Abstract
Background
The mosquito Aedes aegypti (L.) is a major vector of viral diseases like dengue fever, Zika and chikungunya. Aedes aegypti exhibits high morphological and behavioral variation, some of which is thought to be of epidemiological significance. Globally distributed domestic Ae. aegypti have often been grouped into (i) the very pale variety queenslandensis and (ii) the type form. Because the two color forms co-occur across most of their range, there is interest in understanding how freely they interbreed. This knowledge is particularly important for control strategies that rely on mating compatibilities between the release and target mosquitoes, such as Wolbachia releases and SIT. To address this question, we analyzed nuclear and mitochondrial genome-wide variation in the co-occurring pale and type Ae. aegypti from northern Queensland (Australia) and Singapore.
Methods/Findings
We typed 74 individuals at a 1170 bp-long mitochondrial sequence and at 16,569 nuclear SNPs using a customized double-digest RAD sequencing. 11/29 genotyped individuals from Singapore and 11/45 from Queensland were identified as var. queenslandensis based on the diagnostic scaling patterns. We found 24 different mitochondrial haplotypes, seven of which were shared between the two forms. Multivariate genetic clustering based on nuclear SNPs corresponded to individuals’ geographic location, not their color. Several family groups consisted of both forms and three queenslandensis individuals were Wolbachia infected, indicating previous breeding with the type form which has been used to introduce Wolbachia into Ae. aegypti populations.
Conclusion
Aedes aegypti queenslandensis are genomically indistinguishable from the type form, which points to these forms freely interbreeding at least in Australia and Singapore. Based on our findings, it is unlikely that the presence of very pale Ae. aegypti will affect the success of Aedes control programs based on Wolbachia-infected, sterile or RIDL mosquitoes.
Author Summary
Aedes aegypti, the most important vector of dengue and Zika, greatly varies in body color and behavior. Two domestic forms of this mosquito, the very pale queenslandensis and the browner type, are often found together in populations around the globe. Knowing how freely they interbreed is important for the control strategies such as releases of Wolbachia and sterile males. To address this question, we used RAD sequencing to genotype samples of both forms collected in Singapore and northern Queensland. We did not find any association between the mitochondrial or nuclear genome-wide variation and color variation in these populations. Rather, “paleness” is likely to be a quantitative trait under some environmental influence. We also detected several queenslandensis individuals with the Wolbachia infection, indicating free interbreeding with the type form which has been used to introduce Wolbachia into Ae. aegypti populations. Overall, our data show that the very pale queenslandensis are not genomically separate, and their presence is unlikely to affect the success of Aedes control programs based on Wolbachia-infected, sterile or RIDL mosquitoes.
Introduction
The mosquito Aedes aegypti (Linnaeus) is the most important arboviral vector in the tropics and subtropics [1]. Diseases transmitted by Ae. aegypti, like dengue fever and Zika, are on the rise [2], and some are reappearing. For instance, chikungunya has returned to the American tropics in 2013, after being absent for nearly 200 years [3]. Yellow fever was nearly eliminated thanks to an effective vaccine, but is now resurging in central and south Africa [4]. Such epidemiological trends highlight the need to persist with vector control efforts, which requires a thorough understanding of vector biology.
Nearly 60 years ago, Mattingly [5] noted that despite a vast body of literature, few mosquitoes have been”the subject of misconception….in the minds of the general run of entomologists” like Aedes aegypti [5]. The species has a plethora of historical synonyms [6], mainly as a result of having extensive variation in body color and scaling patterns [7] which was also thought to correlate with behavioral differences (e.g. [8]). These issues motivated Mattingly [5] to revise the taxonomy of Ae. aegypti and create a foundation for the modern studies of this disease vector.
Mattingly [5] proposed the intraspecific classification of Ae. aegypti into three forms.
A very dark form that never has pale scales on the first abdominal tergite, avoids biting humans, prefers natural breeding habitats and is confined to sub-Saharan Africa. Mattingly gave this form a subspecies rank, Ae. aegypti spp. formosus (Walker).
Ae. aegypti sensu stricto or the type form, distinctly paler and browner than spp. formosus, with pale scales restricted to the head and the first abdominal tergite. This form prefers to bite humans and to use artificial breeding containers, and is globally distributed.
A very pale form, Ae. aegypti queenslandensis (Theobold), with extension of the pale scaling on the thorax, tergites and legs, that co-occurs with the type form. Mattingly gave this form only a varietal rank (Ae. aegypti var. queenslandensis). This form was very common in the Mediterranean basin before it was eradicated from the region [9]. Aedes aegypti queenslandensis has also been considered the most domestic of the three forms, always breeding and resting very close to humans, both in and outside Africa [5, 8].
A few years later, McClelland [7] reported a high level of variation in color and scaling within and among Ae. aegypti populations, suggesting that subdivision into forms seems oversimplistic and should be abandoned unless correlation between genetic and color variation can be demonstrated. In their latest review of the Ae. aegypti history, Powell and Tabachnick [9] pointed out that McClelland’s recommendations have often been ignored for the past 45 years, despite the fact that multiple genetic marker systems (allozymes, microsatellites, nuclear and mitochondrial SNPs) have failed to find a clear differentiation between forms and markers [10–13].
Recently, Chan et al. [14] suggested that the DNA barcoding technique can be used to distinguish queenslandensis individuals from the type individuals in Singapore. The sequence divergence of 1.5%-1.9% between the two forms [14], although lower than a commonly adopted threshold of 3% for species delineation in insects [15], suggests that the two forms may not freely interbreed. Historical records indicate that the two forms have co-occurred in Singapore and other parts of south-east Asia and Australia for hundreds of generations [5, 8]. In sympatry, genetic isolation can be maintained largely through pre-zygotic isolation mechanisms like incompatibilities in mating behavior [16]. For instance, molecular forms of the malarial mosquito, Anopheles gambiae, fly together in mating swarms but rarely hybridize due to flight-tone matching between males and females of the same form [17]. Similar incompatibilities in Ae. aegypti would have implications for control strategies that rely on successful mating between the release and target mosquitoes, like Wolbachia-based population replacement and suppression [18, 19], releases of sterile males [20] or males with a RIDL construct [21].
To explore this further, we analyzed nuclear and mitochondrial genome-wide variation in the co-occurring pale and type Ae. aegypti from Singapore and northern Queensland (Australia). The RADseq approach we employed allows for detection of genetic structure and ancestry with power unparalleled by previous genetic studies of the Ae. aegypti forms [22]. Any association between genetic structuring (nuclear/mitochondrial) and the mosquito color/scaling would provide support for the hypothesis of restricted interbreeding between the type and the queenslandensis form, with implications for the implementation of biocontrol programs to suppress diseases transmitted by Ae. aegypti.
