Abstract
The survival of the spider Cupiennius salei depends on its hunting success, which largely relies on its immediately paralyzing multicomponent venom. Here, we report on the isolation and characterization of CSTX-13, a neurotoxic enhancer in the spider venom. De novo elucidation of the disulfide bridge pattern of CSTX-13 and the neurotoxin CSTX-1 by tandem MS revealed an identical arrangement. However, in contrast to CSTX-1, CSTX-13 is a two-chain peptide with two interchain and two intrachain disulfide bridges. Furthermore, the insecticidal activity of CSTX-13 is synergistically increased in the presence of K+ ions as well as of the cytolytic peptide cupiennin 1a. We demonstrated that the weakly neurotoxic CSTX-13 enhances the paralytic activity of the neurotoxin CSTX-1 by 65% when it is administered with the latter at its entirely nontoxic physiological concentration, which is 440 times below its LD50 concentration.
Spiders and scorpions use their venom to paralyze prey and/or to defend against predators. These venoms are complex mixtures of different components, and the knowledge about their interactions and role in the envenomation process is still limited. During their evolution, these arthropods have developed a large number of neurotoxins that act simultaneously on various invertebrate and/or vertebrate membrane-bound sodium, potassium, and calcium channels. Also, interactions with acid-sensing ion channels, glutamate receptors, and as yet unidentified targets lead to rapid paralysis or death of the envenomed animals (1, 2).
Cupiennius salei (Keyserling, 1877) is a nocturnal hunting spider living in the Central American rain forest (3). The spider relies on an immediately paralyzing venom activity because, in its arboreal environment, a prey item that escapes is lost. The spider also loses its venom investment and reduces its chance of successfully subduing a subsequent prey item, because its venom storage is limited, regeneration takes ≈16 days (4), and its production involves high metabolic costs. Behavioral, ecological, and biochemical investigations of the venom economy of C. salei indicate that it alters the amount of venom injected according to the size, mobility, and defense behavior of its prey (5–8).
This economical venom use is paralleled on the physiological and biochemical levels by the interactions of different venom components (9). In the venom, low molecular mass compounds such as histamine (5.7 mM) and free amino acids, basically taurine (70 μM) and glycine (43.3 μM), are present. K+, Na+, and Ca2+ ions have also been identified. Remarkably, K+ ions are abundant in the venom and rare in the hemolymph (10, 11). Furthermore, C. salei possesses a complex multicomponent system consisting of a few proteins with molecular masses >10 kDa, among them a highly active hyaluronidase. About 100 different peptides with molecular masses between 2 and 8 kDa have been detected by electrospray ionization (ESI)-MS. The peptides can be roughly divided into two groups. The first group contains the smaller peptides with molecular masses of ≈3–4 kDa, which are mainly highly cationic α-helical peptides without cysteine in their composition. Some peptides have already been characterized and named the cupiennin 1 family. These peptides exhibit strong bactericidal activities in the submicromolar range, but also cytolytic and insecticidal activities. In addition, cupiennin 1a shows a high synergistic effect with the main neurotoxin CSTX-1, facilitating a rapid paralysis (12, 13). Positive insecticidal cooperativity between the cytolytically active oxyopinins and neurotoxins is also reported for the spider Oxyopes kitabensis (14).
The second group involves neurotoxically active peptides with molecular masses of ≈8 kDa, which we named Cupiennius salei toxins (CSTX-1 to -13) (11). There is evidence that CSTX-1 inhibits l-type Ca2+ channels of GH3 cells (J. S. Cruz, personal communication). To date, sequence data for the neurotoxins CSTX-1 and CSTX-9 are available. The four disulfide bridges of CSTX-9 form linkages between C1–C4; C2–C5; C3–C8; and C6–C7 (15–17). This arrangement is also found in other spider neurotoxins belonging to the inhibitor cystine knot (ICK) structural motif (18).
So far, a roughly comparable two-chain peptide structure has only been reported for the spider Agelenopsis aperta: the ω-agatoxins IA and G block presynaptic calcium channels in insect neuromuscular junction (19, 20). A two-chain structure has also been proposed for the Hololena toxin, a presynaptic antagonist of insect neuromuscular transmission (21).
Here, we present the amino acid sequence of CSTX-13, an enhancer peptide from the venom of C. salei, and the de novo determination of the disulfide bridge pattern of CSTX-1 and CSTX-13. Despite its sequence similarity to the neurotoxins CSTX-1 and CSTX-9, CSTX-13 acts as a neurotoxic enhancer. Moreover, its low neurotoxic activity is also augmented by other venom compounds.
Materials and Methods
Chemicals. Chemicals were of analytical grade and purchased from Merck unless otherwise specified.
