in Fig. 3B, M69 was nearly as effective as K61 in blocking MAPK activation. Surprisingly, the cytohesin-1-specific siRNA754, which knocks down cytohesin-1 expression (>80%) but does not significantly affect cytohesin-2 levels (Fig. 3D), left MAPK activation levels at 100%. These results are interesting for two reasons. First, they provide a second confirmation that Sec7-mediated GEF activity is involved in serum-mediated transcriptional activation, independently from the results obtained by the genetic approach (Fig. 2E) and again by applying a domain-specific inhibitor. Second, these data strongly support the notion that in nonimmune cells, cytohesin-2, not cytohesin-1, is involved in MAPK activation. In addition, the finding that K61 did not affect cytohesin-2 GEF activity in vitro (Fig. 2 A and B) but is at least as active as M69 in down-regulating MAPK activation indicates that targeting the N-terminal domain of endogenous cytohesin-2 efficiently abrogates its cellular functionality, even when it possesses an intact Sec7 domain.
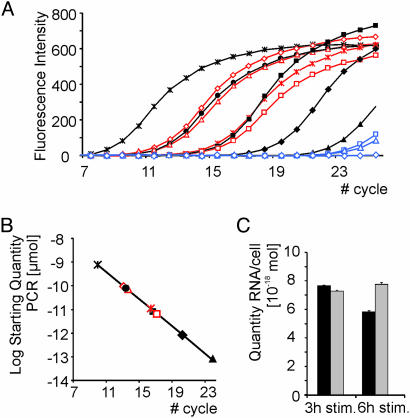
Aptamers Are Highly Stable in Vivo. It is striking that the intracellular aptamer (or intramer) efficiently executed its inhibitory activity with high specificity yet without protection against nuclease degradation. To confirm that the intramer apparently does not require intracellular stabilization, we quantified the amount of intramer and pool-0 negative control RNA isolated from transfected cells 3 and 6 h poststimulation by reverse transcription and subsequent monitoring of cDNA amplification by real-time PCR (Fig. 4A). Calibration curves were generated by using various defined concentrations of K61 or pool-0 DNAs (Fig. 4B). There was no significant degradation of intramer or pool-0 RNA at 3 or 6 h (Fig. 4C). Considering a cell volume of ≈10–11 liters (27), the intracellular aptamer and pool-0 RNA concentrations at these time points range between 0.3 and 1.0 μM (based on ≈3 × 105 cells used per experiment), which is well within the range of the in vitro Kd of the K61/cytohesin-2 complex.
Fig. 4.
Stability of intracellular K61 and pool-0 RNAs. (A) Transfection of HeLa cells with the respective RNA species was followed by quantification by real-time PCR 3 and 6 h after stimulation with FCS. After preparation of total RNA and reverse transcription, a 1:100 or 1:1,000 dilution of cDNA was used in the PCR with the selection primers. The course of DNA amplification was monitored by fluorescence change mediated by the double-stranded DNA-intercalating dye SYBR green. Black curves, standard concentrations of K61 single-stranded DNA: *, 800 amol (10–18 mol); •, 80 amol; ▪, 8 amol; ♦, 0.8 amol; ▴, 0.08 amol. The high accuracy of the PCR is demonstrated by negative controls shown in the blue curves: ⋄, no template; □, no reverse transcription; ▵, no transfection. Red curves, amplification of dilutions of isolated total cellular RNA, reverse transcribed with K61-specific primer 3 and 6 h after stimulation: ⋄, 1:100, 3 h; ▵, 1:100, 6 h; □, 1:1,000, 3 h; *, 1:1,000, 6 h. (B) Standard curve for determining the concentration of isolated K61 RNA: black symbols, standards as in A; red symbols, samples as in A.(C) Quantification of RNAs based on the total number of cells used for their preparation. Neither pool-0 RNA (gray bars) nor K61 (black bars) showed significant degradation 3 or 6 h after stimulation with serum.
Few examples of intramer inhibition of biological targets in vivo are known (3, 28, 29). In these previous studies, efforts were taken to stabilize the intramer or to direct it to the correct cellular compartment. Intramers were expressed by vaccinia virus vectors that are difficult to handle, require certain safety precautions, and have to be genetically engineered for each aptamer that will have to be expressed (3, 28). In another study, transgenic animals had to be engineered for endogenous aptamer expression (29). An alternative strategy uses transfection of expression plasmids that are translocated into the nucleus so that the expression of the aptamers takes place in the nucleus. This method has been used to investigate effects of inhibitory aptamers on nuclear proteins (30); to address proteins that reside in the cytoplasm, it would be necessary to equip the expressed aptamer with an RNA sequence that serves as a nuclear export signal (31). Despite the fact that such signal sequences are scarce, it will have to be ensured that each aptamer remains active within the context of additional RNA sequences. We report here that, surprisingly, inhibitory aptamers can be directly transfected into cultured cells, just like siRNAs, without the need for protecting them against nuclease degradation. This finding represents a significant advance in aptamer technology, and we are optimistic that this method also will be applicable for other eukaryotic systems and aptamers that target cytoplasmic proteins.
