Abstract
CCAAT displacement protein/cut homolog (CDP/cut) is a highly conserved homeodomain protein that contains three cut repeat sequences. CDP/cut is a transcriptional factor for many diverse cellular and viral genes that are involved in most cellular processes, including differentiation, development, and proliferation. Here, we report that CDP/cut interacts with a histone lysine methyltransferase (HKMT), G9a, in vivo and in vitro. The deletion of the cut repeats within CDP/cut abrogates the interaction with G9a. The transcriptional repressor function of CDP/cut is mediated through HKMT activity of G9a associated with CDP/cut. We show that the recruitment of G9a to the human p21waf1/cdi1 promoter is contingent on the interaction with CDP/cut, and CDP/cut is directly associated with an increase in the methylation in vivo of Lys-9 in histone H3 within the CDP/cut-regulatory region of the p21waf1/cdi1 promoter. The endogenous level of p21waf1/cdi1 expression is repressed through CDP/cut and mediated by HKMT activity of G9a. Furthermore, we report the identification of G9a as a component of CDP/cut complex. G9a colocalizes with CDP/cut in the nucleus. These results indicate that G9a functions as a transcriptional corepressor in association with a CDP/cut complex. These studies now reveal the interaction of G9a with a sequence-specific transcription factor that regulates gene repression through CDP/cut.
The CCAAT displacement protein/cut homolog (CDP/cut) is an evolutionarily well conserved transcription factor involved in the regulation of cellular differentiation, development, and cell-cycle progression (1–5). Full-length gene transcripts of CDP/cut (180–190 kDa) encode three cut repeats and a homeodomain that have properties for DNA binding and govern transcriptional regulation. Two N-terminally truncated isoforms, p110 with two cut repeats and a homeodomain generated by proteolytic processing and p75 with a cut repeat and a homeodomain generated by alternate transcription, have been identified, and both isoforms are able to bind to DNA and regulate gene transcription (6, 7). CDP/cut proteins have been characterized to function as transcriptional repressors in a large number of genes including gp91phox, p21waf1/cdi1, c-myc, histones, and the cystic fibrosis transmembrane conductance regulator gene (CFTR) (1, 4, 8–11). Recently, the potential stimulation of transcription from the DNA polymerase α gene promoter by the CDP/cut p110 isoform has been shown (12). In addition, CDP/cut has also been proposed to function as an architectural protein as it binds to nuclear matrix attachment regions associated with gene activity (13, 14).
Repression by CDP/cut functions by at least two mechanisms: competition for binding site occupancy and active repression (15). The evolutionarily conserved domains, three cut repeats and a homeodomain, function as specific DNA-binding domains (16, 17). Posttranslational modifications of CDP/cut have been reported to affect its DNA-binding activity and transcriptional properties. Dephosphorylation of CDP/cut by Cdc25A phosphatase and the proteolytic cleavage of full-length CDP/cut to generate p110 isoform increase stable CDP/cut DNA-binding activity (4, 6). Phosphorylation of CDP/cut by cyclin A–cyclin-dependent kinase 1 within and near the homeodomain and acetylation by CBP/p300 and P/CAF near the homeodomain inhibit CDP/cut DNA-binding activity and prevent transcriptional repression (18, 19). The C-terminal region of CDP/cut regulates gene transcription through active repression (11, 15). We have previously demonstrated that the C-terminal region of CDP/cut including the third cut repeat and the homeodomain associates with histone deacetylase (HDAC)1, and represses gene transcription, suggesting that the recruitment of HDAC activity by CDP/cut occurs to achieve repression of specific target genes (11).
Posttranslational modifications of histone tails including acetylation, phosphorylation, and methylation play important roles in regulating transcription and chromatin structure (20–23). Methylation of histone H3 has been linked to distinct effects on transcription, depending on the enzyme and residue modified (24–27). Of histone H3-specific methyltransferases, Su(var)39H1, the human homolog of Drosophila Su(var)3–9, has been shown to specifically methylate histone H3 at Lys-9 (23). This modification creates a recognition site for the heterochromatin protein 1 (HP1) that is directly associated with DNA methylation and gene silencing (28, 29). The Su(var)39H1-HP1 complex is also involved in repression of genes located within euchromatin (30); however, the function of Su(var)39H1 and HP1 in euchromatin has not yet been determined. G9a is a recently identified Su(var), Enhancer of Zeste, Trithorax (SET) domain-containing protein with histone lysine methyltransferase (HKMT) activity (31). G9a has been characterized to express functions distinct from Su(var)39H1. Compared to Su(var)39H1, G9a specifically methylates Lys-27 as well as Lys-9 residues of histone H3; however, G9a does not colocalize with either HP1α or Su(var)39H1 in the nucleus (31). It has also been reported that G9a is the primary and dominant histone H3 methyltransferase for K9 in euchromatin and essential for early embryogenesis (32). Although the involvement of G9a in transcriptional repression has been shown (32), no sequence-specific transcription factor has yet been reported to target G9a for recruitment to promoters in vivo.
Here, we present that CDP/cut recruits G9a to the human p21waf1/cdi1 promoter (p21pro) and G9a functions as a transcriptional corepressor with CDP/cut. Our data reveal a mechanism for active repression by CDP/cut: recruitment of HKMT activity to the promoter.
