Abstract
Relating thermodynamic parameters to structural and biochemical data allows a better understanding of substrate binding and its contribution to catalysis. The analysis of the binding of carbohydrates to proteins or enzymes is a special challenge because of the multiple interactions and forces involved. Isothermal titration calorimetry (ITC) provides a direct measure of binding enthalpy (ΔHa) and allows the determination of the binding constant (free energy), entropy, and stoichiometry. In this study, we used ITC to elucidate the binding thermodynamics of xylosaccharides for two xylanases of family 10 isolated from Geobacillus stearothermophilus T-6. The change in the heat capacity of binding (ΔCp = ΔH/ΔT) for xylosaccharides differing in one sugar unit was determined by using ITC measurements at different temperatures. Because hydrophobic stacking interactions are associated with negative ΔCp, the data allow us to predict the substrate binding preference in the binding subsites based on the crystal structure of the enzyme. The proposed positional binding preference was consistent with mutants lacking aromatic binding residues at different subsites and was also supported by tryptophan fluorescence analysis.
Enzymes exhibit an exceptional catalytic power with an acceleration rate (kcat/kuncat) that can exceed 1017 (1, 2). Such powerful catalytic action originates, among other things, from the exquisite positional binding of substrates in which the catalytic residues stabilize the charges formed during the transition state (3). In addition, the substrate may be distorted in the binding pocket into a configuration that resembles the transition state. Revealing the fine details of substrate binding, especially the geometry and energetics involved, is essential for understanding the catalytic efficiency of enzymes. Despite the wide diversity in the tertiary structures and binding-site topologies of enzymes, certain common basic features are typically involved in substrate–enzyme interactions. These interactions are reversible, noncovalent, and relatively weak, thus providing enough stability for catalysis, but are also sufficiently low to allow rapid ligand dissociation (1).
Although high-resolution crystal structures of enzyme–substrate complexes provide invaluable insight into the different elements involved in substrate binding, they cannot provide complete information concerning the forces that drive the binding process. In many high-resolution crystal structures, the substrate is held in position by several specific hydrogen bonds. However, the exact contribution of these bonds to binding and/or catalysis often remains obscure. This situation is especially true for cases of relatively weak binding as relevant for most of the enzyme–substrate interactions. Thus, the actual free energy of such binding is typically in the order of ΔG = –10 kcal/mol (Kd in the micromolar range) that can theoretically be satisfied by only about two hydrogen bonds out of the many potential bonds that are usually available. Moreover, the contribution of entropy to binding cannot be easily calculated or determined based on the crystal structure alone. Hence, our understanding of substrate binding and catalysis is significantly improved by exact thermodynamic parameters and their correlation to the corresponding structural and biochemical data. In this regard, thermodynamic measurements are especially valuable in determining the particular driving forces involved in substrate binding and catalysis. The deeper understanding of these enzymatic key parameters can be then used for a rational design of new functionalities.
Isothermal titration calorimetry (ITC) provides a direct measure of binding enthalpy, ΔHa, allowing the simultaneous determination of the binding parameters, Ka, ΔSa, ΔGa, and the binding stoichiometry (4). When the enthalpy of binding is determined at different temperatures, the change in heat capacity (ΔCp = ΔHa/ΔT) for the binding reaction can be extracted. Negative ΔCp values were shown in many systems to be associated with hydrophobic stacking interactions, presumably resulting from the dehydration of highly ordered water molecules surrounding the hydrophobic surfaces (5–8). It should be noted, however, that negative ΔCp can also be obtained in situations where there is cooperative disorder of hydrogen-bonding networks, with no obvious hydrophobic elements, as pointed out by Cooper (9).
Hydrogen bonding and stacking interactions are the dominant interactions in protein–carbohydrate complexes (10, 11). The sugar-hydroxyl groups can serve simultaneously as hydrogen donors and acceptors and may potentially be involved in as many as three hydrogen bonds (as a donor of one hydrogen bond and an acceptor of two) (10). Stacking or hydrophobic interactions of one or both sides of the sugar ring can be formed with aromatic residues at the binding site of the enzyme (10, 12). In this regard, if the sugar ring and the aromatic residue are coplanar, the π electrons of the aromatic residue can form a number of CH–π interactions with the sugar ring (13). It is evident that determining the exact contribution of the various interactions and forces involved in carbohydrate protein binding is a complicated task (10, 11).