Isolation of CSTX-13. Spider maintenance, venom collection, and separation of 425 μl of venom by FPLC and HPLC methods were performed as described (ref. 11; see also Supporting Text, which is published as supporting information on the PNAS web site). Final purification of CSTX-13 was achieved by RP-HPLC on a nucleosil 100–5 C8 column (4 × 250 mm, Macherey & Nagel) using 22% solvent B (0.1% trifluoroacetic acid in acetonitrile) in solvent A (0.1% trifluoroacetic acid in water) with a flow rate of 0.5 ml/min. Directly after injection of the sample, the gradient (22–28% solvent B) was started for 20 min (Fig. 1C). This step was repeated several times to obtain CSTX-13 (variability between different preparations: 0.5–1.3 mg).
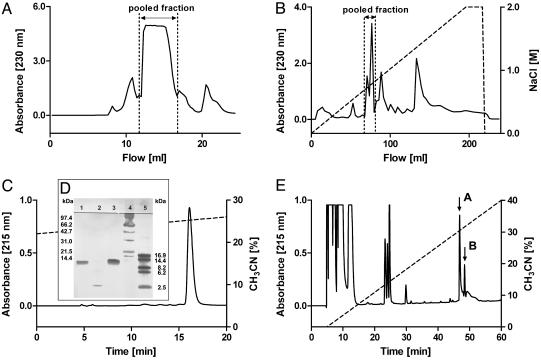
Fig. 1.
Isolation of CSTX-13 from the venom of C. salei.(A) Crude venom was first separated by gel filtration on a Superdex 75 column. (B) Further separation of the pooled fraction was achieved by cationic exchange chromatography on a Mono S HR column. (C) When RP-HPLC was used, CSTX-13 was finally isolated on a nucleosil 100–5C8 column, and the purity was controlled by SDS/PAGE. (D) Lanes: 1, native CSTX-13; 2, reduced CSTX-13; 3, native CSTX-1; 4, molecular mass markers (14.4–97.4 kDa; Bio-Rad); and 5, molecular mass markers (2.5–16.9 kDa; Amersham Pharmacia). (E) RP-HPLC of reduced and alkylated CSTX-13 on a nucleosil 100–5C8 column resulted in the separation of chain A (first arrow) and chain B (second arrow).
PAGE. SDS/PAGE and silver staining of native and reduced (2-mercaptoethanol) CSTX-13 were performed with the Phast-System using high density PhastGel (Amersham Pharmacia).
Reduction and Alkylation. Fifty micrograms of CSTX-13 was reduced and alkylated according to the published procedure (ref. 15, see Supporting Text). Chains A and B were further desalted and separated by RP-HPLC on a nucleosil 100-5 C8 column (4 × 250 mm; Macherey & Nagel) using 100% solvent A with a flow rate of 0.5 ml/min for 0–5 min followed by a 55-min gradient of 0.73% solvent B in solvent A per min (Fig. 1E).
MS. Mass spectrometric analyses were performed on a QSTAR Pulsar hybrid quadrupole time-of-flight mass spectrometer (Applied Biosystems) equipped with a nanoelectrospray ion source. The instrument was tuned for a mass resolving power of 12,000 (m/Δm, full width at half maximum) and calibrated with caesium iodide and reserpine (Sigma). Samples were dissolved in methanol/water (1:1 vol/vol) containing 1% formic acid. The final peptide concentration was 5 pmol/μl. All analyses were performed in the positive ion mode. Numbers represent monoisotopic masses.
For elucidation of the disulfide bridge pattern, 50 μg of native CSTX-13 was cleaved with immobilized trypsin (23 μl wet gel containing 0.5 units of trypsin (Sigma) in 50 μl of 0.1 M Tris·HCl buffer, pH 8.1, and 1.0 mM iodoacetamide, Fluka) under gentle shaking for 17 h at 24°C. The suspension was centrifuged, and the supernatant was recovered for separation by RP-HPLC. The digested and purified peptides were subjected to collision-induced dissociation using nitrogen as the collision gas. Collision energies were in the range of 20–80 eV.
Amino Acid Analysis and Amino Acid Sequence Analysis. Samples were hydrolyzed in the gas phase with 6 M hydrochloric acid containing 0.1% (by volume) phenol for 24 h at 115°C under N2 vacuum according to Chang and Knecht (22). N-terminal sequence analysis was carried out either in a Procise cLC 492 protein sequencer or in a pulsed liquid-phase sequencer 477A, both from Applied Biosystems.
Experiments with Spider Digestive Liquid. Digestive liquid from C. salei was obtained by electrical stimulation and collected in glass capillary tubes, and 8 μl of diluted digestive liquid (1:100 with water) was mixed with 17 μg of CSTX-13 in 8 μl of water. The mixture was kept at 24°C and, after 0.5, 1, and 24 h, aliquots of 2 μl were analyzed by ESI-MS.
For further experiments, 50 μg of CSTX-13 was dissolved in 31.8 μl of water, mixed with 31.8 μl of diluted digestive liquid (1:100 with water), and incubated for 24 h at 24°C, and the fragment was isolated by RP-HPLC on a nucleosil 120–5 C18 column (2 × 125 mm; Macherey & Nagel) using a gradient of 0.2% B in A/min for 200 min and a flow rate of 0.5 ml/min. For ESI-MS analysis, the CSTX-13 fragment was again dissolved in 100 μl of buffer [100 mM Tris·HCl, pH 8.0, containing 135 μM TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone, Sigma) and 220 μM TPCK (N-tosyl-l-phenylalanine chloromethyl ketone, Sigma)], reduced, alkylated, and separated as described above.