Materials and Methods
Ethics statement
The collection of wild mosquitoes in the study areas does not require specific field ethics approval. The sampling was not conducted on protected land, nor did it involve endangered or protected species. Consent was obtained from residents at each location where collections occurred on private property.
Sampling and identification
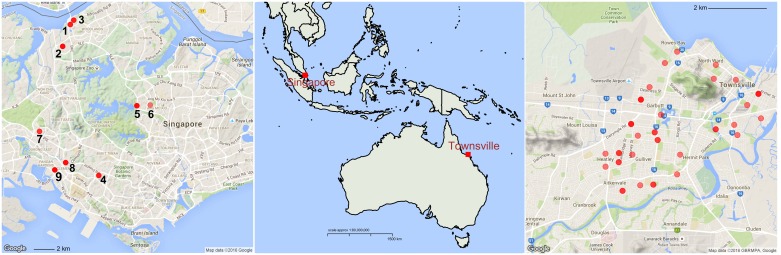
In Singapore, all samples were collected as larvae from the domestic breeding containers at nine locations during the second week of April 2015 (Fig 1, Table 1). These samples were collected during routine inspection by enforcement officers of the National Environment Agency (NEA), Singapore. Larvae were reared to the adult stage under standard laboratory conditions (25° ± 1°C, 80 ± 10% relative humidity and 12 h light/dark cycle). In Townsville (northern Queensland), samples were collected as adults using Biogents Sentinel traps placed at 55 locations in January 2014 (Fig 1, Table 1). Adult mosquitoes were sexed and identified to form based on the key diagnostic color and scaling features, following Mattingly [5] and McClleland [7]. Eleven out of 44 mosquitoes (25%) from Singapore, and seven out of 99 mosquitoes (7%) from Townsville were identified as the queenslandensis form (Table 1). An additional four queenslandensis individuals collected in Cairns (northern Queensland) in December 2014 were included in the analyses (Table 1).
Fig 1. Sampling sites.
In Singapore (left), each sampling point represents one breeding container from which larvae were collected. In Townsville (right), each sampling point represents one BG-Sentinel trap from which adults were collected.
Table 1. Sample information.
Sample ID, region (SNP—Singapore, QLD_T—Townsville, QLD_C—Cairns, Queensland), X, Y (longitude/latitude decimal degrees), collection (method/breeding container), sex (F—female, M—male), form (t–type, q—queenslandensis [5][7]), mitochondrial haplotype (mt hapl, Hap1-24), per individual proportion of heterozygous (het) nuclear loci, average (aver) locus depth, and proportion of missing (miss) loci.
| Sample ID | region | X | Y | collection site | sex | form | mt hapl | het loci | aver depth | miss loci |
|---|---|---|---|---|---|---|---|---|---|---|
| A | SNP | 103.7701 | 1.4418 | Dish tray | F | q | Hap5 | 0.195 | 32.2 | 0.03 |
| B | SNP | 103.7701 | 1.4418 | Dish tray | F | t | Hap5 | 0.149 | 30.9 | 0.03 |
| C | SNP | 103.7701 | 1.4418 | Dish tray | F | q | Hap5 | 0.201 | 33.1 | 0.02 |
| D | SNP | 103.7634 | 1.4228 | Plastic tray | F | t | Hap6 | 0.252 | 34.3 | 0.01 |
| E | SNP | 103.7634 | 1.4228 | Plastic tray | F | t | Hap24 | 0.253 | 30.6 | 0.01 |
| F | SNP | 103.7634 | 1.4228 | Plastic tray | F | t | Hap6 | 0.260 | 26.4 | 0.01 |
| G | SNP | 103.7634 | 1.4228 | Plastic tray | F | t | Hap24 | 0.244 | 32.0 | 0.01 |
| H | SNP | 103.7730 | 1.4456 | Scupper drain | F | t | Hap6 | 0.214 | 28.5 | 0.02 |
| I | SNP | 103.7730 | 1.4456 | Scupper drain | F | q | Hap11 | 0.260 | 33.8 | 0.01 |
| J | SNP | 103.7730 | 1.4456 | Scupper drain | F | t | Hap6 | 0.159 | 35.9 | 0.03 |
| K | SNP | 103.7950 | 1.3099 | Vase | F | t | Hap20 | 0.179 | 23.5 | 0.03 |
| L | SNP | 103.7950 | 1.3099 | Vase | F | t | Hap19 | 0.106 | 29.8 | 0.04 |
| M | SNP | 103.7950 | 1.3099 | Vase | F | q | Hap19 | 0.160 | 26.2 | 0.03 |
| N | SNP | 103.8282 | 1.3709 | Fish tank | F | t | Hap14 | 0.250 | 33.3 | 0.03 |
| O | SNP | 103.8282 | 1.3709 | Fish tank | F | q | Hap6 | 0.209 | 45.1 | 0.01 |
| P | SNP | 103.8282 | 1.3709 | Fish tank | F | t | Hap15 | 0.250 | 29.4 | 0.02 |
| Q | SNP | 103.8282 | 1.3709 | Fish tank | F | t | Hap6 | 0.233 | 22.2 | 0.02 |
| R | SNP | 103.8399 | 1.3714 | Gully trap | F | q | Hap12 | 0.130 | 30.2 | 0.04 |
| S | SNP | 103.7431 | 1.3484 | Flower vase | F | q | Hap6 | 0.256 | 30.6 | 0.01 |
| T | SNP | 103.7431 | 1.3484 | Flower vase | F | t | Hap6 | 0.254 | 29.1 | 0.01 |
| U | SNP | 103.7660 | 1.3211 | Corridor | F | q | Hap6 | 0.264 | 33.4 | 0.01 |
| V | SNP | 103.7660 | 1.3211 | Corridor | F | q | Hap7 | 0.260 | 28.8 | 0.01 |
| W | SNP | 103.7660 | 1.3211 | Corridor | F | q | Hap13 | 0.264 | 37.2 | 0.01 |
| X | SNP | 103.7660 | 1.3211 | Corridor | F | q | Hap6 | 0.262 | 28.2 | 0.02 |
| Y | SNP | 103.7565 | 1.3147 | Flower pot tray | F | t | Hap8 | 0.253 | 26.9 | 0.01 |
| Z | SNP | 103.7565 | 1.3147 | Flower pot tray | F | t | Hap18 | 0.238 | 25.3 | 0.02 |
| AA | SNP | 103.7565 | 1.3147 | Flower pot tray | F | t | Hap16 | 0.240 | 35.7 | 0.01 |
| BB | SNP | 103.7565 | 1.3147 | Flower pot tray | F | t | Hap17 | 0.217 | 13.1 | 0.07 |
| CC | SNP | 103.7565 | 1.3147 | Flower pot tray | F | t | Hap6 | 0.255 | 36.7 | 0.01 |
| F13 | QLD_T | 146.7746 | -19.2788 | BG-Sentinel trap | F | t | Hap22 | 0.195 | 15.9 | 0.04 |
| F19 | QLD_T | 146.7814 | -19.2990 | BG-Sentinel trap | F | t | - | 0.240 | 15.6 | 0.04 |