Materials and Methods
Plasmid Construction. cDNAs for full-length human CDP/cut (amino acids 1–1,505), CDPN1 (amino acids 1–407), CDPN2 (amino acids 1–636), CDPN3 (amino acids 1–1,125), and CDPC (amino acids 1,126–1,505) were PCR-amplified from pMT2-CDP (33) and subcloned into modified pcDNA3 (Invitrogen) containing an N-terminal FLAG-tag (pcDNA3-FLAG) for mammalian expression, pGEX2TK (Amersham Biosciences, Piscataway, NJ) for bacterial expression to generate GST-CDPN1 and GST-CDPC fusion proteins, and pEBVHis (Invitrogen) for expression of (His)6-Xpress-tagged CDPC. Human G9a cDNA was obtained as an EST clone (ID 3912870) from the I.M.A.G.E. Consortium (http://image.llnl.gov). cDNAs encoding full-length hG9a (amino acids 1–1,001) and hG9a (ΔSET) (amino acids 1–758) were PCR-amplified and subcloned into pEGFP (Clontech) for expression of N-terminal enhanced GFP (EGFP)-tagged proteins and pcDNA3-FLAG for expression of FLAG-hG9a. pcDNA3-Gal4DBD-CDPC was constructed by PCR-based strategies by using pSwitch (Invitrogen) as a template for the Gal4 DNA-binding domain (amino acids 1–93) coding region to generate Gal4DBD-CDPC fusion protein. To generate a construct for 6X Gal4 UAS-E1b-luciferase reporter, the PCR product corresponding to the 6X Gal4 UAS-E1b fragment from pGene/V5-His (Invitrogen) was inserted in-frame into pGL2 (Promega). All constructs were verified by DNA sequencing.
Cell Culture and Immunoprecipitation. The 293T cells and IMR90 cells (American Type Culture Collection) were maintained in DMEM supplemented with 10% FBS at 37°C with 5% CO2. For immunoprecipitation, 293T cells were transiently transfected by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were harvested 48 h after transfection in lysis buffer (50 mM Tris·HCl, pH 7.6/120 mM NaCl/0.5% Nonidet P-40) containing protease inhibitor mixture (Roche Applied Sciences). Cleared lysates were immunoprecipitated with M2 agarose beads (Sigma) for FLAG-tagged proteins or anti-GFP Ab (Santa Cruz Biotechnology) and protein G agarose (Roche Applied Sciences) for EGFP-tagged proteins for 4 h at 4°C. Immunoprecipitates were analyzed by Western blot with anti-CDP Ab (Santa Cruz Biotechnology) or anti-GFP Ab. Input lane shows 1% of total. In the case of purified CDPC complex, the eluates were incubated with anti-CDP Ab, anti-G9a Ab (a kind gift from Dr. Y. Nakatani, Dana–Farber Cancer Institute, Boston), or normal serum and protein G agarose.
Protein Expression and Purification of GST Fusion Proteins. GST-CDPN1, GST-CDPC, and GST-histone H3 (amino acids 1–57, WT, and mutants) were expressed in Escherichia coli BL21 strain by standard procedures. Cells were disrupted by sonication in lysis buffer (50 mM Tris·HCl, pH 7.6/120 mM NaCl/1% Triton X-100/5 mM EDTA) containing protease inhibitor mixture and centrifuged. Soluble fractions were incubated with glutathione agarose beads (Sigma) for 1 h at 4°C. For in vitro histone methyltransferase (HMT) assays, GST-histone H3 fusion proteins were eluted with elution buffer (50 mM Tris·HCl, pH 8.0/10 mM glutathione) at 4°C. The purity and the protein concentrations were verified by Coomassie blue staining from SDS/PAGE gels.
GST Pull-Down Assays. [35S]Met, Cys-labeled hG9a was prepared from the EST clone (National Center for Biotechnology Information clone ID 3912870) by using TNT rabbit reticulocyte lysate system (Promega). Equivalent amounts of GST-CDPN1 and -CDPC fusion proteins (≈0.25 μg) immobilized on glutathione agarose beads were incubated with labeled hG9a in GST pull-down buffer (50 mM Tris·HCl, pH 7.6/120 mM NaCl/0.5% Nonidet P-40/1 mM EDTA) for 1 h at 4°C. Bound proteins were resolved by SDS/PAGE and visualized by fluorography. Input lane shows 10% of total.
In Vitro HMT Assays. In vitro HMT assays were performed as described (23). Immunoprecipitates were incubated with 10 μg of histone H3 or H4 (Sigma) in methylase activity buffer containing 250 nCi (1 Ci = 37 GBq) of S-adenosyl-[methyl-14C]-l-methionine for 1 h at 37°C. For assays with GST-histone H3, reactions were performed with 20 μg of GST-histone H3 fusion proteins. Proteins were separated by SDS/PAGE and visualized by Coomassie blue staining and fluorography.
Transfection Assays. The 293T cells in six-well plates (Corning) were cotransfected with 2 μg of pcDNA3-Gal4DBD-CDPC, 0.2–1 μgof pEGFP-hG9a or pEGFP-hG9a (ΔSET), and 1 μg of pGL2-Gal4-E1b-Luc constructs. The total amount of DNA in each transfection was kept constant by addition of pcDNA3. After 48 h, cells were lysed and assayed for luciferase activity by using the Bright-Glo luciferase assay system (Promega). Each transfection was performed in duplicate and repeated five times.