Bioassays and Calculations. Bioassays were performed according to Escoubas et al. (23) using 1- to 3-day-old Drosophila melanogaster female flies. The injected volume was 0.05 μl of 0.1 M ammonium acetate, pH 6.1 (control), and all further injections with different components were carried out in this solution, except nifedipine (Calbiochem), which was injected in ≥0.06 M ammonium acetate, pH 6.1, and ≤5.6 M dimethyl sulfoxide. To estimate the LD50 (24 h after injection), 20 flies were used as control, and 20 flies were used for each concentration.
To investigate synergistic effects between CSTX-13 and further venom components such as cupiennin 1a (9.6 μM) (in nontoxic concentration), histamine (5.7 mM, Sigma), taurine (0.07 mM, Sigma), and KCl (215 mM) (all in physiological venom concentrations), bioassays were performed with 12.6 pmol of CSTX-13 per mg of fly. We tested CSTX-13 alone and in combination with each of the above mentioned venom components [n = 2 × (15 × 5) for each assay]. Venom components in above-mentioned concentrations were injected alone as control (n = 20). The paralytic activity of physiological KCl concentration was measured by comparing the awake time of a control group (n = 20) and a treated group (n = 20) (Mann–Whitney U test, spss 10 software).
To highlight synergistic effects between the neurotoxins CSTX-1 and CSTX-13, corresponding to their molar ratio in the crude venom (9:1), bioassays were performed with 0.315 pmol of CSTX-1 per mg of fly alone and in combination with 0.035 pmol of CSTX-13 per mg of fly, and three further concentrations down to 0.63 fmol of CSTX-13 per mg of fly [n = 2 × (12 × 5) for each assay]. CSTX-13 alone was used in the above mentioned concentrations as a control (n = 2 × 20).
The influence of CSTX-13 on two different Ca2+ channel blockers was further investigated. NiCl2 was administered in a concentration of 5.26 nmol/mg of fly alone and in combination with 0.035 pmol of CSTX-13 per mg of fly [n = 2 × (12 × 5) for each assay]. Nifedipine was tested in a concentration of 0.105 nmol/mg of fly alone and in combination with 0.035 pmol of CSTX-13 per mg of fly [2 × (6 × 5) for each assay], and three further concentrations up to 5.5 pmol of CSTX-13 per mg of fly [n = 6 × 5 for each assay].
The relative mortality of D. melanogaster was arcsin square root-transformed and treated as the dependent variable, whereas the venom components or CSTX-13 were treated as nominal independent variables. The experiment was analyzed by generalized linear models. The means of the nominal independent variables venom components or CSTX-13, respectively, were compared pairwise by the Bonferroni method. Fulfillment of the model assumptions was checked by visual inspection of the residuals distribution for every statistical test conducted. Statistics were performed with s-plus 6.0 professional software.
Results
Purification and Sequence Analysis of CSTX-13. The crude venom (425 μl) was separated in a four-step protocol using gel filtration (Fig. 1 A), cationic exchange chromatography (Fig. 1B), and successive RP-HPLC (Fig. 1C). The retention profile of CSTX-13 revealed no impurities. CSTX-13 was characterized by ESI-MS and amino acid composition. The yield of CSTX-13 obtained by purification of crude venom was 1.2–3.0 μg/μl depending on the separation protocol.
N-terminal sequence analysis of native CSTX-13 provided evidence for a two-chain molecule: Ser/Ala–Asp/Lys–Xaa/Lys–Thr/Glu–Leu/Leu–Arg/Xaa–Asn/Thr–His/Xaa–Asp/Gln–Xaa/Gln. Therefore, CSTX-13 was reduced and alkylated, and chains A and B were separated by RP-HPLC. Both chains were sequenced by Edman degradation from the N to the C termini without any ambiguity. Chain A is composed of 34 residues (measured, 4,342.73 Da; calculated, 4,342.76 Da), and chain B is composed of 29 residues. ESI-MS of chain B gave a monoisotopic mass of 3,475.80 Da, which is one mass unit less than the expected theoretical mass of 3,476.83 Da, thus indicating C-terminal amidation of chain B. The determined amino acid sequences of both chains agree well with the amino acid composition of native CSTX-13 as well as with the individual chains. Taking into account the four disulfide bridges of both peptide chains and the amidation, the calculated monoisotopic mass of CSTX-13 is in agreement with the measured mass of native CSTX-13 (measured, 7,354.51 Da; calculated, 7,354.37 Da) (Fig. 2 and Fig. 5, which is published as supporting information on the PNAS web site).
Fig. 2.
Sequence comparison and disulfide bridge arrangement of CSTX-13, CSTX-1, CSTX-9 (C. salei), and ω-agatoxin IA (A. aperta). Identical amino acids are shaded gray, the disulfide bridges are represented by lines, and the corresponding cysteine residues are shaded black. The disulfide bridge patterns of CSTX-13 and CSTX-1 were determined by nanoelectrospray tandem MS of the corresponding disulfide-linked tryptic fragment.