Glycoside hydrolases hydrolyze one of the most stable bonds in nature, with half-lives of millions years (2, 14). To date, the amino acid sequences of more than 12,600 glycosidases have been identified, and the sequenced-based classification of their catalytic domains into families and clans is available on the continuously updated Carbohydrate-Active Enzymes (CAZy) server (http://afmb.cnrs-mrs.fr/CAZY). Glycoside hydrolases are the subject of intensive research because of their pivotal role in many fundamental biological processes as well as in a wide range of industrial applications (15, 16). Xylanases (EC 3.2.1.8) hydrolyze the β-1-4 xylose backbone of xylan, the most abundant hemicellulose polymer (17, 18), and are found mainly in glycoside hydrolase families 10 and 11 (19, 20).
In the present study, ITC was used to measure the binding thermodynamics of selected xylosaccharides to two family 10 xylanases from Geobacillus stearothermophilus T-6. These ITC measurements provide valuable information concerning the enzyme–substrate interactions and the number of sugar binding sites that are directly involved in each of these enzymes. We also demonstrate herein that the ΔCp of binding can be correlated with the potential stacking interactions at the different subsites as predicted from the crystal structures. Such an approach can be used as a general tool for determining the preference of binding at specific subsites.
Experimental Procedures
Site-Directed Mutagenesis. Site-directed mutagenesis was performed on the xylanase genes xynA (XT6) and xynA2 (IXT6) (GenBank accession nos. AAC98140 and AAC98123, respectively) from G. stearothermophilus T-6 by using the QuikChange site-directed mutagenesis kit (Stratagene). The mutagenic primers for the mutations were as follows (the mutated nucleotides are in bold type): E159Q, 5′-GGGACGTTGTCAATCAGGTTGTGGG-3′ and 5′-CCCACA ACCTGAT TGACA ACGTCCC-3′; Y203A, 5′-GCTTTACATGAATGATGCCAATACAGAAGTCG-3′ and 5′-GCACTTCTGTATTGGCATCATTCATGTAAAGC-3′; W273A, 5′-GATGTGAGCATGTAT-GGTGCACCGCCGCGCGCTTAC-3′ and 5′-GTAAGCGCGGCGGCGGTGCACCATACATGCTCACATC-3′; and E134Q, 5′-GGATGTCATTAATCAAGCGGTAGCCGACGAAGG-3′ and 5′-CCTTCGTCGGCTACCGCTTGATTAATGACATCC-3′. All of the mutated genes were sequenced to confirm that only the desired mutations were inserted.
Protein Production and Purification. All enzymes were overexpressed by using the T-7 expression system in Escherichia coli strain BL21(DE3) and purified as reported in ref. 21.
Microcalorimetry Titration Studies. Titration calorimetry measurements were performed with a VP-ITC calorimeter (Microcal, Northampton, MA) as described by Wiseman et al. in ref. 4. Protein solutions for ITC were dialyzed extensively overnight against buffer A (50 mM Tris·HCl, pH 7.0/100 mM NaCl/0.02% NaN3). Ligand solutions of birch wood and beech wood xylans (Sigma) and xylobiose (X2), xylotriose (X3), xylotetraose (X4), xylopentaose (X5), and xylohexaose (X6) (Megazyme, Wicklow, Ireland) were prepared by diluting with the buffer used for the protein dialysis. Aliquots (10 μl) of the ligand solution at 8.5–20× the binding-site concentration were added by means of a 250-μl rotating stirrer-syringe to the reaction cell, containing 1.41 ml of the 0.1–0.2 mM protein solution. The heat of dilution was determined to be negligible in separate titrations of the ligand into the buffer solution. Calorimetric data analysis was carried out with origin 5.0 software (MicroCal). Binding parameters such as the number of binding sites (n), the binding constant (Ka, M–1), and the binding enthalpy (ΔHa, kcal/mol of bound ligand) were determined by fitting the experimental binding isotherms. Ka was primarily determined by the slope of the isotherm in the equivalence point (22). The unknown molar concentration of binding sites of the high-molecular-weight polysaccharides (xylans) was estimated by altering the concentration of ligand (in the calculation), so as to fit one binding site.
Fluorescence Analysis. Fluorescence measurements were performed with a PerkinElmer luminescence LS50B spectrometer at 25°C with excitation and emission wavelengths of 295 and 345 nm (5-nm slit), respectively. After each ligand addition, the mixture was mixed for 3–5 min to allow the system to reach equilibrium. The binding constants were determined by nonlinear least-squares fit of the data to a one-site binding model by using the program grafit 5.0 (23).