Chromatin Immunoprecipitation (ChIP) Assays. ChIP assays were primarily performed according to the manufacturer's protocol (Upstate Biotechnology, Lake Placid, NY). The chromatin (sonicated to an average DNA size of ≈500 bp) of 20 × 106 transfected 293T cells was used for each immunoprecipitation with preblocked M2 agarose, anti-GFP Ab, anti-dimethyl H3-K9 Ab (Upstate Biotechnology), or normal serum. Immunoprecipitated DNA was analyzed by PCR by using specific primers to the p21pro (5′-GCTGGGCAGCCAGGAGCCTG-3′ and 5′-CTGCTCACACCTCAGCTGGC-3′, 148 bp) and the upstream region (p21up) (5′-ACCTTTGACAGTGGTGGTATCTCC-3′ and 5′-TCCTGGCTCTAACAACATCCCC-3′, 205 bp). PCR conditions were determined to ensure that results were within the linear range of the PCR.
RNA Analysis. Total RNA was isolated from transfected IMR90 cells and used for RT-PCR as described (11). Primers used for RT-PCR were 5′-ACAGATTTCTACCACTCCAAACGC-3′ and 5′-TTAGGAACCTCTCATTCAACCGC-3′ (269 bp).
Purification of CDPC Complex and MS. The 293 Epstein–Barr virus-encoded nuclear antigen (EBNA) cells grown in DMEM with 10% FBS were transfected with a (His)6-Xpress-CDPC expression vector, and stably transfected cells were maintained in the selection medium containing 400 μg/ml hygromycin. Nuclear extracts were prepared from 2 × 109 293 EBNA cells by standard procedures, and ≈4 mg of nuclear extracts was subjected to affinity purification with ProBond resin (Invitrogen), followed by immunoprecipitation with anti-Xpress Ab (Invitrogen) under native conditions according to manufacturer's protocol. Eluates were concentrated with centricon-10 membranes (Amicon), resolved by SDS/PAGE, and visualized by silver staining. Protein bands were excised from SDS/PAGE gels, digested with trypsin, and analyzed by matrix-assisted laser desorption ionization-time-of-flight MS. Abs used for Western blot analysis of eluted materials were anti-CDP, anti-TAFII100 (Santa Cruz Biotechnology), anti-Sp1 (Santa Cruz Biotechnology), and anti-G9a Abs.
Confocal Microscopy. The 293T cells were plated on glass coverslips in 35-mm dishes and transfected with an expression vector for FLAG-hG9a. After 30 h, cells were fixed with 4% paraformaldehyde in PBS for 45 min and permeabilized with 0.2% Triton X-100 in PBS for 5 min. Samples were blocked with 3% BSA in PBS containing 0.1% Tween 20 for 30 min, incubated with mouse monoclonal anti-FLAG M2 Ab (Sigma) and goat polyclonal anti-CDP Ab for 1 h at 37°C, washed, and then incubated with secondary FITC-conjugated anti-mouse Ab (Pierce) and Rhodamine-conjugated anti-goat Ab (Pierce) for 1 h. Coverslips were washed and mounted onto glass slides with Fluoromount-G (Fisher Scientific).
Results
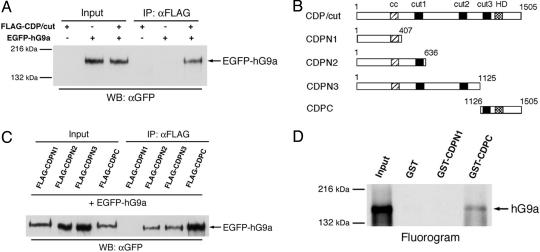
CDP/cut Interacts with hG9a in Vivo and in Vitro. To establish whether CDP/cut associates with G9a in vivo, 293T cells were transfected with expression vectors for FLAG-tagged full-length human CDP/cut and/or EGFP-fused with full-length human G9a. Whole-cell lysates were immunoprecipitated with anti-FLAG Ab and the immunoprecipitates were analyzed for the presence of EGFP-hG9a by Western blot with anti-GFP Ab. Interaction between FLAG-CDP/cut and EGFP-hG9a was detected when FLAG-CDP/cut and EGFP-hG9a, but neither FLAG-CDP/cut alone nor EGFP-hG9a alone, were coexpressed, indicating that hG9a associates with CDP/cut in immunoprecipitates from cotransfected cells (Fig. 1A).
Fig. 1.
CDP/cut interacts with hG9a in vivo and in vitro.(A) hG9a coimmunoprecipitates with FLAG-tagged CDP/cut from cell extracts of transfected 293T cells. Cells were transfected with FLAG-CDP/cut and/or EGFP-hG9a expression vectors as indicated. Whole-cell extracts were immunoprecipitated with anti-FLAG Ab, and immunoprecipitates were analyzed by Western blot with anti-GFP Ab. (B) Schematic representation of CDP/cut with evolutionarily conserved domains (cc, coiled coil; cut, cut repeat; HD, homeodomain) and truncated derivatives. (C) Association with hG9a is disrupted by C-terminal truncation in CDP/cut. Immunoprecipitates from transfected cells with expression vectors for EGFP-hG9a and FLAG-CDP/cut truncated derivatives were analyzed as in A.(D) GST-CDPC (C-terminal region of CDP/cut) fusion protein interacts with in vitro-translated hG9a. GST pull-down assays were performed with in vitro-translated [35S]hG9a and GST fusion proteins as indicated. Input lane shows 10% of total.
