Abstract
Transcription of the gene encoding sterol regulatory element-binding protein 1c (SREBP-1c) is known to be activated by insulin in the liver. The resultant SREBP-1c protein activates transcription of the genes required for fatty acid synthesis. Here, we use SREBP-1c promoter reporter constructs to dissect the mechanism of insulin activation in freshly isolated rat hepatocytes. The data show that a complete insulin response (increase of 6- to 11-fold) requires two binding sites for liver X receptors (LXRs), which are nuclear receptors that are activated by oxygenated sterols. Disruption of these binding sites did not lower basal transcription but severely reduced the response to insulin. In contrast, disruption of the closely linked binding sites for SREBPs and nuclear factor Y lowered basal transcription drastically but still permitted a 4- to 7-fold increase in response to insulin. Arachidonic acid, an inhibitor of LXR activation, blocked the response to insulin. We conclude that insulin activates the SREBP-1c promoter primarily by increasing the activity of LXRs, possibly through production of a ligand that activates LXRs or their heterodimerizing partner, the retinoid X receptor. Nuclear SREBPs and nuclear factor Y play permissive roles.
Elevated hepatic synthesis of fatty acids and triglycerides (lipogenesis) is a troublesome consequence of hyperinsulinemia observed in obesity and type 2 diabetes mellitus. The increased flux of triglycerides from liver to peripheral tissues interferes with glucose usage, thereby accentuating insulin resistance and producing further hyperinsulinemia (1, 2). The elevated hepatic lipogenesis is the result of the elevated insulin levels, which enhance transcription of lipogenic genes in liver. At the same time that the liver is responding to one action of insulin (lipogenesis), the organ becomes resistant to another action of insulin, namely, suppression of gluconeogenesis. The failure to suppress hepatic gluconeogenesis coupled with decreased peripheral glucose uptake accentuates the hyperglycemia and increases insulin secretion, further enhancing lipogenesis and completing a vicious cycle (1, 3). This vicious cycle has been demonstrated in two mouse models of insulin resistance, both of which are associated with a deficiency of the adipocyte hormone, leptin: the ob/ob mouse and the lipodystrophic aP2 sterol regulatory element-binding protein 1c (aP2-SREBP-1c) transgenic mouse (3).
The insulin-responsive transcription factor responsible for insulin-enhanced lipogenesis has been identified as SREBP-1c (4, 5). SREBP-1c is one of three SREBPs that belong to the basic helix–loop–helix-leucine zipper family of transcription factors. Two of the SREBPs (SREBP-1a and SREBP-1c) are derived from one gene through the use of alternative promoters and transcription start sites that produce two isoforms that differ by the first exon (6). In the fed state, SREBP-1c is the main SREBP-1 transcript (mouse liver 1c-to-1a ratio is 9:1). Both of the SREBP-1 isoforms preferentially activate fatty acid and triglyceride synthesis (6). SREBP-2 is encoded by a different gene (4), and it preferentially activates cholesterol synthesis, although functional overlap exists.
Recent reviews have summarized the studies demonstrating that insulin selectively stimulates Srebp-1c gene transcription in liver, and that SREBP-1c in turn mediates insulin-stimulated fatty acid synthesis (4, 5). In brief, insulin was shown to increase SREBP-1c mRNA levels concomitant with its ability to elevate mRNA levels for fatty acid biosynthetic genes in hepatocytes (7), and a dominant-negative form of SREBP-1c blocked the insulin effect on the fatty acid biosynthetic genes (8). The insulin effect on SREBP-1c mRNA was blocked when cAMP levels were elevated by glucagon, and this block was associated with a decline in acetyl-CoA carboxylase and fatty acid synthetase, the two key enzymes of fatty acid biosynthesis (3). Hereafter, we refer to these two mRNAs as lipogenic mRNAs. In the livers of living rodents, SREBP-1c mRNA levels were shown to decline when insulin fell as a result of fasting or treatment with streptozotocin, and this fall was paralleled by decreases in lipogenic mRNAs (7, 9). The opposite result was observed when insulin was administered to the streptozotocin-treated rats.
In livers of lipodystrophic and ob/ob mice, SREBP-1c mRNA levels were elevated as a result of hyperinsulinemia secondary to leptin deficiency (10). Leptin treatment normalized plasma insulin. As a result, SREBP-1c mRNA levels fell, the mRNAs for lipogenic genes declined, and the fatty liver resolved.
The crucial role of SREBP-1c in lipogenesis was demonstrated conclusively by experiments with mice deficient in SREBP cleavage-activating protein (SCAP), a protein that is necessary to process SREBPs to their active nuclear forms (11). When these mice were fasted and then refed a high-carbohydrate diet, they showed a dramatic reduction in the insulin-induced increase in lipogenic mRNA levels. Similar but less dramatic results were observed in knockout mice that lack the SREBP-1c isoform (12).