SDS/PAGE analysis of purified native CSTX-13 revealed a single band at 12 kDa, whereas reduced CSTX-13 revealed a single band at ≈3 kDa, obviously containing the peptide chains A and B. This supports the two-chain structure of the native CSTX-13 (Fig. 1 C and D).
To exclude the possibility that the two-chain structure of CSTX-13 is a proteolytic artifact because of contamination of the venom with digestive liquid (24), CSTX-13 was incubated with fresh digestive liquid. After 0.5, 1, and 24 h of incubation, the obtained mass indicates a proteolytic degradation of the 14 C-terminal amino acid residues of chain B (measured, 5,746.51 Da; calculated, 5,746.51 Da). The CSTX-13 fragment was purified by RP-HPLC, and reduced and alkylated in the presence of two protease inhibitors. ESI-MS analysis of the purified compounds revealed an intact chain A (measured, 4,342.80 Da; calculated, 4,342.76 Da) and a truncated chain B (measured, 1,868.03 Da; calculated, 1,867.98 Da). These findings are in accordance with the result described above and support the assumption of a native two-chain structure of CSTX-13 in the venom. CSTX-13 seems to be present in the venom as a two-chain molecule and to the best of our knowledge does not represent a purification artifact.
Because of the unique amino acid sequences of CSTX-1 and CSTX-13 with cysteine residues arranged in close proximity, classical approaches to determine the disulfide bridge pattern, based on specific enzymatic or chemical cleavages, failed. Consequently, the disulfide bridge patterns of CSTX-1 and CSTX-13 were identified de novo by nanoelectrospray tandem MS. Digestion of the native toxin with immobilized trypsin yielded main fragments consisting of five short peptide chains cross-linked by four disulfide bridges.
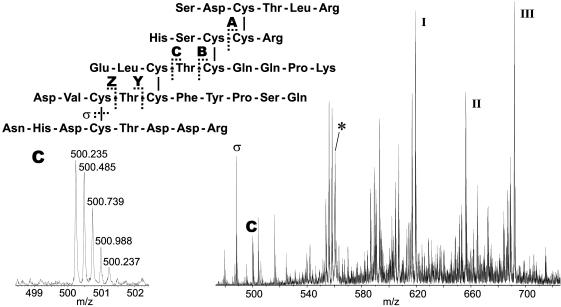
Multiply charged [M + nH]n+ ions (n = 3–9) of the cystine containing tryptic fragment (measured, 4,473.75 Da; calculated, 4,473.76 Da) of CSTX-13 were selected as precursor ions for subsequent collision-induced dissociation (CID). The resulting product ion spectra are characterized by abundant peaks of fragment ions generated by cleavage of the disulfide bridges. These ions define the order of peptide chains. Further abundant peaks indicate repetitive loss of amino acids from the termini of the peptide chains, often occurring in conjunction with the loss of water. Detailed information on the disulfide bridge pattern was obtained by detection of the less abundant fragment ions of mass 968.43 Da (A), 1,895.80 Da (B), 1,995.85 Da (C), 1,289.45 Da (Y), and 1,390.49 Da (Z), generated by cleavage of the peptide backbone between adjacent cystines. The corresponding cleavage sites are indicated in Fig. 3 (see Fig. 6, which is published as supporting information on the PNAS web site). Measurements exhibit a maximum deviation of 0.02 Da from the calculated masses. Fig. 3 also shows a section of the product ion spectrum obtained by CID of the [M + 8H]8+ precursor ion of the tryptic CSTX-13 fragment. The high mass accuracy and resolving power of the tandem mass spectrometer allow unambiguous peak assignment, as demonstrated for the quadruply charged fragment ion C. Additional information was obtained by assigning peaks generated by disulfide bridge cleavage. The same strategy was applied for the elucidation of the disulfide bridge pattern of CSTX-1, which exhibits identical disulfide bridges (Fig. 2).
Fig. 3.
Sequence of the cystine-containing fragment obtained from tryptic digest of native CSTX-13 (4,473.73 Da). The asterisk (*) indicates the [M + 8H]8+ ion with m/z 560.23, which was selected as the precursor for CID. Typical fragmentation pathways include loss of terminal amino acids in conjunction with loss of water (ions I, I, and III), disulfide cleavage (ion σ), and cleavage of peptide bonds between cystines (disulfide bridge-defining ions A–Z). The enlargement shows the isotopic pattern of fragment ion C with m/z 500.22, which defines, in combination with fragment ion A, the Cys 1–Cys 4 and Cys 2–Cys 5 bridges.
Synergistic Insecticidal Effects. To evaluate the biological importance of CSTX-13, comparative bioassays with D. melanogaster were performed. The LD50 of 16.3 pmol/mg of fly (14.5–27.1; 95% confidence limits) indicates a lower toxicity than other neurotoxins of C. salei. CSTX-13 is ≈49 times less toxic than the neurotoxin CSTX-1, and 2.8 times less toxic than the cytolytically active peptide cupiennin 1a, both key components identified in the venom of C. salei (24, 12) (Table 1).