| F20 | QLD_T | 146.7605 | -19.2862 | BG-Sentinel trap | F | t | - | 0.149 | 14.4 | 0.05 |
| F21 | QLD_T | 146.7759 | -19.2639 | BG-Sentinel trap | F | t | - | 0.217 | 20.0 | 0.02 |
| F25 | QLD_T | 146.7833 | -19.2806 | BG-Sentinel trap | F | t | Hap9 | 0.201 | 13.8 | 0.05 |
| F27 | QLD_T | 146.7820 | -19.2774 | BG-Sentinel trap | F | t | Hap22 | 0.255 | 20.6 | 0.02 |
| F28 | QLD_T | 146.7759 | -19.2639 | BG-Sentinel trap | F | t | Hap6 | 0.252 | 12.4 | 0.06 |
| F3 | QLD_T | 146.7759 | -19.2639 | BG-Sentinel trap | F | t | Hap22 | 0.253 | 13.0 | 0.06 |
| F31 | QLD_T | 146.8167 | -19.2743 | BG-Sentinel trap | F | t | Hap23 | 0.260 | 20.4 | 0.03 |
| Mf32 | QLD_T | 146.7918 | -19.2442 | BG-Sentinel trap | M | t | Hap4 | 0.248 | 18.7 | 0.02 |
| Mf33 | QLD_T | 146.8181 | -19.2786 | BG-Sentinel trap | M | t | Hap2 | 0.228 | 16.3 | 0.03 |
| F34 | QLD_T | 146.7663 | -19.2898 | BG-Sentinel trap | F | t | Hap21 | 0.218 | 19.9 | 0.02 |
| F4 | QLD_T | 146.7679 | -19.3015 | BG-Sentinel trap | F | t | Hap22 | 0.255 | 15.1 | 0.05 |
| F5 | QLD_T | 146.8078 | -19.2554 | BG-Sentinel trap | F | t | Hap9 | 0.239 | 14.9 | 0.04 |
| F6 | QLD_T | 146.7864 | -19.2489 | BG-Sentinel trap | F | t | Hap4 | 0.248 | 19.2 | 0.02 |
| F7 | QLD_T | 146.7717 | -19.2740 | BG-Sentinel trap | F | t | Hap22 | 0.221 | 9.8 | 0.13 |
| F9 | QLD_T | 146.7756 | -19.2993 | BG-Sentinel trap | F | t | Hap9 | 0.221 | 14.7 | 0.04 |
| M23 | QLD_T | 146.7663 | -19.2898 | BG-Sentinel trap | M | t | Hap4 | 0.252 | 21.9 | 0.01 |
| M24 | QLD_T | 146.8272 | -19.2616 | BG-Sentinel trap | M | t | Hap9 | 0.260 | 22.1 | 0.01 |
| M27 | QLD_T | 146.7717 | -19.2740 | BG-Sentinel trap | M | t | Hap21 | 0.265 | 21.0 | 0.02 |
| M28 | QLD_T | 146.7717 | -19.2740 | BG-Sentinel trap | M | t | Hap4 | 0.268 | 20.4 | 0.01 |
| M29 | QLD_T | 146.7605 | -19.2916 | BG-Sentinel trap | M | t | Hap21 | 0.259 | 24.2 | 0.01 |
| M30 | QLD_T | 146.7722 | -19.2866 | BG-Sentinel trap | M | t | Hap21 | 0.285 | 22.6 | 0.01 |
| M31 | QLD_T | 146.7814 | -19.2990 | BG-Sentinel trap | M | t | Hap22 | 0.238 | 20.4 | 0.02 |
| M32 | QLD_T | 146.8223 | -19.2662 | BG-Sentinel trap | M | t | Hap21 | 0.243 | 22.0 | 0.03 |
| M33 | QLD_T | 146.8283 | -19.2717 | BG-Sentinel trap | M | t | Hap22 | 0.261 | 27.4 | 0.01 |
| M34 | QLD_T | 146.7797 | -19.2588 | BG-Sentinel trap | M | t | Hap22 | 0.268 | 22.0 | 0.01 |
| M35 | QLD_T | 146.7679 | -19.3015 | BG-Sentinel trap | M | t | Hap4 | 0.262 | 20.3 | 0.02 |
| M36 | QLD_T | 146.8174 | -19.2558 | BG-Sentinel trap | M | t | Hap4 | 0.230 | 15.9 | 0.04 |
| Fm37 | QLD_T | 146.7931 | -19.2866 | BG-Sentinel trap | F | t | Hap22 | 0.257 | 25.5 | 0.01 |
| M38 | QLD_T | 146.8087 | -19.2496 | BG-Sentinel trap | M | t | Hap3 | 0.265 | 20.4 | 0.01 |
| M39 | QLD_T | 146.7820 | -19.2774 | BG-Sentinel trap | M | t | Hap9 | 0.258 | 18.7 | 0.03 |
| M40 | QLD_T | 146.7833 | -19.2806 | BG-Sentinel trap | M | t | Hap22 | 0.227 | 11.6 | 0.08 |
| Fm45 | QLD_T | 146.7917 | -19.2947 | BG-Sentinel trap | F | t | Hap22 | 0.250 | 25.0 | 0.01 |
| Q1 | QLD_T | 146.7847 | -19.2702 | BG-Sentinel trap | F | q | Hap4 | 0.225 | 18.3 | 0.02 |
| Q2 | QLD_T | 146.7664 | -19.2860 | BG-Sentinel trap | F | q | Hap10 | 0.234 | 14.7 | 0.05 |
| Q3 | QLD_T | 146.8272 | -19.2616 | BG-Sentinel trap | F | q | Hap22 | 0.243 | 16.1 | 0.04 |
| Q4 | QLD_T | 146.8086 | -19.2762 | BG-Sentinel trap | F | q | Hap4 | 0.229 | 13.7 | 0.04 |
| Q5 | QLD_T | 146.8272 | -19.2616 | BG-Sentinel trap | F | q | Hap21 | 0.232 | 14.3 | 0.04 |
| Q6 | QLD_T | 146.8086 | -19.2762 | BG-Sentinel trap | F | q | Hap4 | 0.203 | 10.3 | 0.10 |
| Q7 | QLD_T | 146.7664 | -19.2860 | BG-Sentinel trap | F | q | Hap9 | 0.233 | 16.8 | 0.03 |
| Q8 | QLD_T | 145.7488 | -16.9389 | BG-Sentinel trap | F | q | Hap1 | 0.254 | 17.8 | 0.03 |
| Q9 | QLD_C | 145.7562 | -16.9310 | BG-Sentinel trap | F | q | Hap4 | 0.206 | 11.0 | 0.09 |
| Q10 | QLD_C | 145.7562 | -16.9310 | BG-Sentinel trap | F | q | Hap4 | 0.163 | 13.0 | 0.05 |
| Q11 | QLD_C | 145.7562 | -16.9310 | BG-Sentinel trap | F | q | Hap4 | 0.194 | 13.9 | 0.05 |
RADseq genotyping
DNA was extracted from 29 individuals collected in Singapore (18 female type, 11 female queenslandensis) and 45 individuals from northern Queensland (17 male type, 17 female type, 11 female queenslandensis) (Table 1). Qiagen Blood and Tissue DNA kit (Venlo, Limburg, NL) was used to extract DNA from a whole adult mosquito. 100 ng of DNA from each individual was used to construct the double-digest RAD library following a previously validated protocol [22]. In short, 100 units of the two frequently cutting enzymes (MluCI and NlaIII, New England Biolabs, Beverly MA, USA) were used to digest 100 ng of DNA during three hours of incubation at 37°C. 100 pM P1 and 300 pM P2 Illumina adapters with customized barcode sequences were ligated to the genomic fragments using 100 units of T4 ligase at 16°C overnight (New England Biolabs, Beverly, MA, USA). Pooled ligations were purified and size selected for fragments 300-450bp in length, using the 2% Pippin Prep cassette (Sage Sciences, Beverly, MA, USA). The final libraries (one for each geographic region) were enriched with 12 PCR cycles with standard Illumina primers and then sequenced in two HiSeq2500 lanes with the 100 bp paired-end chemistry.