Results and Discussion
Thermodynamics of Substrate Binding. The aim of this study was to characterize the binding thermodynamics of xylooligosaccharides and xylans to family 10 xylanases by using ITC. Two xylanases from G. stearothermophilus T-6 were used, a 43,808-Da extracellular xylanase, XT6, and a 38,639-Da intracellular xylanase, IXT6. The 3D structures of both enzymes were recently determined by x-ray crystallography (XT6, PDB ID code 1HIZ (24); IXT6, PDB ID code 1N82). Both proteins are made of a single TIM barrel catalytic domain and share ≈40% sequence identity. For the ITC measurements, we used inactive mutants [XT6(E159Q) and IXT6(E134Q)], lacking the catalytic acid base residues to prevent significant hydrolysis of the substrate during the titration experiments.
Table 1 and Table 2 summarize the thermodynamic parameters and the binding constants extracted from the binding isotherms. These parameters are given for both enzymes at a temperature range of 20–50°C. All of the binding interactions were exothermic, and the enthalpy values ranged from –3.2 to –23.7 kcal/mol. For XT6, entropy values were negative and for IXT6, entropy values ranged from TΔSa = –6.2 to +2.7 kcal/mol. All of the titration curves fit very well into a single binding model with a calculated n (stoichiometry) of 1 ± 0.05. For both enzymes, the association constants (Ka) for the different xylosaccharides ranged from ≈104 M–1 for the hydrolytic product X2 to 106 M–1 for the substrates X6 and xylans. These values reflect the different affinities of the substrates and products to the binding site of the enzyme and are well within the range of typical enzyme–substrate binding constants, corresponding to Kd (≈Km in most cases) in the range of 0.4 to 40 μM (1). For XT6, the maximum Ka values were with X6 and xylans, suggesting a binding site of six sugar subsites in this enzyme. For IXT6, similar Ka values were obtained for X5, X6, and xylans suggesting a binding pocket of five subsites. Elongated binding sites are relatively common for glycoside hydrolases, as many of them that act on high-molecular-weight polysaccharides have evolved extended binding pockets that accommodate several sugar units. These binding subsites are labeled, according to convention, from –n to +n, with –n at the nonreducing end and +n at the reducing end, whereas the cleavage occurs between the –1 and +1 subsites (25).
Table 1. Thermodynamic parameters for xylosaccharide and xylan binding to XT6(E159Q).
| Ligand | T, °C | Ka × 104, M-1 | Kd (1/Ka), μM | ΔHa, kcal/mol | TΔSa, kcal/mol | ΔGa, kcal/mol | ΔCp, cal/mol·K |