To map the region in CDP/cut responsible for the interaction with hG9a, FLAG-tagged CDP/cut derivatives and EGFP-hG9a were coexpressed in 293T cells and the immunoprecipitates with anti-FLAG Ab were probed with anti-GFP Ab. Deletion of the N-terminal region, generating CDPC (amino acids 1,126–1,505), had no effect on the interaction with hG9a (Fig. 1 B and C). Thus, the C-terminal region of CDP/cut that contains the cut3 repeat and the homeodomain is sufficient for effective interaction with hG9a. Interestingly, the N-terminal derivative CDPN3, in which the C-terminal region corresponding to the CDPC fragment is deleted, showed the interaction with hG9a, and another N-terminal derivative CDPN2 that contains the cut1 repeat, retained the ability to interact with hG9a. However, further truncation, generating CDPN1, in which all three cut repeats are deleted, but the coiled-coil domain remains, resulted in abrogation of the interaction with hG9a. Both CDPN2 and CDPN3 exhibited the weak interaction with hG9a as hG9a coimmunoprecipitated less effectively, compared to CDPC. These results indicate that cut repeat(s) mediate the interaction with hG9a, whereas the coiled-coil domain is not responsible for this interaction. The immunoprecipitation experiments demonstrated that the C-terminal region of CDP/cut associates with hG9a in vivo.
Next, GST pull-down assays were performed to demonstrate the direct interaction between CDP/cut and hG9a in vitro. GST-CDPN1 and GST-CDPC fusion proteins were incubated with in vitro translated 35S-labeled hG9a. GST-CDPC could associate with hG9a, whereas GST-CDPN1 failed (Fig. 1D). Thus, it was demonstrated that the C-terminal region of CDP/cut is involved in the interaction with hG9a in vivo and in vitro.
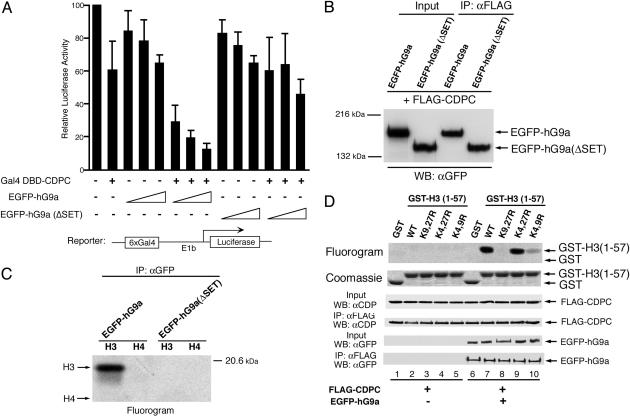
CDP/cut Represses Transcription Through HKMT Activity of hG9a. G9a has been suggested to be involved in transcriptional repression through its HKMT activity (32). It raises the possibility that G9a may be involved in CDP/cut-mediated transcriptional regulation. We next investigated whether hG9a functions as a transcriptional corepressor with CDP/cut through its HKMT activity. In our reporter gene assays, a Gal4-driven luciferase reporter together with combinations of expression vectors for Gal4DBD-CDPC, EGFP-hG9a, and SET domain deletion mutant EGFP-hG9a (ΔSET) were transfected into 293T cells. EGFP-hG9a significantly repressed the transcription of the reporter gene in a dose-dependent manner only in the presence of Gal4DBD-CDPC (Fig. 2A). However, when the methylase domain was deleted, the resulting EGFP-hG9a (ΔSET) did not repress transcription effectively compared to WT EGFP-hG9a in cooperation with Gal4DBD-CDPC. Because the interaction between CDPC and hG9a does not require the SET domain of hG9a (Fig. 2B), we conclude that transcriptional repression is mediated through HMT activity of hG9a. This was verified by in vitro HMT assays with WT hG9a and hG9a (ΔSET) immunoprecipitated from transfected cells (Fig. 2C).
Fig. 2.
CDP/cut represses transcription through HKMT activity of hG9a. (A) hG9a corepresses transcription with CDP/cut. The 293T cells were transfected with a Gal4-driven luciferase reporter with combinations of the expression vector for Gal4DBD-CDPC and increasing amounts of the EGFP-hG9a or EGFP-hG9a (ΔSET) expression vector. Luciferase activity was measured 48 h after transfection. The means ± SD of duplicate determinations from five separate experiments are shown. (B) hG9a (ΔSET) interacts with CDPC. The 293T cells were transfected with FLAG-CDPC and EGFP-hG9a or EGFP-hG9a (ΔSET) expression vectors. Whole-cell extracts were immunoprecipitated with anti-FLAG Ab, and immunoprecipitates were analyzed by Western blot with anti-GFP Ab. (C) Immunoprecipitated EGFP-hG9a possesses histone H3 methyltransferase activity, but EGFP-hG9a (ΔSET) does not. The 293T cells were transfected with EGFP-hG9a or EGFP-hG9a (ΔSET) expression vector, and the whole-cell extracts were immunoprecipitated with anti-GFP Ab. Immunoprecipitates were assayed for in vitro HMT activity by using histone H3 and H4 as substrates. (D) CDP/cut-mediated repression is associated with H3-K9 and H3-K27 methyltransferase activity by G9a. The 293T cells were transfected, and the whole-cell extracts were immunoprecipitated as in B. Immunoprecipitates were assayed for in vitro HMT activity by using GST-histone H3 (amino acids 1–57) fusion proteins (WT and mutants) as substrates. Also shown are Coomassie blue staining of purified GST-histone H3 fusion proteins introduced into assays as well as Western blots with Abs against CDP and GFP of inputs and immunoprecipitates with anti-FLAG Ab.