Raw fastq sequences were processed within our customized pipeline. First, all reads were trimmed to the same length of 90 bp and removed if the base quality score was below 13 (FASTX Toolkit, http://hannonlab.cshl.edu/fastx_toolkit/index.html). High quality reads were then aligned to the reference mitochondrial genome [23] and the nuclear genome version AaegL1 [24] using the aligner Bowtie [25]. Uniquely aligned reads were passed to the refmap.pl program that runs the Stacks v.1.35 pipeline [26]. In addition to the samples from Singapore, Townsville and Cairns, we included previously sequenced individuals: 15 from Rio de Janeiro (Brazil) [27], 15 from Gordonvale (northern Queensland), and 15 from Ho Chi Minh City (Vietnam) (S1 Table). This was done to compare the extent of genetic structuring within and among samples at a regional and global scale. Sexing of the larval samples from Brazil and Vietnam could not be done based on the external morphological features, so we employed a genetic sexing method based on the presence/absence of the male-specific RAD tags [28]. All 119 individuals were included in the creation of the RAD tag catalogues using the default Stacks parameters in the maximum likelihood model of SNP and genotype calling. The populations module was used to filter the catalogues and export data in the FASTA format (for the mitochondrial variation) and the variant calling format (VCF, for the nuclear variation).
Analyses of genetic diversity and structure
The mitochondrial haplotype richness within and among groups (Ae. aegypti forms and geographic regions) was calculated using the rarefaction method implemented in the program HP-rare [29]. Phylogenetic relationship among mitochondrial haplotypes was estimated with the maximum likelihood approach in the program RAxML (GTRM + G, rapid bootstrap heuristic algorithm and thorough ML search) [30]. Haplotypes of three related Aedes species, for which the whole mitochondrial genome sequences were available, served as outgroups: Ae. albopictus (NCBI: NC_006817.1), Ae. notoscriptus (NC_025473.1) [31] and Ae. vigilax (KP995260.1) [32]. Haplotype sequence of the Ae. aegypti reference line (Liverpool, NC_010241.1) was also included in the analysis.
Parameters of data quality and diversity (RAD tag depth, percentage of missing data, heterozygosity averaged per individual) were compared between females of the two co-occurring forms using independent sample t-test. The level of nuclear genetic structuring was estimated using the non-spatial multivariate method DAPC [33] in the R package adegenet [34]. Rousset’s genetic distance (â) and geographic distance between pairs of individuals were calculated in the program spagedi [35]. Color distance between pairs of individuals was treated as a binary value: 0 (same color/form) and 1 (different color/form).
Results & Discussion
Variation and phylogenetic relationship among mitochondrial haplotypes
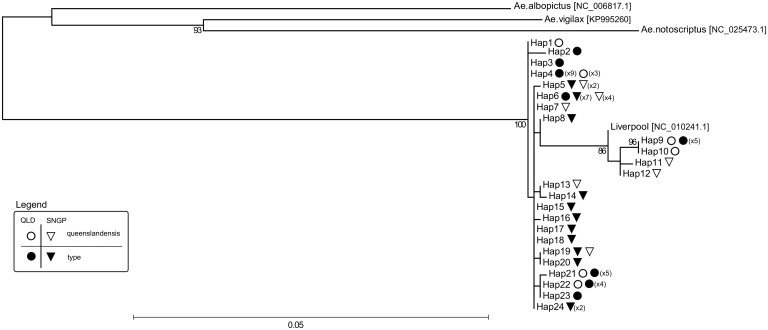
From the mitochondrial RAD tag catalogue, we extracted 13 polymorphic tags that were shared between at least 80% of individuals (60/74, Table 1). Tags were distributed across eight different mitochondrial genes (COXI, Cytb, ATP6, ND1-2, ND4-6; S1 File). All 13 tags were concatenated into a final 1170 bp long sequence that was treated as a mitochondrial haplotype. We found 24 different haplotypes in samples from Singapore and Townsville. Haplotype richness did not differ between the two forms in either location (Singapore type = 5.13, queenslandensis = 5.07; Townsville type = 4.19, queenslandensis = 5.0). Moreover, seven haplotypes were shared between the two forms (Table 1, Fig 2).
Fig 2. Mitochondrial Maximum likelihood phylogeny.
Twenty-four different mitochondrial haplotypes (Hap1-24) found in Aedes aegypti type and var. queenslandensis that co-occur in Singapore and northern Queensland, Australia. Sequences of the three outgroups (Ae. albopictus, Ae. vigilax, Ae. notoscriptus) and Ae. aegypti Liverpool strain were obtained from the NCBI nucleotide sequence/genome database, with the NCBI accession numbers listed in square brackets. The number of Ae. aegypti individuals with a given mitochondrial haplotype is listed in parentheses. A circle designates haplotypes found in Queensland, and a triangle those found in Singapore. Open symbols designate haplotypes found in the queenslandensis form, and filled symbols those found in the type form.