Table 1. Insecticidal activity of spider venom components.
| Venom components | Physiological venom concentration, mM | LD50 concentration | Confidence limits, 95% | Synergistically tested concentration |
|---|---|---|---|---|
| CSTX-1 | 1.4-3.3 | 6.3 μM | 5.2-7.6 | |
| CSTX-9 | 0.2-1.1 | 211 μM | 190-273 | |
| CSTX-13 | 0.2-0.4 | 309 μM | 275-514 | 0.01-0.7 μM |
| Cupiennin 1a | ≈1.2 | 106 μM | 75-149 | 9.6 μM |
| Taurine | 0.07 | >160 mM | 0.07 mM | |
| Histamine | 5.7 | 917 mM | 794-1,100 | 5.7 mM |
| KCl | 215 | 1,845 mM | 1,650-2,131 | 215 mM |
Estimation of the lethal doses (LD50) in a Drosophila bioassay, where 50% of the test flies died of intoxication 24 h after injection. Different amounts of peptides, histamine, taurine, and KCl were dissolved in 0.1 M ammonium acetate, at a pH of 6.1, and 0.05 μl was injected into the flies. The physiological concentrations of CSTX-1 (13), taurine (13), histamine (13), KCl (13), CSTX-9 (15), and cupiennin 1a (12) in the venom were reported.
Synergistic interactions of different venom components (taurine, histamine, KCl) with the paralytic activity of CSTX-13 were analyzed. Taurine itself was not toxic to D. melanogaster up to 8.9 nmol/mg of fly. The LD50 of histamine was 51.0 nmol/mg of fly (44.2–61.2; 95% confidence limits) and KCl showed a LD50 of 102.5 nmol/mg of fly (91.7–118.4; 95% confidence limits) (Table 1).
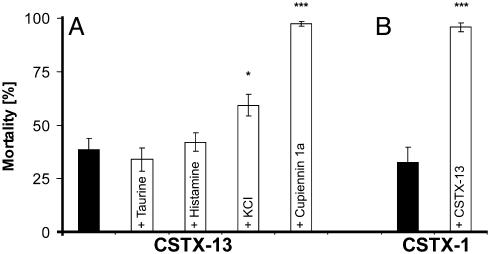
When tested alone at physiological concentrations, taurine (3.88 pmol/mg of fly) and histamine (316.67 pmol/mg of fly) showed no effects in a Drosophila bioassay. Injection of KCl (11.94 nmol/mg of fly) showed a short significant paralytic effect (564.7 s ± SD 288.9 s; P < 0.001) when compared with the control group (234.5 s ± SD 116.5 s). An injection of 12.6 pmol of CSTX-13/mg of fly resulted in a mortality of 39%. No statistically significant differences were observed by coinjection of CSTX-13 with taurine (mortality of 34%) or with histamine (mortality of 42%). In contrast, coinjection of CSTX-13 with KCl significantly increased the mortality to 59% (P < 0.05) (Fig. 4A). When injected alone, the cytolytic cupiennin 1a is not toxic to D. melanogaster in a concentration of 0.53 pmol/mg of fly, but it increases the mortality of CSTX-13 from 39% to 97% (P < 0.001) (Fig. 4A).
Fig. 4.
Synergistic effects between CSTX-13 and venom components. (A) Synergistic effects between CSTX-13 and low molecular venom components. In a Drosophila bioassay, the lethal effect of CSTX-13 injected alone (239.4 μM) was compared with the lethal effect of coinjected CSTX-13 (239.4 μM) with taurine (0.07 mM; not significant), histamine (5.7 mM; not significant), KCl (215 mM; *, P < 0.05), or cupiennin 1a (9.6 μM; ***, P < 0.001). As controls, taurine, histamine, KCl, and cupiennin 1a showed no toxic effect when administered alone. (B) Synergistic effects of CSTX-13 on the toxicity of CSTX-1. The lethal effect of CSTX-1 (5.99 μM) was compared with the lethal effect of coinjected CSTX-1 (5.99 μM) with CSTX-13 (0.67 μM/mg of fly; ***, P < 0.001) (molar ratio of 9:1) corresponding to their concentrations in the venom. CSTX-13 was also injected alone as control and showed no toxic effect on the flies. Statistical analysis was done by using the Bonferroni method. Standard error bars are shown for every treatment.
In addition, we investigated the synergistic effect of CSTX-13 on the toxicity of CSTX-1. At physiological concentrations in the venom, the molar ratio of CSTX-1 and CSTX-13 is 9:1. With administration of one peptide alone, 0.315 pmol of CSTX-1 per mg of fly caused a mortality of 31%, and injection of 0.035 pmol of CSTX-13 per mg of fly had no effect. Surprisingly, coinjection of CSTX-1 and CSTX-13 in the above mentioned molar ratio of 9:1 significantly increased the mortality to 96% (P < 0.001) (Fig. 4B), and, even in a molar ratio of 500:1, the enhancing effect of CSTX-13 was observed (45%, not significant).