G9a has been reported to specifically methylate Lys-9 and Lys-27 of histone H3 (31). To confirm that hG9a retains the specific HKMT activity with CDPC, in vitro HMT assays were performed. FLAG-CDPC alone or FLAG-CDPC and EGFP-hG9a were expressed in 293T cells. The cell lysates were immunoprecipitated with anti-FLAG Ab, and the immunoprecipitates were introduced into in vitro HMT assays by using GST-histone H3 (amino acids 1–57) fusion proteins with or without double lysine mutations as substrates. Among substrates, WT (Fig. 2D, lane 7), K4, 27R in which Lys-4 and Lys-27 are replaced with Arg (lane 9), and K4, 9R in which Lys-4 and Lys-9 are replaced with Arg (lane 10) were methylated by CDPC-associated hG9a, whereas K9, 27R in which Lys-9 and Lys-27 are mutated (lane 8) was not methylated. Therefore, the interaction of hG9a with CDPC retained the specific HKMT activity for Lys-9 and Lys-27, but not for Lys-4 of histone H3, which is consistent with previously reported results of in vitro HKMT assays by using immunoprecipitated hG9a (31). Endogenous HKMT activity associated with CDPC was not detected from transfected cells with the FLAG-CDPC expression vector (lanes 1–5). Taken together, these results indicate that hG9a functions as a transcriptional corepressor through its HKMT activity when brought to a promoter region through the interaction with a DNA-binding transcription factor.
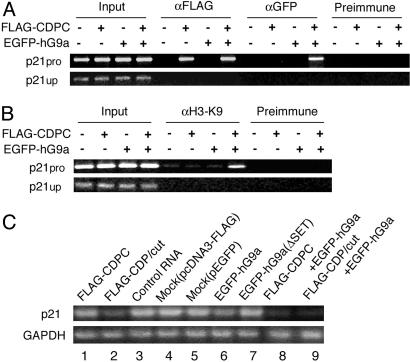
CDP/cut Recruits hG9a to Repress p21waf1/cdi1. The finding that hG9a functions as a corepressor through the interaction with CDPC suggests that CDP/cut recruits hG9a to a CDP/cut-regulated promoter. To test this hypothesis, ChIP analysis of the cyclin-dependent kinase inhibitor p21pro, which is a natural target for CDP/cut-mediated regulation, was performed. Cross-linked chromatin fragments from 293T cells that were transfected with combinations of expression vectors for FLAG-CDPC and EGFP-hG9a were immunoprecipitated with anti-FLAG Ab or anti-GFP Ab. The p21pro was associated with FLAG-CDPC in transfected cells with and without EGFP-hG9a (Fig. 3A). When EGFP-hG9a alone was expressed, the p21pro was not associated with EGFP-hG9a, whereas when FLAG-CDPC and EGFP-hG9a were coexpressed, the promoter region was associated with EGFP-hG9a. In contrast, a distal region ≈4 kb upstream of the p21waf1/cdi1 transcription initiation site (p21up) was not associated with FLAG-CDPC or EGFP-hG9a.
Fig. 3.
CDP/cut recruits hG9a to repress p21waf1/cdi1. (A) hG9a recruitment to the p21pro is CDP/cut dependent. ChIPs from transfected 293T cells with combinations of FLAG-CDPC and EGFP-hG9a expression vectors as indicated were performed with anti-FLAG Ab, anti-GFP Ab, or normal serum. Immunoprecipitates were analyzed for the presence of the p21pro and the upstream region (p21up) by PCR amplification. Results are representative of three independent experiments. (B) hG9a recruitment elevates dimethylated H3-Lys-9 in the p21pro. ChIPs were performed as in A except that experiments were carried out with anti-dimethyl H3-Lys-9 Ab. (C) p21waf1/cdi1 expression is repressed by CDP/cut mediated through hG9a. Total RNA was isolated from human diploid fibroblast IMR90 cells that were transfected with expression vectors as indicated and analyzed for the expression of p21waf1/cdi1 by RT-PCR. Results are representative of three independent experiments.
We next investigated whether the level of methylated histone H3-Lys-9 is changed in this promoter region by using Ab specific for H3 dimethylated at Lys-9 (α H3-K9). When FLAG-CDPC and EGFP-hG9a were coexpressed, histone H3-Lys-9 methylation was significantly increased (Fig. 3B). The histone H3-Lys-9 methylation was not detected in the p21waf1/cdi1 upstream region. These results indicate that hG9a is directed to the p21pro through interaction with CDP/cut and the recruitment of hG9a to the p21pro is associated with a substantial increase in methylation of Lys-9 in histone H3.
To determine whether CDP/cut cooperates with hG9a to repress endogenous p21waf1/cdi1, we performed RT-PCR analysis by using IMR90 cells that were transfected with combinations of expression vectors for FLAG-CDPC/-CDP/cut and EGFP-hG9a/-hG9a (ΔSET). FLAG-CDPC, FLAG-CDP/cut, and EGFP-hG9a were able to reduce p21waf1/cdi1 expression individually, but EGFP-hG9a (ΔSET) was not (Fig. 3C, lanes 1–7). When EGFP-hG9a and FLAG-CDPC or FLAG-CDP/cut were coexpressed, endogenous p21waf1/cdi1 was significantly reduced (lanes 8 and 9). Thus, CDP/cut recruits hG9a to repress p21waf1/cdi1.