There were 207 distinctive alignment patterns and 8.17% of undetermined characters in the dataset consisting of 24 haplotypes from Singapore and Queensland, one from the Liverpool strain and three from other Aedes species (outgroups). A phylogeny based on maximum likelihood revealed two highly statistically supported maternal lineages in Ae. aegypti: a basal clade (more similar to the outgroups) and a clade arising from it (a derived clade) (Fig 2). Nucleotide distance (p-distance) between the two clades ranged from 1.2% to 1.6% (S2 File). Importantly, haplotypes of the two Ae. aegypti forms were found in both clades, indicating no association between mitochondrial variation and color variation (Fig 2).
While our results do not support the tentative patterns suggested by Chan et al. [14], they match those from the most comprehensive mitochondrial phylogeny of the African and global Ae. aegypti generated to date [10]. Using the ND4 variation, Moore et al. [10] showed that Ae. aegypti populations outside Africa represent “mixtures” of mosquitoes from the basal clade and the derived clade, with the basal clade likely originating from West Africa and the derived clade mainly from East Africa. Our analyses of the mitochondrial genome-wide variation revealed the same matrilineage structure in populations from Singapore and northern Queensland (Fig 2). A lack of mitochondrial distinctiveness between the queenslandensis and the type form is also in line with the findings of Moore et al. [10], who could not separate the type and formosus forms into distinct mitochondrial clades despite their assumed subspecies rank.
Nuclear genetic structuring
We extracted nuclear RAD tags that were shared between at least 80% of individuals in the entire dataset (Singapore, Townsville, Gordonvale, Ho Chi Minh City and Rio de Janeiro). To avoid redundant information from the highly linked markers, we randomly selected one SNP per tag with a minor allele frequency greater than 5%, which gave a total of 16,569 markers for downstream analyses.
Parameters of data quality and diversity did not significantly differ between the co-occurring queenslandensis and type individuals, including the average: percentage of reads uniquely aligned to the reference genome (Singapore: t df,27 = 1.46, p = 0.15; Townsville: t df,26 = 0.782, p = 0.44), locus depth (Singapore: t df,27 = 1.66, p = 0.11; Townsville: t df,26 = -1.73, p = 0.095), percentage of missing data (Singapore: t df,27 = -0.67, p = 0.51; Townsville: t df,26 = 0.951, p = 0.35), or heterozygosity (Singapore: t df,27 = 0.46, p = 0.65; Townsville: t df,26 = -2.42, p = 0.023) (Table 1, S1 Fig).
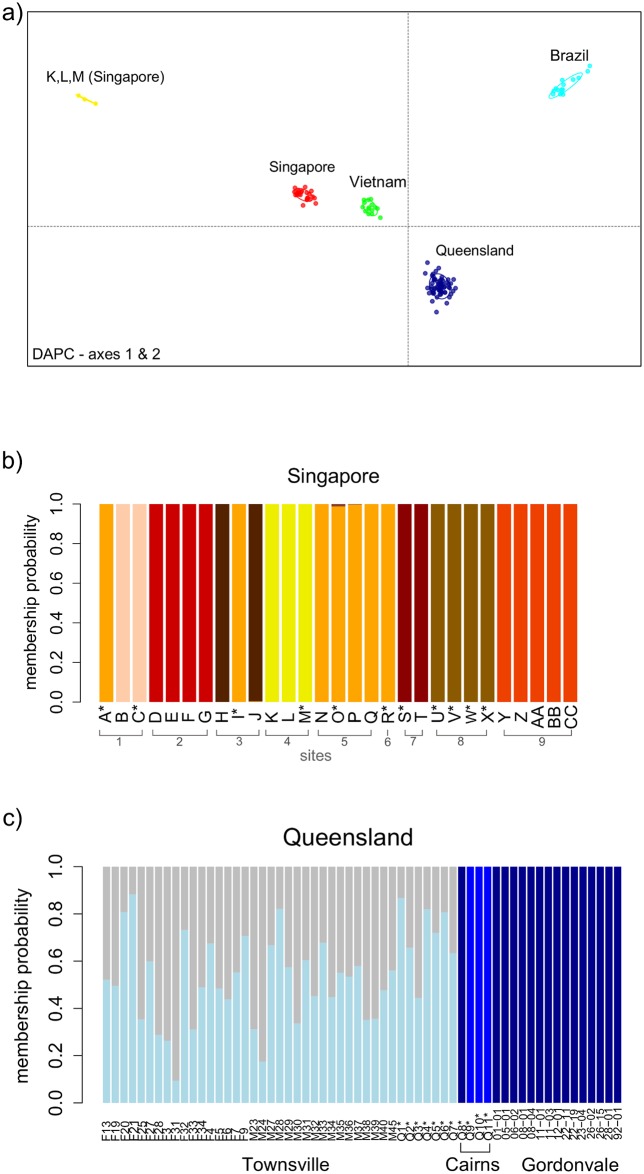
Discriminant analysis of principal components (DAPC) showed a clear-cut differentiation of mosquitoes based on their geographic origin and not their color. When the entire dataset was considered, Ae. aegypti individuals formed genetic clusters that corresponded to their sampling region (i.e. Rio de Janeiro, Ho Chi Minh City, Singapore and northern Queensland) (Fig 3a). The only exceptions were three individuals in Singapore (K-M) that formed a distinct genetic group (Fig 3a). They were collected as larvae from the same breeding container, and two were identified as the type form and one as the queenslandensis form (Table 1, Fig 3b). Given their high relatedness (Supplemental file 4) and shared mitochondrial haplotype, as well as high nuclear differentiation from other mosquitoes in the region, it is likely that individuals K, L and M are offspring of an incursion female(s) not local to Australia and Vietnam. These individuals were found near the city port, suggesting a possible route of introduction.
Fig 3. Nuclear genetic structuring (DAPC).
Individuals marked with an asterisk (*) in their sample ID were identified as Aedes aegypti queenslandensis based on diagnostic scaling patterns [7]. (a) Scatterplot summarizing the individual DAPC scores (axes 1 and 2) in Aedes aegypti samples collected in Singapore, Queensland, Ho Chi Minh City (Vietnam) and Rio de Janeiro (Brazil); (b) Individual membership probability to genetic groups in Singapore; (c) Individual membership probability to genetic groups in northern Queensland.