In view of the fact that CSTX-1 inhibits l-type Ca2+ channels and that, in Drosophila muscle, a 1,4-dihydropyridine-sensitive (25) homolog of the mammalian l-type/α1D (Dmca1D) subunit gene is expressed (26), the influence of CSTX-13 on the activity of the l-type calcium channel blocker nifedipine (27) as well as NiCl2, a general inhibitor of calcium channels (28), was investigated. No synergistic effects between NiCl2 or nifedipine and CSTX-13 were detected.
Discussion
The Structure of CSTX-13. In CSTX-13, we have characterized a two-chain peptide from the venom of C. salei. It paralyzes flies only in high concentrations (LD50 309 μM) when applied alone, but synergistically enhances the paralytic activity of the main neurotoxin CSTX-1 at low concentrations (0.7 μM).
In the venom, CSTX-13 is constitutively present at a 7–8 times lower concentration than the main neurotoxin CSTX-1 and in an up to 2.8 times lower concentration than a further neurotoxin CSTX-9. Similarly, its insecticidal activity, expressed as a LD50 value, is 49 times lower than the activity of CSTX-1 and 1.5 times lower than that of CSTX-9 (Table 1) (24). Protein database search using blastp 2.2.8 (29) resulted in a high sequence identity of 56% (70% similarity) between CSTX-1 and CSTX-9, but in lower sequence identities of 35% between CSTX-13 and CSTX-1 (51% similarity), and of 31% between CSTX-9 and CSTX-13 (49% similarity). Nevertheless, all three peptides exhibit identical disulfide bridge patterns (15–17) (Fig. 2). Sequence comparison implies that, upon processing, a short peptide is excised in the loop forming the disulfide bridge C6–C7, thus leading to the two-chain structure of CSTX-13.
No sequence similarities were detected with further neurotoxins and the two other sequenced two-chain calcium channel blockers ω-agatoxin IA (66 and 3 residue chains) and ω-agatoxin G (62 and 3 residue chains) (20) from the spider Agelenopsis aperta. In contrast to CSTX-13, which contains two interchain and two intrachain disulfide linkages, these two neurotoxins possess four intrachain and one interchain disulfide linkage. ω-agatoxin IA is formed from its precursor by excision of an internal heptapeptide leading to a major peptide chain that is connected to the minor peptide chain (three residues) by one disulfide bridge (19) (Fig. 2).
Unlike the above mentioned ω-agatoxins, CSTX-13 is neurotoxic by itself only at a high micromolar concentration. This circumstance raised the question of whether the two-chain structure of CSTX-13 might be the result of a purification artifact caused by contamination with proteases. The purification protocols of CSTX-13 over the last 5 years have always resulted in a pure peptide with identical molecular masses. Experiments with spider digestive liquid, which could be the major source of protease contaminations, resulted only in a C-terminal truncation of 14 residues of chain B. As shown previously, C-terminal proteolytic degradation of CSTX-1 by spider digestive liquid stopped at position 49 (Gly) (24). Therefore, we conclude that the two-chain structure of CSTX-13 is not a purification artifact or protease degradation product, but a valid constitutive component of the C. salei venom. Whether the two-chain structure is posttranslationally generated, however, remains to be investigated.
Biological Function of CSTX-13 in the Venom. To analyze the biological function of CSTX-13, we investigated possible interactions between CSTX-13 and different venom components in a Drosophila bioassay. Previously, we have shown that the neurotoxicity of the main neurotoxin CSTX-1 to blow flies (Protophormia sp.) could be increased when coinjected with taurine and histamine (9). However, coinjection of CSTX-13 with taurine or histamine in its physiological venom concentration did not increase its insecticidal activity. Nevertheless, histamine as a neurotransmitter and taurine as a neuromodulator play an important role in the insect nerve system (30–33).
Remarkably, the venom of C. salei exhibits a very high K+ ion concentration that is 32-fold higher than in the hemolymph, and even 2.7-fold higher than in the prevenom of the scorpion Parabuthus transvaalicus (11, 34). Hammock and coworkers (34) suggest an economically motivated strategy in venom utilization for this scorpion. P. transvaalicus first secretes a prevenom containing a high K+ ion concentration at a low protein content, whereas the subsequently secreted venom is characterized by a high protein content and a 15-fold lower K+ ion concentration. The synergistic activity in the prevenom between the “inexpensive” K+ ion and the assumed inhibitors of rectifier K+ channels is proposed as a means of conserving metabolically expensive neuropeptides in the venom (34). In part, C. salei also uses this strategy to enhance its venom efficacy. Coinjection of CSTX-13 with K+ ions increases the mortality of the flies by 20%. The synergistic cooperation of K+ ions is also detectable when applied together with CSTX-1, a suggested l-type Ca2+ channel blocker (B.W. and L.K.-N., unpublished data). The high K+ concentration in the venom alone caused an immediate short paralysis, and there seems to be a general cooperation between K+ ions and various ion channel blockers described here for a labidognath spider.