In addition to regulation by insulin and glucagon, hepatic SREBP-1c mRNA levels are increased by agonists of liver X receptors (LXRs), nuclear hormone receptors that form active heterodimers with retinoid X receptors (RXRs) (13–15). The primary activators of LXRs are oxygenated intermediates in cholesterol synthesis (16). LXR activity is inhibited competitively by polyunsaturated fatty acids (17). A synthetic LXR ligand, T0901317, increases hepatic SREBP-1c mRNA levels and thereby increases mRNAs for lipogenic enzymes when administered to mice. The result is fatty liver and hypertriglyceridemia (13, 14). Similar results are obtained with agonists of RXR, the heterodimerizing partner of the LXRs (13). Gene-knockout mice lacking SREBP-1c show a blunted increase in lipogenic mRNAs in response to T0901317, confirming the role of SREBP-1c in this response (12).
The above studies point to regulation of Srebp-1c transcription as a central event in lipid homeostasis, and studies have begun to delineate the cis elements that are responsible for this regulation. Relevant elements of the mouse (18, 19) and rat (20) proximal SREBP-1c promoter have been mapped by gene reporter studies in cultured cells. An analysis of the mouse SREBP-1c promoter revealed two putative LXR elements (LXREs), one putative nuclear factor Y (NF-Y) binding site, and one sterol regulatory element (SRE) (18, 19). All of these elements are highly conserved in the human and rat genomes. The presence of an SRE suggests that SREBP-1c mRNA can increase in response to nuclear SREBPs, creating a feed-forward activation. This view was supported by studies showing that the SREBP-1c reporter construct was stimulated by cotransfection of plasmids encoding truncated nuclear forms of SREBP-1c, SREBP-1a, or SREBP-2 (19). Thus far, SREBPs always activate target gene transcription in concert with a constitutively active transcription factor, typically NF-Y or Sp1, which binds at a nearby site (21–23). In the case of the SREBP-1c promoter, the SRE is surrounded by a 5′ NF-Y site and a 3′ Sp1 site. The NF-Y site was shown to be required for the SREBP-1c promoter to be stimulated by nuclear SREBPs (19).
The role of the LXREs in Srebp-1c transcription was established by the finding of a blunted response to T0901317 when these elements had been scrambled by in vitro mutagenesis (13). The conclusion of all of these studies is that Srebp-1c transcription can be regulated positively by nuclear SREBPs (together with NF-Y and/or Sp1) and by LXR agonists through identifiable elements in the SREBP-1c proximal promoter. None of the earlier studies addressed the mechanism of the insulin response.
The current studies were designed to explore the mechanism by which insulin stimulates the SREBP-1c promoter. We were unable to find a cultured hepatocyte cell line that retains the ability to respond to insulin by up-regulating SREBP-1c mRNA levels. Therefore, we performed these studies in freshly isolated rat hepatocytes. The results indicate that the SREBP-1c promoter does not have a separate insulin response element, and that insulin acts primarily through the LXR elements in a reaction that requires the NF-Y site and the SRE.
Methods
Materials. We obtained bovine insulin (catalog no. I 6634), dexamethasone (no. D 4902), 3,3′,5-triiodo-l-thyronine (no. T 5516), and arachidonic acid (no. A 9673) from Sigma; lipofectin (no. 18292-037) and medium 199 (no. 11150-059) from Invitrogen; PBS (no. 21-031-CV), DMEM (no. 10-014-CV), and RPMI medium 1640 (no. 10-040-CV) from Mediatech (Herndon, VA); 60-mm collagen-I-coated dishes (no. 356401) from Becton Dickinson Labware; jetPEI transfection reagent (no. GDSP10110) from Qbiogene (Carlsbad, CA); T0901317 compound (lot no. 54-143-1) from J-Star Research (South Plainfield, NJ); and FCS from Atlanta Biologicals (Norcross, GA). A stock solution of sodium arachidonate bound to defatted BSA was prepared as described in ref. 17.
Insulin and LXR Agonist. Stock solutions of 0.1 mM insulin were prepared in distilled water adjusted to pH 4.5 with glacial acetic acid, stored at 4°C, and used within 3 months. Stock solutions of 1 mM T0901317 (synthetic LXR agonist) were prepared in DMSO, stored at –20°C, and used within 1 month. Stock solutions of 10 mM compactin were prepared as described (24), stored at –80°C, and used fresh for each experiment.