Further analysis of genetic structuring within Singapore revealed that family groups were sampled within the breeding containers, some of which had both color forms (Fig 3b). Highly related queenslandensis and type pairs were found at four locations (Fig 3b), including the incursion family group (K,L,M). Most of the related individuals (24/28 pairs), however, had the same color (Fig 4). These results suggest that the color/scaling pattern is likely to represent a quantitative trait under some environmental influence (e.g. temperature, humidity, light, nutrient availability). The frequency of the color forms has been shown to vary between the dry and the wet season in Ae. aegypti populations from Surabaya, Indonesia [36]. Also, the dorsal abdominal scaling pattern responds to artificial selection [36, 37] and multiple QTLs associated with this trait have been recently reported [37]. Individuals with the color/scalling patterns corresponding to the queenslandensis form have also been observed (albeit rarely) in our laboratory colonies which are maintained by occasional crossing to field-caught type Ae. aegypti (Jason Axford, personal communication).
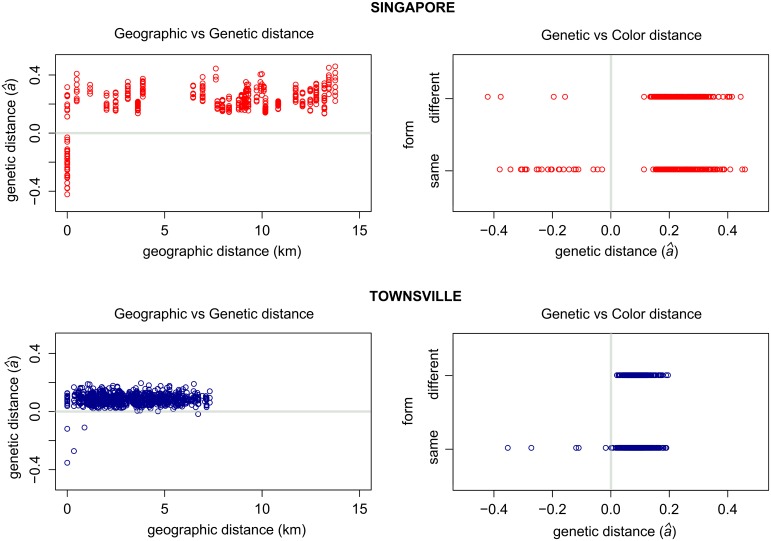
Fig 4. Pairwise genetic versus geographic and color distance.
Pairs of Aedes aegypti collected in Singapore (upper graphs) and Townsville (lower graphs). A value of zero for Rousset’s genetic distance (â) indicates a distance between a pair of individuals randomly drawn from a given sample, while a negative value indicates lower than average genetic distance between a pair (i.e. their higher relatedness). Color distance between pairs of individuals was treated as a binary value: 0 (same color/form) and 1 (different color/form).
Individuals from northern Queensland were grouped into three clusters corresponding to the three towns where the sampling occurred (Fig 3c). An exception was one queenslandensis individual from Cairns that was grouped with the type individuals from Gordonvale (Fig 3c). The two forms in Townsville could not be distinguished based on their nuclear genome-wide variation (Fig 3c). We found four pairs of closely related individuals: two queenslandensis and two type pairs (Fig 4, S3 File). In other words, all related pairs detected in Townsville were of the same form.
A lower frequency of related individuals in Townsville when compared to Singapore is not surprising given that different sampling methods were employed in these locations. Collection of multiple larvae from the same breeding container increases the chance of sampling family groups, as seen in Singapore and parts of Rio de Janeiro [22]. On the other hand, when BG-sentinel traps are used, the likelihood of related individuals being collected is low. In Townsville, 12.5% of pairs from the same trap were close relatives. Sampling effects are reflected in an elevated level of pairwise genetic distance over geographic distance for mosquitoes from Singapore when compared to Townsville (Fig 4). Such differences in genetic patterns could be erroneously interpreted as differences in the underlying processes (e.g. different dispersal rates), and highlight that sampling methods are crucial when inferring processes within and among Ae. aegypti populations.
In summary, we did not find any association between nuclear genetic variation and color/scaling variation in Ae. aegypti from Singapore and northern Queensland. Our results are unlikely to be caused by a lack of power to detect genetic structure, given that more than 16,000 genome-wide SNPs allowed us to delineate family groups at a very fine spatial scale. In fact, several families had the queenslandensis and type members. A recent study of global Ae. aegypti populations at 12 microsatellite loci found that at least in one locality in Africa (Senegal) the two established forms (formosus and type) are interbreeding with no sign of genetic subdivision when brought into sympatry [11], so it is not surprising that the type and queenslandensis variety also form one genetic cluster. Our results also help explain the similar vectorial capacity for a dengue 2 viral strain of type and queenslandensis females originating from wild Ae. aegypti in Thailand [38].
Wolbachia infection
In addition to an absence of genetic structuring between the two Ae. aegypti aegypti forms, another line of evidence in support of ongoing interbreeding is the presence of Wolbachia in both forms. We detected this endosymbiotic bacterium in three (out of four) queenslandensis individuals from Cairns and 14 (out of 15) type individuals from Gordonvale, using a light-cycler assay for Wolbachia detection [39]. Wolbachia is not naturally found in Ae. aegypti, but was introduced into the populations in Gordonvale in 2011 and Cairns in 2013 in an effort to reduce dengue transmission [40, 41]. This was done by releasing Wolbachia-infected females and males from a colony that originated from type Ae. aegypti [42]. Because the infection is transmitted from mother to offspring, the only way queenslandensis individuals could have become infected by Wolbachia is by mating with infected, type females. Given the high Wolbachia frequency (> 85%) in Cairns and Gordonvale at the time of our sampling [41], the presence of the infection in 3/4 individuals caught in Cairns, and 14/15 individuals caught in Gordonvale was expected.
Conclusion
Our analyses of mitochondrial and nuclear genome-wide variation and the Wolbachia infection indicate that Ae. aegypti queenslandensis and Ae. aegypti type mosquitoes interbreed freely, at least in Singapore and northern Queensland. These findings are of practical importance for control strategies that rely on successful mating between the released and target mosquitoes. Our results also re-enforce the recommendations by the early taxonomic authorities (Mattingly and McClelland) that the extant Ae. aegypti queenslandensis should not be ranked as a subspecies.
Supporting Information
Mosquitoes from Rio de Janeiro (Brazil), Gordonvale (northern Queensland), Ho Chi Minh city (Vietnam), used in the DAPC analysis.
(PDF)
FASTA file with mitochondrial sequences from Aedes aegypti (Hap1-24), the Liverpool strain, and three outgroups used in the RAxML phylogenetic reconstruction. Mitochondrial haplotypes were generated by concatenating 90 bp RAD sequences from: ND2 (1–90 bp), COXI (91–270 bp), ATP6 (271–450 bp), ND5 (451–630 bp), ND4 (631–720 bp), ND6 (721–810 bp), cytB (811–1080 bp), ND1 (1081–1170 bp).