Conclusion
In conclusion, by applying an inhibitor specific for cytohesin-2, a member of the class of small GEFs, we showed that cytohesin-2 participates in regulating gene expression via the MAPK signaling pathway (Fig. 3E). Our results provide evidence that the activity of cytohesin-2, but not of cytohesin-1, is required for transcriptional activation in nonimmune cells. Although the two proteins are both expressed in HeLa cells (Fig. 3D), the consequences of specific inhibition of cytohesin-2 by the discriminatory aptamer K61 or dominant negative expression of its E156K mutant derivative (Fig. 2E), in contrast to knockdown of cytohesin-1 expression by its specific siRNA (Fig. 3), indicate that the two proteins are not redundant, despite their 90% identity. Cytohesin-1 was recently shown to be involved in activating the IL-2 gene promoter in T cells (13). Taken together, this finding and our results support the hypothesis that transcriptional activation by cytohesins may be a much more common activity among the small GEFs than previously assumed and may be assigned unambiguously to individual members of the cytohesin family within one cell type, possibly through binding to cell type-specific interacting molecules.
In this context, our result suggesting that the coiled-coil domain of cytohesin-2 is important for its function in cells is noteworthy. Binding of intramer K61 to this domain has the same inhibitory effect on serum-induced MAPK activation as the expression of the GEF-deficient E156K mutant of cytohesin-2. The dominant negative effect of this mutant has been ascribed to a competition with endogenous cytohesin-2 for its substrate ARF, but this interpretation is not consistent with biochemical and structural studies indicating that ARNO (E156K) cannot form a complex with ARF at physiological Mg2+ concentrations (26, 32). Thus, our study supports an alternative explanation for the dominant negative effect of the E156K mutant, namely a competition with endogenous cytohesin-2 for the coiled-coil domain-binding protein or cellular partners other than ARF (32).
More generally, we show here that aptamers can be directly transfected into cultured cells without the need for protecting them against nuclease degradation. Inhibitory intramers can be an extremely valuable complement to loss-of-function phenotypic knockdown approaches using siRNA technology (33) for elucidating the biological function of proteins and for assigning novel activities to highly homologous family members. Our study provides a direct comparison between aptamers and siRNAs for investigating the function of a protein and shows that intramers can be as effective in down-regulating protein function as siRNAs, the difference being not that the gene is knocked out, but the expressed protein is inhibited. There also may be cases in which only partial knockdown of target proteins is achieved by an siRNA because residual amounts of proteins can be present even days after transfection with an siRNA because of stability of the endogenous protein. In these cases, combination with an inhibitory intramer might be useful for achieving complete knockdown of protein function.
Moreover, we show that individual domains of highly homologous proteins can be selectively targeted by inhibitory intramers. This finding is useful in our case not only for assigning specific biological functions to individual members of the interesting small GEF protein family but also for exploring novel strategies for developing GEF-selective drugs, the importance of which is stressed in ref. 32. Inhibitory aptamers might be directly converted into lead compounds by using assays that screen small molecule libraries for compounds that displace the aptamer from its target and adopt its inhibitory activity (34).
Acknowledgments
We thank J. Goldberg (Memorial Sloan–Kettering Cancer Center, New York) for cytohesin-2 cDNA plasmids, V. Fieberg for technical assistance, I. Grüne for purification of M69 RNA, and D. U. Gommel for help with GDP/GTP exchange assays. This work was supported by the Deutsche Forschungsgemeinschaft.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ARF, ADP-ribosylation factor; ARNO, ARF nucleotide-binding site opener; EGFP, enhanced GFP; GEF, guanine nucleotide exchange factor; PH, pleckstrin homology; siRNA, small interfering RNA; SRE, serum-response element.