Enhancement of insecticidal efficacy through the cooperative interaction of different venom peptide neurotoxins in spiders (35) and scorpions (36, 37) has been well investigated. Additionally, synergistic interactions between acylpolyamines and cysteine-rich peptide neurotoxins (38) as well as between cytolytic peptides and neurotoxins have been described (12–14). These positive interactions were principally demonstrated by applying both components in toxic concentrations.
In contrast, a nontoxic concentration of the cytolytically active cupiennin 1a (20 times lower than its LD50) dramatically enhances the efficacy of CSTX-1 (12, 13). The same effect was observed when testing CSTX-13 and cupiennin 1a. It is assumed that, in both cases, mainly through the nonspecific cytolytic activity of cupiennin 1a, CSTX-1 and CSTX-13 have better access to their targets.
Surprisingly, when CSTX-1 and CSTX-13 were administered together at their venom concentrations, a strong positive cooperation was found. The data presented here on the two-chain neurotoxic enhancer CSTX-13 show that it enhances the efficacy of the neurotoxin CSTX-1 at a concentration of 440 times below its LD50. Tests with different concentrations of CSTX-13 revealed a positive correlation between the amount of CSTX-13 and the efficacy of CSTX-1. The cooperation between CSTX-1 and CSTX-13 seems to be highly specific, because no synergistic interactions between CSTX-13 and other Ca2+ channel blockers, such as nifedipine and NiCl2, were found.
Conclusions
In summary, the structural and biological characterization of CSTX-13 provide further insight into the complexity of C. salei venom as more multiple interactions between different venom components become apparent. After venom injection into a prey animal, the hyaluronidase seems to act as a spreading factor, followed by the dual cytolytic activity of the cupiennins. They facilitate the activity of the neurotoxins and at the same time protect the venom duct and glands against bacterial invasion by membrane disturbance and pore building. Additionally, antimicrobial peptides may also modulate intracellular signaling by increasing intracellular Ca2+, as reported for parabutoporin and opistoporin from scorpion venoms (39). Simultaneously, the inhibition of ion channels by the neurotoxins is further enhanced by the high K+ ion concentration in the venom, shifting the K+ equilibrium potential (34). Finally, the neurotoxins act on different ion channels with a concomitant enhancement by CSTX-13.
Supplementary Material
Acknowledgments
We thank Dr. Patrik Kehrli and Dr. Sven Bacher for statistical advice, Dr. Heather Murray for critical comments on the manuscript, and the Swiss National Science Foundation for funding.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: ESI, electrospray ionization
Data deposition: The sequences reported in this paper have been deposited in the Swiss-Prot and TrEMBL databases [accession nos. P83919 (CSTX-13 chain A) and P83920 (CSTX-13 chain B)].
References
- 1.Loret, E. & Hammock, B. (2001) in Scorpion Biology and Research, eds. Brownell, P. & Polis, G. (Oxford Univ. Press, New York), pp. 204–233.
- 2.Escoubas, P., Diochot, S. & Corzo, G. (2000) Biochimie 82, 893–907. [DOI] [PubMed] [Google Scholar]
- 3.Barth, F. G. (2002) A Spider's World: Senses and Behavior (Springer, New York).
- 4.Boevé, J.-L., Kuhn-Nentwig, L., Keller, S. & Nentwig, W. (1995) Toxicon 33, 1347–1357. [DOI] [PubMed] [Google Scholar]
- 5.Malli, H., Imboden, H. & Kuhn-Nentwig, L. (1998) Toxicon 36, 1959–1969. [DOI] [PubMed] [Google Scholar]
- 6.Malli, H., Kuhn-Nentwig, L., Imboden, H. & Nentwig, W. (1999) J. Exp. Biol. 202, 2083–2089. [DOI] [PubMed] [Google Scholar]
- 7.Wigger, E., Kuhn-Nentwig, L. & Nentwig, W. (2002) Toxicon 40, 749–752. [DOI] [PubMed] [Google Scholar]
- 8.Wullschleger, B. & Nentwig, W. (2002) Funct. Ecol. 16, 802–807. [Google Scholar]
- 9.Kuhn-Nentwig, L., Bücheler, A., Studer, A. & Nentwig, W. (1998) Naturwissenschaften 85, 136–138. [DOI] [PubMed] [Google Scholar]