(TXT)
The number of base differences per site from between sequences are shown. The analysis involved 28 nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1170 positions in the final dataset. Evolutionary analyses were conducted in MEGA6.
(TXT)
Estimates of relatedness (r) by Wang (2002) for Aedes aegypti pairs in Singapore and Townsville. Reference: Wang, J. 2002. An estimator for pairwise relatedness using molecular markers. Genetics 160: 1203–1215.
(TXT)
Boxplots of per-individual values for the proportion of uniquely aligned reads, RAD tag read depth, proportion of heterozygous loci, proportion of missing data for Aedes aegypti from Singapore (left) and Queensland (right).
(PDF)
Acknowledgments
We thank Carly Herbertson, Melanie Commerford and the Eliminate Dengue Program for providing Aedes aegypti from Townsville and Cairns. We also thank the NeCTAR Cloud for providing computational resources.
Data Availability
Raw sequence files (fasq) for all samples included in this work can be accessed via NCBI Accession PRJNA348645. DOI of a pre-print version of this manuscript deposited with the bioRxiv is: doi: http://dx.doi.org/10.1101/063792
Funding Statement
This work is funded by the National Health and Medical Research Council and the National Environment Agency, Singapore. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kraemer MUG, Sinka ME, Duda KA, Mylne AQN, Shearer FM, Barker CM, et al. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. eLife. 2015;4:e08347 10.7554/eLife.08347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benelli G, Mehlhorn H. Declining malaria, rising of dengue and Zika virus: insights for mosquito vector control. Parasitology Research. 2016;115(5):1747–54. 10.1007/s00436-016-4971-z [DOI] [PubMed] [Google Scholar]
- 3.Halstead SB. Reappearance of chikungunya, formerly called dengue, in the Americas. Emerging infectious diseases. 2015;21(4):557–61. Epub 2015/03/31. 10.3201/eid2104.141723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan M. Yellow fever: the resurgence of a forgotten disease. The Lancet. 387(10034):2165–6. 10.1016/S0140-6736(16)30620-1 [DOI] [PubMed] [Google Scholar]
- 5.Mattingly PF. Genetical aspects of the Aedes aegypti problem. I. Taxonom: and bionomics. Annals of tropical medicine and parasitology. 1957;51(4):392–408. Epub 1957/12/01. . [PubMed] [Google Scholar]
- 6.Howard LO, Dyar HG, Knab F. The mosquitoes of North and Central America and the West Indies. Washington, D. C.: Carnegie Institution of Washington; 1912. [Google Scholar]
- 7.McClelland GAH. A worldwide survey of variation in scale pattern of the abdominal tergum of Aedes aegypti (L.) (Diptera: Culicidae). Transactions of the Royal Entomological Society of London. 1974;126(2):239–59. 10.1111/j.1365-2311.1974.tb00853.x [DOI] [Google Scholar]
- 8.Hill GF. Notes on some unusual breeding-places of Stegomyia fasciata, Fabr., in Australia. Annals of Tropical Medicine & Parasitology. 1921;15(1):91–2. 10.1080/00034983.1921.11684253 [DOI] [Google Scholar]
- 9.Powell JR, Tabachnick WJ. History of domestication and spread of Aedes aegypti—a review. Memorias do Instituto Oswaldo Cruz. 2013;108 Suppl 1:11–7. Epub 2014/01/30. 10.1590/0074-0276130395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore M, Sylla M, Goss L, Burugu MW, Sang R, Kamau LW, et al. Dual African origins of global Aedes aegypti s.l. populations revealed by mitochondrial DNA. PLoS Negl Trop Dis. 2013;7(4):e2175 10.1371/journal.pntd.0002175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown JE, Evans BR, Zheng W, Obas V, Barrera-Martinez L, Egizi A, et al. Human impacts have shaped historical and recent evolution in Aedes aegypti, the dengue and yellow fever mosquito. Evolution. 2014;68(2):514–25. 10.1111/evo.12281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown JE, McBride CS, Johnson P, Ritchie S, Paupy C, Bossin H, et al. Worldwide patterns of genetic differentiation imply multiple 'domestications' of Aedes aegypti, a major vector of human diseases. Proceedings Biological sciences / The Royal Society. 2011;278(1717):2446–54. Epub 2011/01/14. 10.1098/rspb.2010.2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gloria-Soria A, Ayala D, Bheecarry A, Calderon-Arguedas O, Chadee DD, Chiappero M, et al. Global Genetic Diversity of Aedes aegypti. Molecular Ecology. 2016:n/a–n/a. 10.1111/mec.13866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan A, Chiang LP, Hapuarachchi HC, Tan CH, Pang SC, Lee R, et al. DNA barcoding: complementing morphological identification of mosquito species in Singapore. Parasites & vectors. 2014;7:569 Epub 2014/12/17. 10.1186/s13071-014-0569-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kress WJ, Erickson DL. DNA barcodes: methods and protocols. Methods in molecular biology (Clifton, NJ). 2012;858:3–8. Epub 2012/06/12. 10.1007/978-1-61779-591-6_1 . [DOI] [PubMed] [Google Scholar]
- 16.Mayr E. Animal species and evolution: Belknap Press of Harvard University Press Cambridge, Massachusetts; 1963. [Google Scholar]
- 17.Pennetier C, Warren B, Dabire KR, Russell IJ, Gibson G. "Singing on the wing" as a mechanism for species recognition in the malarial mosquito Anopheles gambiae. Current biology: CB. 2010;20(2):131–6. Epub 2010/01/05. 10.1016/j.cub.2009.11.040 . [DOI] [PubMed] [Google Scholar]
- 18.Axford JK, Ross PA, Yeap HL, Callahan AG, Hoffmann AA. Fitness of wAlbB Wolbachia infection in Aedes aegypti: parameter estimates in an outcrossed background and potential for population invasion. The American journal of tropical medicine and hygiene. 2016;94(3):507–16. Epub 2015/12/30. 10.4269/ajtmh.15-0608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chambers EW, Hapairai L, Peel BA, Bossin H, Dobson SL. Male mating competitiveness of a Wolbachia-introgressed Aedes polynesiensis strain under semi-field conditions. PLoS Neglected Tropical Diseases. 2011;5(8):e1271 10.1371/journal.pntd.0001271. PMC3149012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang D, Lees RS, Xi Z, Gilles JR, Bourtzis K. Combining the sterile insect technique with Wolbachia-based approaches: II—a safer approach to Aedes albopictus population suppression programmes, designed to minimize the consequences of inadvertent female release. PLoS One. 2015;10(8):e0135194 Epub 2015/08/08. 