|---|---|---|---|---|---|---|---|
| Xylosaccharides | |||||||
| X2 | 20 | 4.1 ± 0.1 | 24 | -6.8 ± 0.1 | -0.6 | -6.2 | |
| 30 | 2.6 ± 0.3 | 38 | -6.7 ± 0.5 | -0.6 | -6.1 | -2 ± 5 | |
| 40 | 1.2 ± 0.1 | 83 | -6.9 ± 0.3 | -1.1 | -5.8 | ||
| 50 | 2.5 ± 0.3 | 40 | -6.8 ± 0.2 | -0.6 | -6.2 | ||
| X3 | 20 | 12 ± 0.5 | 8.3 | -9.0 ± 0.1 | -2.2 | -6.8 | |
| 30 | 11 ± 2 | 9.4 | -10.5 ± 0.1 | -3.5 | -7.0 | -94 ± 19 | |
| 40 | 4.9 ± 0.2 | 20 | -10.9 ± 0.1 | -4.2 | -6.7 | ||
| 50 | 2.9 ± 0.7 | 34 | -12.0 ± 0.2 | -5.1 | -6.9 | ||
| X4 | 20 | 50 ± 2.3 | 2.0 | -13.4 ± 0.1 | -5.8 | -7.6 | |
| 30 | 29 ± 5 | 3.5 | -14.4 ± 0.3 | -6.8 | -7.6 | -233 ± 24 | |
| 35 | 17 ± 1.7 | 5.9 | -15.6 ± 0.3 | -8.2 | -7.4 | ||
| 40 | 15 ± 1.7 | 6.7 | -17.9 ± 0.3 | -10.4 | -7.5 | ||
| 50 | 8.5 ± 0.6 | 12 | -20.0 ± 1.2 | -12.7 | -7.3 | ||
| X5 | 20 | 280 ± 27 | 0.36 | -13.2 ± 0.1 | -4.6 | -8.6 | |
| 30 | 110 ± 14 | 0.93 | -13.6 ± 1.0 | -5.3 | -8.3 | -257 ± 27 | |
| 40 | 84 ± 5 | 1.2 | -16.8 ± 0.1 | -8.3 | -8.5 | ||
| 50 | 41 ± 2.4 | 2.4 | -21.1 ± 0.2 | -12.8 | -8.3 | ||
| X6 | 20 | 760 ± 100 | 0.13 | -12.8 ± 0.1 | -3.6 | -9.2 | |
| 30 | 290 ± 23 | 0.35 | -15.0 ± 0.2 | -6.2 | -8.8 | -380 ± 35 | |
| 40 | 200 ± 23 | 0.5 | -20.3 ± 0.2 | -11.2 | -9.1 | ||
| 50 | 140 ± 2.3 | 0.74 | -23.7 ± 0.2 | -14.7 | -9.0 | ||
| Xylans | |||||||
| Beech wood | 20 | 160 ± 22 | 0.61 | -14.4 ± 0.2 | -6.1 | -8.3 | |
| 30 | 110 ± 19 | 0.89 | -15.9 ± 0.9 | -7.6 | -8.3 | -330 ± 32 | |
| 40 | 92 ± 5.8 | 1.1 | -20.3 ± 0.2 | -11.8 | -8.5 | ||
| 50 | 89 ± 1.6 | 1.1 | -23.2 ± 0.9 | -14.4 | -8.8 | ||
| Birch wood | 20 | 130 ± 16 | 0.79 | -10.0 ± 0.1 | -1.8 | -8.2 | |
| 30 | 150 ± 20 | 0.65 | -12.6 ± 1.0 | -4.2 | -8.4 | -360 ± 29 | |
| 35 | 160 ± 17 | 0.64 | -14.4 ± 0.1 | -5.6 | -8.8 | ||
| 40 | 98 ± 8 | 1.0 | -16.4 ± 0.2 | -7.8 | -8.6 | ||
| 50 | 130 ± 16 | 0.75 | -20.3 ± 0.2 | -11.3 | -9.0 |
Each set of results is based on the average of three independent measurements. The stoichiometry in all experiments with xylosaccharides was very close to 1 (±0.05). The size of a single “binding unit” for xylans was adjusted to fit n = 1 and was 17 xylose units for beech and birch wood xylans.
Table 2. Thermodynamic parameters for xylosaccharide and xylan binding to IXT6(E134Q).
| Ligand | T, °C | Ka × 104, M-1 | Kd (1/Ka), μM | ΔHa, kcal/mol | TΔSa, kcal/mol | ΔGa, kcal/mol | ΔCp, cal/mol K |
|---|---|---|---|---|---|---|---|
| Xylosaccharides | |||||||
| X2 | 20 | 2.5 ± 0.2 | 40 | -3.9 ± 0.09 | 2.0 | -5.9 | |
| 30 | 1.6 ± 0.6 | 63 | -3.2 ± 0.8 | 2.7 | -5.9 | -1 ± 14 | |
| 40 | 1.3 ± 0.8 | 77 | -3.6 ± 0.1 | 2.3 | -5.9 | ||