- 10.Loewe, R., Linzen, B. & von Stackelberg, W. (1970) Z. Vergl. Physiol. 66, 27–34. [Google Scholar]
- 11.Kuhn-Nentwig, L., Schaller, J. & Nentwig, W. (1994) Toxicon 32, 287–302. [DOI] [PubMed] [Google Scholar]
- 12.Kuhn-Nentwig, L., Müller, J., Schaller, J., Walz, A., Dathe, M. & Nentwig, W. (2002) J. Biol. Chem. 277, 11208–11216. [DOI] [PubMed] [Google Scholar]
- 13.Kuhn-Nentwig, L., Schaller, J. & Nentwig, W. (2004) Toxicon 43, 543–553. [DOI] [PubMed] [Google Scholar]
- 14.Corzo, G., Villegas, E., Gómez-Lagunas, F., Possani, L. D., Belokoneva, O. S. & Nakajima, T. (2002) J. Biol. Chem. 277, 23627–23637. [DOI] [PubMed] [Google Scholar]
- 15.Schaller, J., Kämpfer, U., Schürch, S., Kuhn-Nentwig, L., Haeberli, S. & Nentwig, W. (2001) Cell. Mol. Life Sci. 58, 1538–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaller, J., Kuhn-Nentwig, L., Schürch, S., Kämpfer, U., Müller, J. & Nentwig, W. (2001) Chimia 55, 1058–1062. [Google Scholar]
- 17.Schürch, S., Schaller, J., Kämpfer, U., Kuhn-Nentwig, L. & Nentwig, W. (2001) Chimia 55, 1063–1066. [Google Scholar]
- 18.Norton, R. S. & Pallaghy, P. K. (1998) Toxicon 36, 1573–1583. [DOI] [PubMed] [Google Scholar]
- 19.Santos, A. D., Imperial, J. S., Chaudhary, T., Beavis, R. C., Chait, B. T., Hunsperger, J. P., Olivera, B. M., Adams, M. E. & Hillyard, D. R. (1992) J. Biol. Chem. 267, 20701–20705. [PubMed] [Google Scholar]
- 20.Saccomano, N. A. & Ahlijanian, M. K. (1994) Drug Dev. Res. 33, 319–343. [Google Scholar]
- 21.Bowers, C. W., Phillips, H. S., Lee, P., Jan, Y. N. & Jan, L. Y. (1987) Proc. Natl. Acad. Sci. USA 84, 3506–3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang, J.-Y. & Knecht, R. (1991) Anal. Biochem. 197, 52–58. [DOI] [PubMed] [Google Scholar]
- 23.Escoubas, P., Palma, M. F. & Nakajima, T. (1995) Toxicon 33, 1549–1555. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn-Nentwig, L., Schaller, J., Kämpfer, U., Imboden, H., Malli, H. & Nentwig, W. (2000) Arch. Insect Biochem. Physiol. 44, 101–111. [DOI] [PubMed] [Google Scholar]
- 25.Gielow, M. L., Gu, G.-G. & Singh, S. (1995) J. Neurosci. 15, 6085–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren, D., Xu, H., Eberl, D. F., Chopra, M. & Hall, L. M. (1998) J. Neurosci. 18, 2335–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen, C. J., Ertel, E. A., Smith, M. M., Venema, V. J., Adams, M. E. & Leibowitz, M. D. (1992) Mol. Pharmacol. 42, 947–951. [PubMed] [Google Scholar]
- 28.Wakamori, M., Strobeck, M., Niidome, T., Teramoto, T., Imoto, K. & Mori, Y. (1998) J. Neurophysiol. 79, 622–634. [DOI] [PubMed] [Google Scholar]
- 29.Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng, Y., Hirschberg, B., Yuan, J., Wang, A. P., Hunt, D. C., Ludmerer, S. W., Schmatz, D. M. & Cully, D. F. (2002) J. Biol. Chem. 277, 2000–2005. [DOI] [PubMed] [Google Scholar]
- 31.Witte, I., Kreienkamp, H.-J., Gewecke, M. & Roeder, T. (2002) J. Neurochem. 83, 504–514. [DOI] [PubMed] [Google Scholar]
- 32.Buchner, E., Buchner, S., Burg, M. G., Hofbauer, A., Pak, W. L. & Pollack, I. (1993) Cell Tissue Res. 273, 119–125. [DOI] [PubMed] [Google Scholar]
- 33.Bicker, G. (1991) Brain Res. 560, 201–206. [DOI] [PubMed] [Google Scholar]
- 34.Inceoglu, B., Lango, J., Jing, J., Chen, L., Doymaz, F., Pessah, I. N. & Hammock, B. D. (2003) Proc. Natl. Acad. Sci. USA 100, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bindokas, V. P., Venema, V. J. & Adams, M. E. (1991) J. Neurophysiol. 66, 590–601. [DOI] [PubMed] [Google Scholar]
- 36.Herrmann, R., Moskowitz, H., Zlotkin, E. & Hammock, B. D. (1995) Toxicon 33, 1099–1102. [DOI] [PubMed] [Google Scholar]
- 37.Regev, A., Rivkin, H., Inceoglu, B., Gershburg, E., Hammock, B. D., Gurevitz, M. & Chejanovsky, N. (2003) FEBS Lett. 537, 106–110. [DOI] [PubMed] [Google Scholar]
- 38.Adams, M. E., Herold, E. E. & Venema, V. J. (1989) J. Comp. Physiol. A 164, 333–342. [DOI] [PubMed] [Google Scholar]
- 39.Moerman, L., Verdonck, F., Willems, J., Tytgat, J. & Bosteels, S. (2003) Biochem. Biophys. Res. Commun. 311, 90–97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.