Mouse SREBP-1c Promoter–Luciferase Constructs. A series of plasmids, constructed by standard techniques of recombinant DNA engineering (25), contained fragments of the mouse SREBP-1c promoter, extending for various distances in the 5′ direction and terminating either at the A of the initiation codon in exon 1c (plasmids A–C) or at the end of the first codon of exon 2 (plasmids D–I), as shown in Fig. 1. These fragments were cloned by PCR amplification using mouse SM-1 embryonic stem cell genomic DNA as the template (12). Primers used for these PCRs were based on mouse sequences obtained from the Celera database (www.celera.com) and confirmed by sequencing (12). The various amplified fragments were inserted into the pGL3 luciferase reporter vector (Promega).
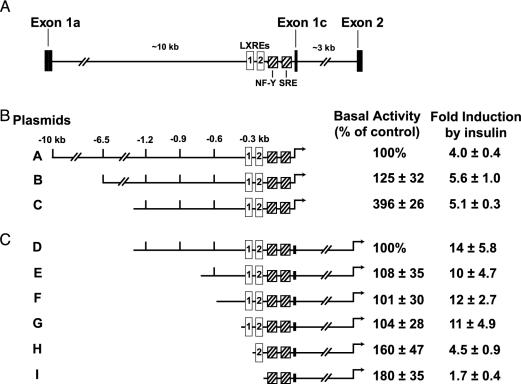
Fig. 1.
Activation of mouse SREBP-1c promoter in rat primary hepatocytes. (A) Schematic of partial mouse Srebp-1c gene, illustrating the upstream regulatory elements and the intron between exons 1c and 2. The proximal promoter regulatory region is shown with its cis-acting elements: two LXREs (open boxes) and one NF-Y and one SRE (hatched boxes). (B and C) Deletion analysis of SREBP-1c promoter region and its effects on basal and insulin-induced luciferase activities. The indicated plasmids were transfected into primary hepatocytes as described in Methods. Six hours after transfection, the medium was switched to medium B with or without 100 nM insulin and incubated for 21 h, after which the cells were harvested and assayed for dual luciferase activities as described in Methods. The 100% values for basal activity correspond to the normalized luciferase activity obtained with plasmid A (B) or plasmid D (C) in the absence of insulin. The fold induction in B and C was calculated as the ratio of normalized luciferase activity in the presence of insulin to that in the absence of insulin. Each value represents the mean ± SEM of three independent transfection experiments (each assayed in duplicate).
To construct plasmids m1–m3 (Fig. 2A), we cloned double-stranded oligonucleotides containing the indicated scramble mutation (9–12 bp) into plasmid D (Fig. 1D), thus replacing the corresponding WT sequence. Scramble mutations in plasmids m4–m30 (Fig. 2 A) were generated with the QuickChange site-directed mutagenesis kit (Stratagene) using pBP1c(1271)-luciferase plasmid (13) as the template. Mutagenic oligonucleotides 52 bases in length were synthesized so that the 9- to 12-base sequence to be scrambled was located in the center of the oligonucleotide. The nucleotide sequence of the 9- to 12-bp scramble was designed to retain the length and base composition of the WT sequence and to contain novel SmaIor PstI restriction sites. Plasmids containing the desired scramble mutations were identified by restriction digestions (XhoI/SmaI or XhoI/PstI) and DNA sequencing, and the fragments containing the scramble mutations were excised and cloned into plasmid D. To construct plasmid m31, a double-stranded oligonucleotide containing the LXRE1 mutation in m9 was cloned into plasmid m13 (LXRE2 mutant), thus generating m31 with mutations in both LXRE1 and LXRE2. To construct plasmids m32–m34, double-stranded oligonucleotides containing mutations in LXRE1 (m9), LXRE2 (m13), or LXRE1 + LXRE2 (m31) were cloned into plasmid m24 (SRE mutant), thus generating plasmids with mutations in LXRE1 + SRE (m32), LXRE2 + SRE (m33), or LXRE1 + LXRE2 + SRE (m34), respectively.
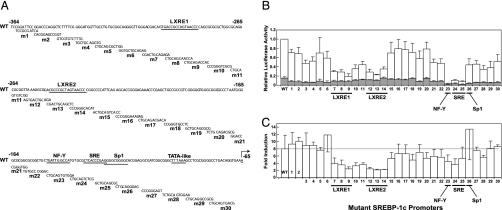
Fig. 2.