References
- 1.Geiger, C., Nagel, W., Boehm, T., van Kooyk, Y., Figdor, C. G., Kremmer, E., Hogg, N., Zeitlmann, L., Dierks, H., Weber, K. S., et al. (2000) EMBO J. 19, 2525–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank, S. R., Hatfield, J. C. & Casanova, J. E. (1998) Mol. Biol. Cell 9, 3133–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mayer, G., Blind, M., Nagel, W., Böhm, T., Knorr, T., Jackson, C. L., Kolanus, W. & Famulok, M. (2001) Proc. Natl. Acad. Sci. USA 98, 4961–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donaldson, J. G. & Jackson, C. L. (2000) Curr. Opin. Cell Biol. 12, 475–482. [DOI] [PubMed] [Google Scholar]
- 5.Novick, P., Ferro, S. & Schekman, R. (1981) Cell 25, 461–469. [DOI] [PubMed] [Google Scholar]
- 6.Jackson, C. L. & Casanova, J. E. (2000) Trends Cell Biol. 10, 60–67. [DOI] [PubMed] [Google Scholar]
- 7.Chardin, P., Paris, S., Antonny, B., Robineau, S., Beraud-Dufour, S., Jackson, C. L. & Chabre, M. (1996) Nature 384, 481–484. [DOI] [PubMed] [Google Scholar]
- 8.Kolanus, W., Nagel, W., Schiller, B., Zeitlmann, L., Godar, S., Stockinger, H. & Seed, B. (1996) Cell 86, 233–242. [DOI] [PubMed] [Google Scholar]
- 9.Strausberg, R. L., Feingold, E. A., Grouse, L. H., Derge, J. G., Klausner, R. D., Collins, F. S., Wagner, L., Shenmen, C. M., Schuler, G. D., Altschul, S. F., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 16899–16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank, S., Upender, S., Hansen, S. H. & Casanova, J. E. (1998) J. Biol. Chem. 273, 23–27. [DOI] [PubMed] [Google Scholar]
- 11.Zuker, M. (2003) Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mathews, D. H., Sabina, J., Zuker, M. & Turner, D. H. (1999) J. Mol. Biol. 288, 911–940. [DOI] [PubMed] [Google Scholar]
- 13.Perez, O. D., Mitchell, D., Jager, G. C., South, S., Murriel, C., McBride, J., Herzenberg, L. A., Kinoshita, S. & Nolan, G. P. (2003) Nat. Immunol. 4, 1083–1092. [DOI] [PubMed] [Google Scholar]
- 14.Klug, S. J., Hüttenhofer, A., Kromayer, M. & Famulok, M. (1997) Proc. Natl. Acad. Sci. USA 94, 6676–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao, J., Hodge, C., Meyer, D., Ho, P. S., Rosenspire, K. & Schwartz, J. (1997) J. Biol. Chem. 272, 25951–25958. [DOI] [PubMed] [Google Scholar]
- 16.Ellington, A. D. & Szostak, J. W. (1990) Nature 346, 818–822. [DOI] [PubMed] [Google Scholar]
- 17.Tuerk, C. & Gold, L. (1990) Science 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 18.Famulok, M., Blind, M. & Mayer, G. (2001) Chem. Biol. 8, 931–939. [DOI] [PubMed] [Google Scholar]
- 19.Famulok, M. & Verma, S. (2002) Trends Biotechnol. 20, 462–466. [DOI] [PubMed] [Google Scholar]
- 20.Macia, E., Chabre, M. & Franco, M. (2001) J. Biol. Chem. 276, 24925–24930. [DOI] [PubMed] [Google Scholar]
- 21.Kruljac-Letunic, A., Moelleken, J., Kallin, A., Wieland, F. & Blaukat, A. (2003) J. Biol. Chem. 278, 29560–29570. [DOI] [PubMed] [Google Scholar]
- 22.Cherfils, J., Menetrey, J., Mathieu, M., Le Bras, G., Robineau, S., Beraud-Dufour, S., Antonny, B. & Chardin, P. (1998) Nature 392, 101–105. [DOI] [PubMed] [Google Scholar]
- 23.Weber, K. S., Weber, C., Ostermann, G., Dierks, H., Nagel, W. & Kolanus, W. (2001) Curr. Biol. 11, 1969–1974. [DOI] [PubMed] [Google Scholar]
- 24.Santy, L. C. & Casanova, J. E. (2002) Curr. Biol. 12, R360–R362. [DOI] [PubMed] [Google Scholar]
- 25.Hill, C. S., Wynne, J. & Treisman, R. (1995) Cell 81, 1159–1170. [DOI] [PubMed] [Google Scholar]
- 26.Beraud-Dufour, S., Robineau, S., Chardin, P., Paris, S., Chabre, M., Cherfils, J. & Antonny, B. (1998) EMBO J. 17, 3651–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan, M. A., Thrower, D. & Wilson, L. (1991) Cancer Res. 51, 2212–2222. [PubMed] [Google Scholar]
- 28.Blind, M., Kolanus, W. & Famulok, M. (1999) Proc. Natl. Acad. Sci. USA 96, 3606–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi, H., Hoffman, B. E. & Lis, J. T. (1999) Proc. Natl. Acad. Sci. USA 96, 10033–10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Good, P. D., Krikos, A. J., Li, S. X., Bertrand, E., Lee, N. S., Giver, L., Ellington, A., Zaia, J. A., Rossi, J. J. & Engelke, D. R. (1997) Gene Ther. 4, 45–54. [DOI] [PubMed] [Google Scholar]
- 31.Hamm, J., Huber, J. & Lührmann, R. (1997) Proc. Natl. Acad. Sci. USA 94, 12839–12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Renault, L., Guibert, B. & Cherfils, J. (2003) Nature 426, 525–530. [DOI] [PubMed] [Google Scholar]
- 33.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- 34.Hartig, J. S., Najafi-Shoushtari, S. H., Grüne, I., Yan, A., Ellington, A. D. & Famulok, M. (2002) Nat. Biotechnol. 20, 717–722. [DOI] [PubMed] [Google Scholar]