| 50 | 1.6 ± 0.2 | 63 | -3.9 ± 0.2 | 2.3 | -6.2 | ||
| X3 | 20 | 5.8 ± 0.3 | 17 | -4.1 ± 0.04 | 2.3 | -6.4 | |
| 30 | 4.3 ± 0.5 | 23 | -4.8 ± 0.3 | 1.7 | -6.5 | -90 ± 14 | |
| 40 | 3.6 ± 0.3 | 28 | -5.6 ± 0.1 | 0.9 | -6.5 | ||
| 50 | 3.3 ± 0.7 | 30 | -6.9 ± 0.2 | -0.2 | -6.7 | ||
| X4 | 20 | 28 ± 1.2 | 3.6 | -5.7 ± 0.02 | 1.6 | -7.3 | |
| 30 | 20 ± 0.2 | 5.0 | -8.2 ± 0.3 | -0.8 | -7.4 | -253 ± 18 | |
| 40 | 11 ± 0.3 | 9.1 | -10.4 ± 0.05 | -3.2 | -7.2 | ||
| 50 | 7.5 ± 0.4 | 13 | -13.4 ± 0.1 | -6.2 | -7.2 | ||
| X5 | 20 | 78 ± 7.6 | 1.3 | -6.7 ± 0.06 | -1.2 | -7.9 | |
| 30 | 47 ± 5 | 2.1 | -7.9 ± 0.1 | 0.0 | -7.9 | -242 ± 28 | |
| 40 | 30 ± 1.8 | 3.3 | -11.5 ± 0.1 | -3.6 | -7.9 | ||
| 50 | 19 ± 0.7 | 5.3 | -13.6 ± 0.06 | -5.8 | -7.8 | ||
| X6 | 20 | 112 ± 10 | 0.89 | -6.4 ± 0.04 | -1.8 | -8.2 | |
| 30 | 49 ± 5.8 | 2.0 | -7.4 ± 0.1 | 0.5 | -7.9 | -244 ± 34 | |
| 40 | 39 ± 3.5 | 2.6 | -11.4 ± 0.1 | -3.4 | -8.0 | ||
| 50 | 17 ± 0.5 | 5.9 | -13.2 ± 0.07 | -5.4 | -7.8 | ||
| Xylans | |||||||
| Beech wood | 30 | 42 ± 2.7 | 2.4 | -6.2 ± 0.04 | 1.5 | -7.9 | |
| Birch wood | 30 | 42 ± 4.2 | 2.4 | -4.7 ± 0.03 | 3.1 | -7.8 |
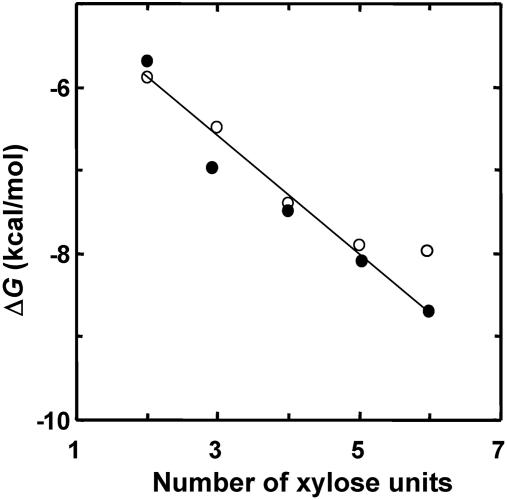
At 30°C, the free energy of binding of X2 was –6.1 kcal/mol, and each additional sugar unit resulted in an average free-energy gain (relative to X2) of about –0.7 kcal/mol (Fig. 1). These results, together with the one-to-one stoichiometry obtained for X2, strongly suggest that X2 has high preference to a invariable site and probably also controls the binding preference of longer xylosaccharides.
Fig. 1.
Free energy of xylosaccharide binding to XT6(E159Q) (•) and to IXT6(E134Q) (○) vs. the number of xylose units in the saccharide chain. Note that for IXT6, X6 and X5 give similar values, indicating that in IXT6 there are only five binding subsites.
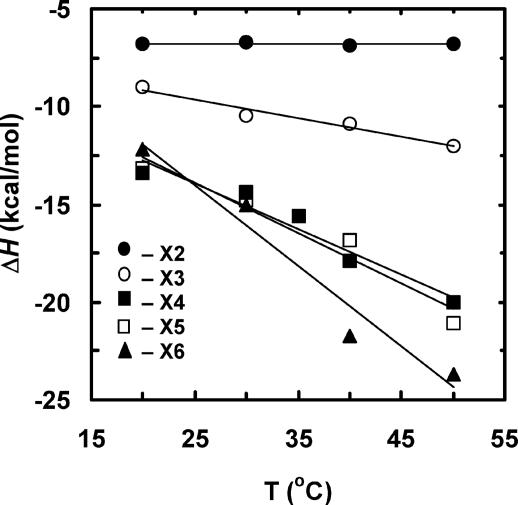
Binding Preference of Xylosaccharides in the Active Site of XT6. From the thermodynamic parameters obtained at different temperatures, the heat capacity change (ΔCp = δΔHa/δT) for the binding reaction could be extracted. Plotting ΔHa of binding vs. temperature for the various xylosaccharides gave a linear correlation, indicating that ΔCp for binding is constant at the temperature range used (Fig. 2). The change in heat capacity (ΔCp) for the interactions of XT6 with different xylosaccharides becomes more negative as the size of the substrate increases (Table 1). The ΔCp for X2 is negligible, that for X3 is –94 cal/mol·K, and those for X4 and X5 are about –245 cal/mol·K, and those for X6 and xylans are about –360 cal/mol·K (Table 1).