Nucleotide sequences (A), relative transcriptional activities (B), and fold induction (C) of WT and mutant mouse SREBP-1c promoters in primary rat hepatocytes. (A) The sequence of a portion of the normal SREBP-1c promoter is shown at the top and numbered according to a convention in which the A of the ATG initiation codon of mouse SREBP-1c is + 1. The putative binding sites for LXR (LXRE1 and LXRE2), NF-Y, SREBP (SRE), and Sp1 and the TATA-like sequence are underlined. The transcription initiation site is indicated by an arrow. Below the sequence of the WT promoter are shown 30 overlapping scramble mutations that are separately introduced into the DNA by site-directed oligonucleotide mutagenesis as described in Methods. The names of the mutant plasmids, m1–m30, are denoted on the left. The mutant scrambled sequence that was introduced is shown below the normal promoter sequence. (B) The height of each bar represents the relative luciferase activity of plasmid D (WT) or the indicated mutant plasmid (m1–m30) in transfected hepatocytes cultured in the absence (shaded bar) or presence (open bar) of insulin as described in Methods. The relative luciferase activity of the WT promoter (plasmid D) in the presence of insulin (mean of 12 independent experiments) was arbitrarily assigned a value of 1.0. Each bar for mutant promoters m1–m30 represents mean ± SEM of three to six independent transfections. (C) The data in B were replotted to show the fold induction of the various promoters by insulin. The fold induction was calculated as the ratio of normalized luciferase activity in the presence of insulin to that in the absence of insulin. The luciferase activity of each promoter in the absence of insulin was arbitrarily assigned a value of 1.0.
All plasmid constructs were verified by restriction endonuclease mapping and DNA sequencing.
Primary Rat Hepatocytes. Male Sprague–Dawley rats (250–350 g; obtained from Harlan Breeders, Indianapolis) were housed in colony cages, maintained on a 12-h light/12-h dark cycle, and fed 4% Mouse/Rat Diet no. 7001 (Harlan Teklad, Madison, WI). Nonfasted rats were killed 30 min before the dark cycle, and primary hepatocytes were isolated by the collagenase method (26) with modifications as described in ref. 7. The isolated hepatocytes were plated onto 60-mm collagen-I-coated dishes in 4 ml of medium A (DMEM supplemented with 5% (vol/vol) FCS, 100 units/ml sodium penicillin, and 100 μg/ml streptomycin sulfate). The cells were incubated at 37°C in 5% CO2. After 3–4 h, the attached cells were washed once with 4 ml of PBS, incubated for 14–16 h in medium B (medium 199 supplemented with 100 nM dexamethasone, 100 nM 3,3′,5-triiodo-l-thyronine, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate) plus 1 nM insulin, and then used for transfection experiments.
Plasmid Transfection and Luciferase Assays. Duplicate dishes of hepatocytes were washed once with 4 ml of PBS, incubated in serum-free RPMI medium 1640 at pH 7.55, and cotransfected with 2 μg per dish of WT or mutant firefly luciferase reporter plasmids and 0.5 μg per dish of a control plasmid phRL-TK (catalog no. E6241, Promega) encoding the synthetic Renilla luciferase gene. In all experiments, except that in Fig. 1B, transfections were performed with Lipofectin (GIBCO), in which the plasmids were incubated with 2 ml of RPMI medium 1640 for 6 h at 37°C in a 5% CO2 incubator. In Fig. 1B, transfections were performed with 0.2 ml of jetPEI reagent and 1.8 ml of RMPI medium 1640. After transfection, cells were washed once with 4 ml of PBS and switched to medium B containing various reagents as described in the figure legends. Twenty-one hours later, cells were washed once with 4 ml of PBS and harvested by scraping in 0.4 ml of 1× passive lysis buffer (no. E1941, Promega). Firefly and Renilla luciferase activities in the cell lysates were measured with the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's protocol. Photon production was detected as relative light units by using an Optima II luminometer (MGM Instruments, Hamden, CT). All values represent the mean of duplicate transfections, each assayed in triplicate. The amount of firefly luciferase activity in transfected cell lysates was normalized to the amount of Renilla luciferase activity from the same test tube.
Results
Fig. 1 A shows a schematic of the 5′ end of the mouse Srebp-1 gene, including exons 1a, 1c, and 2. The location of the two previously identified LXREs, the NF-Y site, and the SRE proximal to the first exon of SREBP-1c are denoted by the open and hatched bars, respectively (13, 18). These sites are 10 kb downstream of exon 1a, the first exon of the SREBP-1a transcript, which is not regulated by insulin. As a first step in identifying the DNA elements that mediate insulin induction of the SREBP-1c transcript, we introduced the 10-kb promoter fragment between exons 1a and 1c and various truncations into the pGL3-Basic reporter vector to generate plasmids A (10 kb), B (6.5 kb), and C (1.3 kb), each driving production of firefly luciferase. To assay for insulin action, we introduced plasmids A, B, and C into primary rat hepatocytes by transfection together with the Renilla luciferase vector driven by the thymidine kinase promoter (phRL-TK) as a control. Luciferase activities were measured after incubation of the cells for 21 h in the presence or absence of 100 nM insulin. As shown in Fig. 1B, insulin induced 4-, 5.6-, and 5.1-fold activations of plasmids A, B, and C, respectively.