10.1371/journal.pone.0135194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massonnet-Bruneel B, Corre-Catelin N, Lacroix R, Lees RS, Hoang KP, Nimmo D, et al. Fitness of transgenic mosquito Aedes aegypti males carrying a dominant lethal genetic system. PLoS ONE. 2013;8(5):e62711 10.1371/journal.pone.0062711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rašić G, Filipović I, Weeks AR, Hoffmann AA. Genome-wide SNPs lead to strong signals of geographic structure and relatedness patterns in the major arbovirus vector, Aedes aegypti. BMC Genomics. 2014;15(1):275 10.1186/1471-2164-15-275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behura SK, Lobo NF, Haas B, deBruyn B, Lovin DD, Shumway MF, et al. Complete sequences of mitochondria genomes of Aedes aegypti and Culex quinquefasciatus and comparative analysis of mitochondrial DNA fragments inserted in the nuclear genomes. Insect biochemistry and molecular biology. 2011;41(10):770–7. Epub 2011/06/07. 10.1016/j.ibmb.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu Z, et al. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316(5832):1718–23. 10.1126/science.1138878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10(3):R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Molecular ecology. 2013;22(11):3124–40. 10.1111/mec.12354. PMC3936987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasic G, Schama R, Powell R, Maciel-de Freitas R, Endersby-Harshman NM, Filipovic I, et al. Contrasting genetic structure between mitochondrial and nuclear markers in the dengue fever mosquito from Rio de Janeiro: implications for vector control. Evol Appl. 2015;8(9):901–15. Epub 2015/10/27. 10.1111/eva.12301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontaine A, Filipović I, Fansiri T, Hoffmann AA, Rašić G, Lambrechts L. Cryptic genetic differentiation of the sex-determining chromosome in the mosquito Aedes aegypti. bioRxiv. 2016. 10.1101/060061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalinowski ST. hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Molecular Ecology Notes. 2005;5(1):187–9. 10.1111/j.1471-8286.2004.00845.x [DOI] [Google Scholar]
- 30.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–90. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- 31.Hardy CM, Court LN, Morgan MJ, Webb CE. The complete mitochondrial DNA genomes for two lineages of Aedes notoscriptus (Diptera: Culicidae). Mitochondrial DNA Part A, DNA mapping, sequencing, and analysis. 2016;27(3):2024–5. Epub 2014/10/29. 10.3109/19401736.2014.974171 . [DOI] [PubMed] [Google Scholar]
- 32.Hardy CM, Court LN, Morgan MJ. The complete mitochondrial DNA genome of Aedes vigilax (Diptera: Culicidae). Mitochondrial DNA Part A. 2016;27(4):2552–3. 10.3109/19401736.2015.1038800 [DOI] [PubMed] [Google Scholar]
- 33.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics. 2010;11(1):94 10.1186/1471-2156-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24(11):1403–5. 10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- 35.Hardy OJ, Vekemans X. spagedi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Molecular Ecology Notes. 2002;2(4):618–20. 10.1046/j.1471-8286.2002.00305.x [DOI] [Google Scholar]
- 36.Tsuda Y, Yotopranoto S, Bendryman SS, Rosmanida R, Dachlan YP, Takagi M. Seasonal changes in variation of dorsal scale pattern of Aedes aegypti (L.) (Diptera: Culicidae) in Surabaya, Indonesia. Medical Entomology and Zoology. 2003;54(1):73–80. 10.7601/mez.54.73 [DOI] [Google Scholar]
- 37.Mori A, Tsuda Y, Takagi M, Higa Y, Severson DW. Multiple QTL determine dorsal abdominal scale patterns in the mosquito Aedes aegypti. J Hered. 2016;107(5):438–44. Epub 2016/05/01. 10.1093/jhered/esw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wasinpiyamongkol L, Thongrungkiat S, Jirakanjanakit N, Apiwathnasorn C. Susceptibility and transovarial transmission of dengue virus in Aedes aegypti: a preliminary study of morphological variations. The Southeast Asian journal of tropical medicine and public health. 2003;34 Suppl 2:131–5. Epub 2003/01/01. . [PubMed] [Google Scholar]
- 39.Lee SF, White VL, Weeks AR, Hoffmann AA, Endersby NM. High-throughput PCR assays to monitor Wolbachia infection in the dengue mosquito (Aedes aegypti) and Drosophila simulans. Applied and Environmental Microbiology. 2012;78(13):4740–3. 10.1128/aem.00069-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476(7361):454–7. Epub 2011/08/26. 10.1038/nature10356 . [DOI] [PubMed] [Google Scholar]
- 41.Hoffmann AA, Iturbe-Ormaetxe I, Callahan AG, Phillips BL, Billington K, Axford JK, et al. Stability of the wMel Wolbachia Infection following Invasion into Aedes aegypti populations. PLoS Negl Trop Dis. 2014;8(9):e3115 10.1371/journal.pntd.0003115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476(7361):450–3. 10.1038/nature10355 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mosquitoes from Rio de Janeiro (Brazil), Gordonvale (northern Queensland), Ho Chi Minh city (Vietnam), used in the DAPC analysis.
(PDF)
FASTA file with mitochondrial sequences from Aedes aegypti (Hap1-24), the Liverpool strain, and three outgroups used in the RAxML phylogenetic reconstruction. Mitochondrial haplotypes were generated by concatenating 90 bp RAD sequences from: ND2 (1–90 bp), COXI (91–270 bp), ATP6 (271–450 bp), ND5 (451–630 bp), ND4 (631–720 bp), ND6 (721–810 bp), cytB (811–1080 bp), ND1 (1081–1170 bp).
(TXT)
The number of base differences per site from between sequences are shown. The analysis involved 28 nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1170 positions in the final dataset. Evolutionary analyses were conducted in MEGA6.
(TXT)
Estimates of relatedness (r) by Wang (2002) for Aedes aegypti pairs in Singapore and Townsville. Reference: Wang, J. 2002. An estimator for pairwise relatedness using molecular markers. Genetics 160: 1203–1215.
(TXT)
Boxplots of per-individual values for the proportion of uniquely aligned reads, RAD tag read depth, proportion of heterozygous loci, proportion of missing data for Aedes aegypti from Singapore (left) and Queensland (right).
(PDF)
Data Availability Statement
Raw sequence files (fasq) for all samples included in this work can be accessed via NCBI Accession PRJNA348645. DOI of a pre-print version of this manuscript deposited with the bioRxiv is: doi: http://dx.doi.org/10.1101/063792