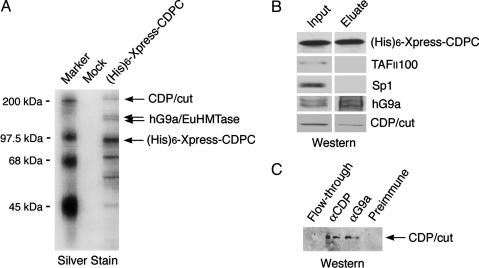
hG9a Is a Component of the CDP/cut Complex. To substantiate the physiological relevance of the interaction between CDP/cut and hG9a, a (His)6-Xpress-tagged CDPC complex was purified from nuclear extract of 293 EBNA cells that stably express (His)6-Xpress-CDPC by using nickel-chelating resin, followed by immunoprecipitation with anti-Xpress Ab. Eluates were resolved by SDS/PAGE, and the CDPC-associated proteins were identified by MS. In this purification, hG9a and its related protein EuHMTase (34) were identified in addition to full-length CDP/cut (Fig. 4A). Specific association of endogenous hG9a with CDPC complex was confirmed by Western blot analysis. hG9a was detected in the CDP/cut complex, whereas TAFII100 and Sp 1 were not detected (Fig. 4B). Eluates were further subjected to immunoprecipitation with anti-CDP Ab or anti-G9a Ab and Western blot analysis with anti-CDP Ab. CDP/cut coimmunoprecipitated with hG9a (Fig. 4C), indicating that endogenous hG9a associates with endogenous CDP/cut. Taken together, these results indicate that hG9a is a component of the CDP/cut complex.
Fig. 4.
hG9a is a component of CDP/cut complex. (A) Endogenous hG9a is a component of purified CDP/cut complex. (His)6-Xpress-CDPC complex was purified from 293 EBNA cells stably expressing (His)6-Xpress-CDPC by using Ni+-agarose, followed by immunoprecipitation with anti-Xpress Ab, and analyzed by SDS/PAGE and silver staining. Identities of protein bands were determined by MS. As a control, mock purification was performed from 293 EBNA cells generated with an empty (His)6-Xpress-tagged protein expression vector. (B) Endogenous hG9a specifically associates with the CDP/cut complex. The eluates of (His)6-Xpress-CDPC complex in A were analyzed by Western blots with Abs against CDP, TAFII100, Sp1, or G9a. (C) Endogenous hG9a associates with endogenous CDP/cut. The eluates of (His)6-Xpress-CDPC complex in A were immunoprecipitated with anti-CDP Ab, anti-G9a Ab, or normal serum, and the immunoprecipitates were analyzed by Western blot with anti-CDP Ab.
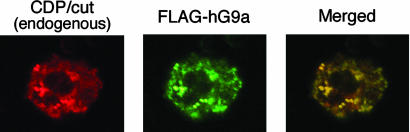
hG9a Colocalizes with CDP/cut in the Nucleus. We examined the nuclear localization of hG9a and CDP/cut by using confocal microscopy. The 293T cells were transfected with an expression vector for FLAG-hG9a and ectopically expressed FLAG-hG9a was detected by immunofluorescent labeling with anti-FLAG Ab in combination with FITC-conjugated secondary Ab. Endogenous CDP/cut was detected with anti-CDP Ab in combination with Rhodamine-conjugated secondary Ab. In most cells examined, both endogenous CDP/cut (red) and FLAG-hG9a (green) were clearly localized in nucleus and they showed a similar punctate distribution (Fig. 5). The nuclear foci of FLAG-hG9a were observed as well as endogenous CDP/cut. The merged image showed significant overlap between endogenous CDP/cut and FLAG-hG9a, and a good correspondence between foci enriched in endogenous CDP/cut and FLAG-hG9a was observed. These results indicate that hG9a colocalizes with endogenous CDP/cut in the nucleus.
Fig. 5.
hG9a colocalizes with CDP/cut in the nucleus. The 293T cells were transfected with an expression vector for FLAG-hG9a and double-stained with anti-FLAG and anti-CDP Abs. FLAG-hG9a was detected with FITC-conjugated secondary Ab, and endogenous CDP/cut was detected with Rhodamine-conjugated secondary Ab. Representative red immunofluorescent image for endogenous CDP/cut (Left), green immunofluorescent image for FLAG-hG9a (Center), and merged image (Right) are shown. The yellow dots in the merged images indicate colocalizaion of hG9a and CDP/cut.
Discussion
We present evidence that the HKMT hG9a functions as a transcriptional corepressor for a DNA-binding transcription factor. We established that hG9a interacts with the transcription factor CDP/cut and hG9a is recruited to the p21pro through the interaction with CDP/cut to function as a transcriptional corepressor requiring HKMT activity. Furthermore, we identified hG9a as a component of CDP/cut complex. Our studies provide evidence for a unique mechanism of CDP/cut transcriptional repression: recruitment of HKMT activity to the promoter.