To mimic the endogenous Srebp-1c gene structure more completely, we included the 3-kb intron between exons 1c and 2 in a series of deletion reporter plasmids designated as D (–1271), E (–694), F (–570), G (–368), H (–278), and I (–170), as shown in Fig. 1C. When we included this intron, the relative induction by insulin increased from 5.1-fold (plasmid C) to 14-fold (plasmid D). Plasmids E, F, and G showed the same level of basal transcription and fold induction by insulin as did plasmid D. Deletions in plasmids H and I significantly reduced the fold induction by insulin from 14 to 4.5 and 1.7, respectively. These results indicate that the 368-bp promoter fragment in plasmid G, containing the two LXREs, the NF-Y site, and the SRE, is sufficient to mediate insulin-induced Srebp-1c transcription.
To map the insulin-responsive elements more precisely, we introduced a series of overlapping 9- to 12-bp scramble mutations covering the nucleotide sequence from –364 to –65 into plasmid D to generate plasmids m1–m30, as shown in Fig. 2 A. Fig. 2B shows the transcription of the WT plasmid D and mutant plasmids m1–m30 in the absence or presence of insulin. Fig. 2C shows the fold induction by insulin. The scramble mutations in plasmids m7–m9 and m12–m14 destroyed the first and second LXREs, respectively. These mutations decreased insulin-induced transcription by >60% without affecting basal transcription significantly. The scramble mutation in plasmid m23 destroyed the NF-Y site, and mutations m24 and m25 destroyed the SRE. Each of these mutations decreased basal transcription to <10% of the WT values, but they still permitted at least a 4-fold induction by insulin. Mutation of the adjacent Sp1 site (m26) reduced basal transcription slightly but permitted a robust response to insulin. No other DNA region appeared to be required for insulin-mediated induction.
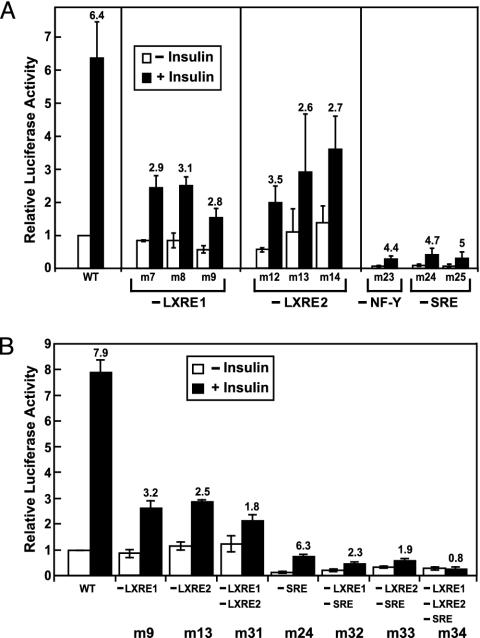
The aforementioned nine scramble mutation plasmids were studied in different transfection experiments. To circumvent any interexperiment variability, we measured the responses to insulin in a single transfection experiment in which all of the plasmids were studied. These experiments were repeated three times, and the mean results are shown in Fig. 3A. Scramble mutations in plasmids m7–m9 (LXRE1) and m12–m14 (LXRE2) reduced insulin-induced luciferase activity from 6.4 in the WT plasmid to 1.5–3.6 without changing the basal level of luciferase activity. Mutations in plasmids m23 (NF-Y), m24 (SRE), and m25 (SRE + Sp1) reduced the basal promoter activity by >90%, but insulin was able to increase the activity by 3- to 5-fold.
Fig. 3.
Mutations of LXRE1, LXRE2, and SRE in SREBP-1c promoter sequence reduce insulin-induced activation in primary rat hepatocytes. The indicated WT or scramble mutant plasmid was transfected into hepatocytes, incubated with or without 100 nM insulin, and assayed for dual luciferase activity as described in the legend to Fig. 1. Each value represents the mean ± SEM of three independent transfection experiments (each assayed in duplicate). In each experiment, all of the mutant plasmids were studied. The normalized luciferase activity of the WT promoter in the absence of insulin was arbitrarily assigned a value of 1.0. The numbers above the bars refer to the fold increase by insulin.
Because mutations in LXRE1 (m9), LXRE2 (m13), or SRE (m24) all affected transcription of the SREBP-1c promoter, we next studied plasmids containing combinations of mutations disrupting these three elements (Fig. 3B). Mutating both LXRE1 and LXRE2 in the same plasmid (m31) had minimal effects on basal activity but reduced the induction by insulin to 1.8-fold vs. 7.9-fold for WT. As before, destruction of the SRE (m24) reduced basal transcription but permitted a 6.3-fold induction by insulin. Mutations disrupting LXRE1, LXRE2, and SRE in the same plasmid (m34) severely reduced basal activity and abolished insulin-mediated activation of the SREBP-1c promoter.
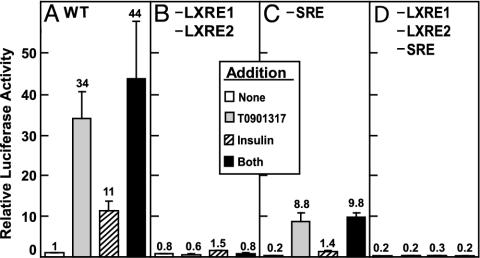
In light of the requirement for the LXREs in insulin activation of the SREBP-1c promoter, we next examined insulin-mediated activation in the absence or presence of T0901317, a nonsterol synthetic LXR ligand (Fig. 4). With the WT promoter, T0901317, insulin, and the combination of T0901317 plus insulin increased transcription by 34-, 11-, and 44-fold, respectively. Mutating both LXREs (m31) abolished activation by T0901317 and severely reduced the insulin response without affecting the basal activity. In contrast, mutating the SRE (m24) reduced the basal and insulin-stimulated SREBP-1c promoter activities by >80% but reduced only slightly the fold induction by insulin (1.4/0.2 = 7-fold vs. 11-fold for WT). Despite the reduction in basal activity, the SRE mutant showed a normal relative induction by T0901317 (44-fold vs. 34-fold for WT) and the insulin–T0901317 combination (49-fold induction). Mutating both LXREs and SRE in the same plasmid (m34) reduced the basal promoter activity to 20% of WT and abolished activation by insulin or T0901317 or the combination.
Fig. 4.
Maximal activation of SREBP-1c promoter by LXR agonist (T0901317) requires intact SRE as well as LXRE1 and LXRE2. The WT plasmid D (A) or the indicated mutant plasmid (B, plasmid m31; C, m24; D, m34) was transfected into primary rat hepatocytes as described in Fig. 1 and incubated in the absence or presence of 1 μM T0901317 or 100 nM insulin or both as indicated. After incubation for 21 h, the cells were harvested and assayed for dual luciferase activities. Relative luciferase activity was calculated as described in Fig. 3. The numbers above the bars refer to the fold increase relative to the WT value in the absence of addition. Each value represents the mean ± SEM of three independent transfection experiments (each assayed in duplicate).
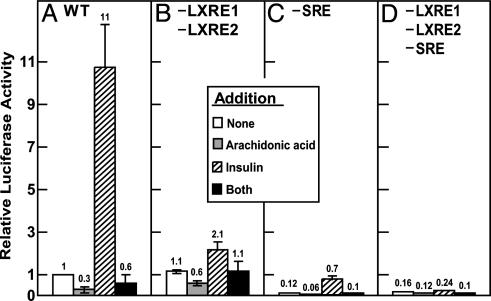
Polyunsaturated fatty acids, such as arachidonic acid, have the potential to reduce Srebp-1c transcription by two mechanisms: (i) they bind to LXRα, where they function as competitive antagonists (17), and (ii) they inhibit the proteolytic activation of SREBP-1 isoforms, thereby potentially interfering with feed-forward stimulation of Srebp-1c transcription through the SRE (27). Fig. 5 shows an experiment in which we tested the effect of 100 μM arachidonic acid on basal and insulin-stimulated transcription driven by the SREBP-1c promoter in hepatocytes. With the WT promoter, arachidonic acid reduced basal transcription by 70% and severely blunted the response to insulin (Fig. 5A). When the two LXREs were disrupted (Fig. 5B), basal transcription was unaffected. Arachidonic acid reduced transcription to a somewhat lesser extent (40%). As expected, with this plasmid the response to insulin was blunted (only 2-fold), and this was slightly reduced by arachidonic acid. As seen previously, the SRE(–) plasmid had reduced basal transcription and a preserved relative response to insulin (Fig. 5C). This response was nearly abolished by arachidonic acid. Because this plasmid does not respond to SREBPs, the ability of arachidonic acid to block insulin activation must be attributable to its inhibition of LXRs. When the LXREs and the SRE were both disrupted, basal and insulin-stimulated transcription were severely reduced and there was little effect of arachidonic acid (Fig. 5D).
Fig. 5.
Suppression of SREBP-1c promoter activity by arachidonic acid is mediated through the LXR elements. The WT plasmid D (A) or the indicated mutant plasmid (B, plasmid m31; C, m24; D, m34) was transfected into primary rat hepatocytes as described in Fig. 1 and incubated without or with 100 μM sodium arachidonate in the absence or presence of 100 nM insulin in medium B containing 0.1% (wt/vol) BSA. After incubation for 21 h, the cells were harvested and assayed for dual luciferase activities. Relative luciferase activity was calculated as described in Fig. 3. The numbers above the bars refer to the fold increase relative to the WT value in the absence of addition. Each value represents the mean ± SEM of three independent transfection experiments (each assayed in duplicate).