We observed that both CDPN2 that is the N-terminal fragment containing the cut1 repeat and CDPN3 containing the cut1 and cut2 repeat domains coimmunoprecipitated with hG9a, but less effectively than CDPC, which is the C-terminal fragment containing the cut3 repeat and the homeodomain (Fig. 1C). CDPN1 that does not include any cut repeat did not coimmunoprecipitate with hG9a. These results indicate that (i) cut repeat(s) mediate the interaction with hG9a, and (ii) CDPN2 and CDPN3 exhibited the interaction with hG9a due to the presence of cut repeat(s) (cut1 repeat in CDPN2 and cut1/2 repeats in CDPN3) as three cut repeats (≈70 aa) are closely related to each other. We have previously shown that the C-terminal region encoding the cut3 domain through the end of CDP/cut interacts with HDAC1, whereas the truncation of the C terminus, generating the fragment that encodes the cut3 repeat and the homeodomain, impairs the ability to interact with HDAC1 (11). Therefore, it is likely that the C terminus downstream of the homeodomain is crucial for HDAC1 association. In contrast, we observed that the C terminus truncated fragments such as CDPN2 and CDPN3 retained the ability to interact with hG9a to some extent. This difference in the region responsible for the interaction with hG9a from that for HDAC1 interaction suggests that HDACs and HKMTs may have additive or synergetic effects on CDP/cut gene repression. We demonstrated that the C-terminal region (CDPC) overlapping the cut3 repeat through the carboxyl end brings hG9a HKMT activity to the CDP/cut-regulated p21pro to repress the p21waf1/cdi1 gene. Furthermore, we identified hG9a in the purified CDPC complex. Taken together, the C-terminal region of CDP/cut encoding the cut3 repeat through the carboxyl end has a role in transcriptional repression in cooperation with hG9a.
Su(var)39H1 is enriched at heterochromatin region (35), however, Su(var)39H1 has been suggested to be involved in repression of euchromatic genes as well as heterochromatic silencing (30, 36). In contrast, G9a localization is mostly excluded from pericentric heterochromatin region in the nucleus (31). Methylated H3-Lys-9 is abolished mostly in euchromatin region in G9a deficient cells (32). These reports suggest that the role of G9a appears to be restricted to euchromatic gene regulation. The results presented here provide evidence that G9a plays a role in repression of specific euchromatic genes as a transcriptional corepressor in complex with CDP/cut. We have observed the interaction between CDP/cut and Su(var)39H1 (data not shown), suggesting that CDP/cut may function for gene silencing in heterochromatin region in cooperation with Su(var)39H1 as well as for transcriptional regulation in euchromatin region with G9a. It is plausible to consider that posttranslational modifications of CDP/cut may affect the ability of CDP/cut to recognize and associate with either G9a or Su(var)39H1 and facilitate gene regulation and/or chromatin organization in euchromatin or heterochromatin regions through CDP/cut-dependent chromatin modification, depending on the specific HKMT associated with CDP/cut.
It has been reported that CDP/cut binds to matrix attachment regions and also interacts with its recognition motif within a nucleosomal context without displacing the nucleosome core histones (13, 14, 37). These reports suggest that CDP/cut may function as an architectural protein to organize chromatin structure. Thus, the role of CDP/cut may not be restricted to transcriptional regulation at local chromatin level, but may be extended to higher order chromatin structure organization, both of which are mediated at least in part through histone modifications by enzymes targeted to complex with CDP/cut. The ability of CDP/cut to associate with different histone modifying enzymes indicates that CDP/cut may play multiple roles in regulating gene transcription and organizing chromatin structure.
Acknowledgments
We thank Dr. E. Neufeld (University of California, Los Angeles) for the pMT2-CDP, Dr. Y. Shinkai (Kyoto University, Kyoto) for the GST-histone tail plasmids, and Dr. Y. Nakatani for the anti-G9a Ab. This work was supported by National Institutes of Health Public Health Service Award HL67099 (to M.J.W.) and a grant from the Cystic Fibrosis Foundation (to M.J.W.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CDP/cut, CCAAT displacement protein/cut homolog; HKMT, histone lysine methyltransferase; HDAC, histone deacetylase; ChIP, chromatin immunoprecipitation; EGFP, enhanced GFP; p21pro, p21waf1/cdi1 promoter; HMT, histone methyltransferase; SET, Su(var), Enhancer of Zeste, Trithorax; EBNA, Epstein–Barr virus-encoded nuclear antigen.
References
- 1.Skalnik, D. G., Strauss, E. C. & Orkin, S. H. (1991) J. Biol. Chem. 266, 16736–16744. [PubMed] [Google Scholar]
- 2.van Wijnen, A. J., Choi, T. K., Owen, T. A., Wright, K. L., Lian, J. B., Jaenisch, R., Stein, J. L. & Stein, G. S. (1991) Proc. Natl. Acad. Sci. USA 88, 2573–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Wijnen, A. J., Cooper, C., Odgren, P., Aziz, F., De Luca, A., Shakoori, R. A., Giordano, A., Quesenberry, P. J., Lian, J. B., Stein, G. S., et al. (1997) J. Cell. Biochem. 66, 512–523. [DOI] [PubMed] [Google Scholar]