Discussion
The data in this paper indicate that the insulin induction of Srebp-1c transcription in rat hepatocytes requires a cooperative interaction between the SRE and the two LXR binding sites. The SRE appears to play a permissive role. Disruption of this element severely reduces basal levels of transcription, but insulin is still able to increase this low level of activity by 4- to 7-fold (Figs. 2, 3, 4, 5). On the other hand, disruption of both LXREs lowers basal transcription rates only slightly but severely disrupts the response to insulin. This observation is illustrated by the results with mutant m31 in Figs. 3B, 4, and 5. The slight insulin stimulation of this mutant plasmid might be attributable to the increase in nuclear SREBPs that is to be expected from the effect of insulin on endogenous SREBPs in these hepatocytes. This conclusion is supported by the finding that the insulin response is abolished when the SRE and the LXREs are all mutated. These data suggest that insulin acts primarily by increasing the ability of LXRα or LXRβ to activate the SREBP-1c promoter. This conclusion is supported by the observation that arachidonic acid, an LXR antagonist, blocks insulin activation even when the SRE is mutated. The activation of LXRs by insulin appears to be relatively specific for the SREBP-1c promoter because insulin does not increase transcription of other LXR-responsive genes such as those encoding the sterol transporters Abcg5 and Abcg8. In vivo studies in livers of rats and mice also have shown that insulin does not increase expression of Abcg5 and Abcg8 (28, 29), whereas it markedly increases expression of Srebp-1c (7).
In principle, insulin might activate LXRs by stimulating the production of an activating ligand such as an oxygenated sterol (15, 16) by increasing the activity of a coactivator such as PGC-1 (30, 31) or ASC-2 (32), or by increasing the activity of LXR itself. The current data provide a clue that insulin may act by producing an activating ligand. This clue comes from the result in Fig. 4, in which we added T0901317, a strong LXR agonist. This compound activated the SREBP-1c reporter construct by a massive 34-fold, and there was little further activation when insulin was added on top of T0901317 (Fig. 4A). If insulin were acting by increasing the intrinsic activity of LXR or by activating a coactivator, we would have expected that insulin would have synergized with T0901317. The fact that insulin did not further activate when the LXR sites are already occupied suggests that insulin itself creates a ligand for LXR or for its heterodimerizing partner, RXR, which is known to be activated by 9-cis-retinoic acid (33). Further studies will be necessary to test this hypothesis.
Acknowledgments
We thank our colleagues Young-Ah Moon and Jay Horton for helpful suggestions, Scott Clark for excellent technical assistance, Richard Gibson for invaluable help with animals, and Jeff Cormier and Erin Friedman for DNA sequencing. This work was supported by National Institutes of Health Grant HL-20948, the Perot Family Foundation, and the Moss Heart Foundation.
Abbreviations: LXR, liver X receptor; LXRE, LXR element; NF-Y, nuclear factor Y; RXR, retinoid X receptor; SRE, sterol regulatory element; SREBP, sterol regulatory element-binding protein; SCAP, SREBP cleavage-activating protein.
References
- 1.McGarry, J. D. (1992) Science 258, 766–770. [DOI] [PubMed] [Google Scholar]
- 2.Spiegelman, B. M. & Flier, J. S. (1996) Cell 87, 377–389. [DOI] [PubMed] [Google Scholar]
- 3.Shimomura, I., Matsuda, M., Hammer, R. E., Bashmakov, Y., Brown, M. S. & Goldstein, J. L. (2000) Mol. Cell 6, 77–86. [PubMed] [Google Scholar]
- 4.Horton, J. D., Goldstein, J. L. & Brown, M. S. (2002) J. Clin. Invest. 109, 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foufelle, F. & Ferre, P. (2002) Biochem. J. 366, 377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimomura, I., Shimano, H., Horton, J. D., Goldstein, J. L. & Brown, M. S. (1997) J. Clin. Invest. 99, 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimomura, I., Bashmakov, Y., Ikemoto, S., Horton, J. D., Brown, M. S. & Goldstein, J. L. (1999) Proc. Natl. Acad. Sci. USA 96, 13656–13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foretz, M., Guichard, C., Ferre, P. & Foufelle, F. (1999) Proc. Natl. Acad. Sci. USA 96, 12737–12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horton, J. D., Bashmakov, Y., Shimomura, I. & Shimano, H. (1998) Proc. Natl. Acad. Sci. USA 95, 5987–5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimomura, I., Hammer, R. E., Ikemoto, S., Brown, M. S. & Goldstein, J. L. (1999) Nature 401, 73–76. [DOI] [PubMed] [Google Scholar]
- 11.Matsuda, M., Korn, B. S., Hammer, R. E., Moon, Y.-A., Komuro, R., Horton, J. D., Goldstein, J. L., Brown, M. S. & Shimomura, I. (2001) Genes Dev. 15, 1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang, G., Yang, J., Horton, J. D., Hammer, R. E., Goldstein, J. L. & Brown, M. S. (2002) J. Biol. Chem. 277, 9520–9528. [DOI] [PubMed] [Google Scholar]
- 13.Repa, J. J., Liang, G., Ou, J., Bashmakov, Y., Lobaccaro, J.-M. A., Shimomura, I., Shan, B., Brown, M. S., Goldstein, J. L. & Mangelsdorf, D. J. (2000) Genes Dev. 14, 2819–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz, J. R., Tu, H., Luk, A., Repa, J. J., Medina, J. C., Li, L., Schwendner, S., Wang, S., Thoolen, M., Mangelsdorf, D. J., et al. (2000) Genes Dev. 14, 2831–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeBose-Boyd, R. A., Ou, J., Goldstein, J. L. & Brown, M. S. (2001) Proc. Natl. Acad. Sci. USA 98, 1477–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janowski, B. A., Grogan, M. J., Jones, S. A., Wisely, G. B., Kliewer, S. A., Corey, E. J. & Mangelsdorf, D. J. (1999) Proc. Natl. Acad. Sci. USA 96, 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ou, J., Tu, H., Shan, B., Luk, A., DeBose-Boyd, R. A., Bashmakov, Y., Goldstein, J. L. & Brown, M. S. (2001) Proc. Natl. Acad. Sci. USA 98, 6027–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshikawa, T., Shimano, H., Amemiya-Kudo, M., Yahagi, N., Hasty, A. H., Matsuzaka, T., Okazaki, H., Tamura, Y., Iizuka, Y., Ohashi, K., et al. (2001) Mol. Cell. Biol. 21, 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amemiya-Kudo, M., Shimano, H., Yoshikawa, T., Yahagi, N., Hasty, A. H., Okazaki, H., Tamura, Y., Shionoiri, F., Iizuka, Y., Ohashi, K., et al. (2000) J. Biol. Chem. 275, 31078–31085. [DOI] [PubMed] [Google Scholar]
- 20.Deng, X., Cagen, L. M., Wilcox, H. G., Park, E. A., Raghow, R. & Elam, M. B. (2002) Biochem. Biophys. Res. Commun. 290, 256–262. [DOI] [PubMed] [Google Scholar]
- 21.Dawson, P. A., Hofmann, S. L., van der Westhuyzen, D. R., Brown, M. S. & Goldstein, J. L. (1988) J. Biol. Chem. 263, 3372–3379. [PubMed] [Google Scholar]
- 22.Jackson, S. M., Ericsson, J., Mantovani, R. & Edwards, P. A. (1998) J. Lipid Res. 39, 767–776. [PubMed] [Google Scholar]
- 23.Dooley, K. A., Millinder, S. & Osborne, T. F. (1998) J. Biol. Chem. 273, 1349–1356. [DOI] [PubMed] [Google Scholar]
- 24.Brown, M. S., Faust, J. R., Goldstein, J. L., Kaneko, I. & Endo, A. (1978) J. Biol. Chem. 253, 1121–1128. [PubMed] [Google Scholar]
- 25.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 3rd Ed.
- 26.Berry, M. N. & Friend, D. S. (1969) J. Cell Biol. 43, 506–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hannah, V. C., Ou, J., Luong, A., Goldstein, J. L. & Brown, M. S. (2001) J. Biol. Chem. 276, 4365–4372. [DOI] [PubMed] [Google Scholar]
- 28.Kok, T., Wolters, H., Bloks, V. W., Havinga, R., Jansen, P. L. M., Staels, B. & Kuipers, F. (2003) Gastroenterology 124, 160–171. [DOI] [PubMed] [Google Scholar]
- 29.Bloks, V. W., Bakker-van Waarde, W. M., Verkade, H. J., Kema, I. P., Wolters, H., Vink, E., Groen, A. K. & Kuipers, F. (2004) Diabetologia 47, 104–112. [DOI] [PubMed] [Google Scholar]
- 30.Puigserver, P. & Spiegelman, B. M. (2003) Endocr. Rev. 24, 78–90. [DOI] [PubMed] [Google Scholar]
- 31.Kelly, D. P. & Scarpulla, R. C. (2004) Genes Dev. 18, 357–368. [DOI] [PubMed] [Google Scholar]
- 32.Kim, S.-W., Park, K., Kwak, E., Choi, E., Lee, S., Ham, J., Kang, H., Kim, J. M., Hwang, S. Y., Kong, Y.-Y., et al. (2003) Mol. Cell. Biol. 23, 3583–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Repa, J. J. & Mangelsdorf, D. J. (2000) Annu. Rev. Cell Dev. Biol. 16, 459–481. [DOI] [PubMed] [Google Scholar]