- 4.Coqueret, O., Berube, G. & Nepveu, A. (1998) EMBO J. 17, 4680–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Gurp, M. F., Pratap, J., Luong, M., Javed, A., Hoffmann, H., Giordano, A., Stein, J. L., Neufeld, E. J., Lian, J. B., Stein, G. S., et al. (1999) Cancer Res. 59, 5980–5988. [PubMed] [Google Scholar]
- 6.Moon, N. S., Premdas, P., Truscott, M., Leduy, L., Berube, G. & Nepveu, A. (2001) Mol. Cell. Biol. 21, 6332–6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goulet, B., Watson, P., Poirier, M., Leduy, L., Berube, G., Meterissian, S., Jolicoeur, P. & Nepveu, A. (2002) Cancer Res. 62, 6625–6633. [PubMed] [Google Scholar]
- 8.Dufort, D. & Nepveu, A. (1994) Mol. Cell. Biol. 14, 4251–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.el-Hodiri, H. M. & Perry, M. (1995) Mol. Cell. Biol. 15, 3587–3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Last, T. J., Birnbaum, M., van Wijnen, A. J., Stein, G. S. & Stein, J. L. (1998) Gene 221, 267–277. [DOI] [PubMed] [Google Scholar]
- 11.Li, S., Moy, L., Pittman, N., Shue, G., Aufiero, B., Neufeld, E. J., LeLeiko, N. S. & Walsh, M. J. (1999) J. Biol. Chem. 274, 7803–7815. [DOI] [PubMed] [Google Scholar]
- 12.Truscott, M., Raynal, L., Premdas, P., Goulet, B., Leduy, L., Berube, G. & Nepveu, A. (2003) Mol. Cell. Biol. 23, 3013–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banan, M., Rojas, I. C., Lee, W. H., King, H. L., Harriss, J. V., Kobayashi, R., Webb, C. F. & Gottlieb, P. D. (1997) J. Biol. Chem. 272, 18440–18452. [DOI] [PubMed] [Google Scholar]
- 14.Wang, Z., Goldstein, A., Zong, R.-T., Lin, D., Neufeld, E. J., Scheuermann, R. H. & Tucker, P. W. (1999) Mol. Cell. Biol. 19, 284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mailly, F., Berube, G., Harada, R., Mao, P. L., Phillips, S. & Nepveu, A. (1996) Mol. Cell. Biol. 16, 5346–5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aufiero, B., Neufeld, E. J. & Orkin, S. H. (1994) Proc. Natl. Acad. Sci. USA 91, 7757–7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moon, N. S., Berube, G. & Nepveu, A. (2000) J. Biol. Chem. 275, 31325–31334. [DOI] [PubMed] [Google Scholar]
- 18.Santaguida, M., Ding, Q., Berube, G., Truscott, M., Whyte, P. & Nepveu, A. (2001) J. Biol. Chem. 276, 45780–45790. [DOI] [PubMed] [Google Scholar]
- 19.Li, S., Aufiero, B., Schiltz, R. L. & Walsh, M. J. (2000) Proc. Natl. Acad. Sci. USA 97, 7166–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grunstein, M. (1997) Nature 389, 349–352. [DOI] [PubMed] [Google Scholar]
- 21.Wei, Y., Yu, L., Bowen, J., Gorovsky, M. A. & Allis, C. D. (1999) Cell 97, 99–109. [DOI] [PubMed] [Google Scholar]
- 22.Zhang, Y. & Reinberg, D. (2001) Genes Dev. 15, 2343–2360. [DOI] [PubMed] [Google Scholar]
- 23.Rea, S., Eisenhaver, F., O'Carroll, D., Strahl, B. D., Sun, Z. W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C. P., Allis, C. D., et al. (2000) Nature 406, 593–599. [DOI] [PubMed] [Google Scholar]
- 24.Wang, H., Cao, R., Xia, L., Erdjument-Bromage, H., Borchers, C., Tempst, P. & Zhang, Y. (2001) Mol. Cell 8, 1207–1217. [DOI] [PubMed] [Google Scholar]
- 25.Nishioka, K., Chuikov, S., Sarma, K, Erdjument-Bromage, H., Allis, C. D., Tempst, P. & Reinberg, D. (2002) Genes Dev. 16, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zegerman, P., Canas, B., Pappin, D. & Kouzarides, T. (2002) J. Biol. Chem. 277, 11621–11624. [DOI] [PubMed] [Google Scholar]
- 27.Chen, D., Ma, H., Hong, H., Koh, S. S., Huang, S. M., Schurter, B. T., Aswad, D. W. & Stallcup, M. R. (1999) Science 284, 2174–2177. [DOI] [PubMed] [Google Scholar]
- 28.Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. (2001) Nature 410, 116–120. [DOI] [PubMed] [Google Scholar]
- 29.Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C. & Kouzarides, T. (2001) Nature 410, 120–124. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen, S. J., Schneider, R., Bauer, U. M., Bannister, A. J., Morrison, A., O'Carroll, D., Firestein, R., Cleary, M., Jenuwein, T., Herrera, R., et al. (2001) Nature 412, 561–565. [DOI] [PubMed] [Google Scholar]
- 31.Tachibana, M., Sugimoto, K., Fukushima, T. & Shinkai, Y. (2001) J. Biol. Chem. 276, 25309–25317. [DOI] [PubMed] [Google Scholar]
- 32.Tachibana, M., Sugimoto, K., Nozaki, M., Ueda, J., Ohta, T., Ohki, M., Fukuda, M., Takeda, N., Niida, H., Kato, H., et al. (2002) Genes Dev. 16, 1779–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neufeld, E. J., Skalnik, D. G., Lievens, P. M. & Orkin, S. H. (1992) Nat. Genet. 1, 50–55. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa, H., Ishiguro, K., Gaubatz, S., Livingston, D. M. & Nakatani, Y. (2002) Science 296, 1132–1136. [DOI] [PubMed] [Google Scholar]
- 35.Melcher, M., Schmid, M., Aagaard, L., Selenko, P., Laible, G. & Jenuwein, T. (2000) Mol. Cell. Biol. 20, 3728–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, J., Lin, Q., Yoon, H., Huang, Z., Strahl, B. D., Allis, C. D. & Wong, J. (2002) Mol. Cell. Biol. 22, 5688–5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Last, T. J., van Wijnen, A. J., de Ridder, M. C., Stein, G. S. & Stein, J. L. (1999) Mol. Biol. Rep. 26, 185–194. [DOI] [PubMed] [Google Scholar]