Fig. 2.
Changes in the binding enthalpies (ΔHa) at different temperatures for interaction of xylosaccharides with XT6(E159Q). Values of ΔCp were derived from the slopes of the linear correlation.
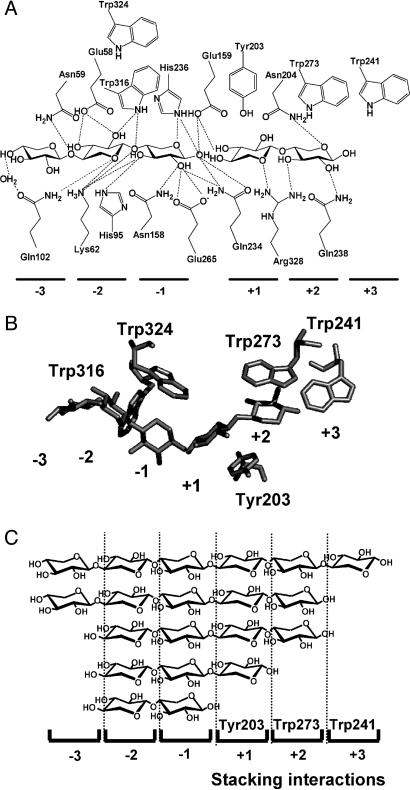
How do the measured ΔCp values match the 3D structure of the relevant complexes? Such a correlation can be made on the basis of several crystal structures of enzyme–substrate complexes of XT6, which were recently analyzed. In the crystal structures of XT6 (PDB ID codes 1HIZ in ref. 24 and 1R85) and its complex with its products, cleaved X5 (PDB ID code 1R87), it is evident that X3 and X2 occupy subsites –3 to –1 and +1 to +2, respectively (Fig. 3). These structures indicate that there are five conserved aromatic residues in the vicinity of the active site that potentially can be involved in stacking interactions with the bound sugars at the different subsites: Trp-316 at subsite –2, Trp-324 at –1, Tyr-203 at +1, Trp-273 at +2, and Trp-241 at +3. A close examination of these residues reveals that in the –2 and –1 subsites, the sugar rings and the aromatic residues have small contact surface area (10–20 Å2) and are not parallel to each other, a geometry that does not support stacking interactions (Fig. 3B). On the other hand, the aromatic residues at the +1 and +2 subsites are oriented parallel to the sugar rings with a much larger contact surface area (>50 Å2), making stacking interactions much more favorable (Fig. 3B). These overall arrangements of aromatic residues (lacking apparently stacking interactions in subsites –2 to –1 and with stacking interaction in subsite +1) are found also in many other family 10 xylanases, for which the structures of xylosaccharide–enzyme complexes are available. These structures include the xylanases from Cellulomonas fimi (PDB ID codes 1FH7–1FH10), Cellvibrio japonicus (PDB ID code 1E5N), Penicillium simplicissimum (PDB ID codes 1B3Y and 1B3Z), Streptomyces lividans (PDB ID codes 1EOX and 1EOY), Streptomyces olivaceoviridis E86 (PDB ID codes 1ISW and 1ISX), and Thermoascus aurantiacus (PDB ID codes 1GOQ and 1GOR) (26–32). Interestingly, a recent structure of Cellvibrio japonicus xylanase in its complex with X4 (PDB ID code 1US2) indicates a potential stacking interaction at the –3 subsite (33). Taking together the ΔCp values and the 3D structure of XT6, we can now predict the binding preference of XT6 toward xylosaccharides. The ΔCp for X2 was negligible, thus, it is most likely that X2 binds to the –2 and –1 subsites, where there are no stacking interactions (Fig. 3C). X3 has one stacking interaction and probably occupies the three subsites between –2 and +1 (a single stacking interaction with Tyr-203 at the +1 subsite). X4 and X5 seem to have two stacking interactions, and presumably their binding occurs at the –2 to +2 and the –3to +2 subsites, respectively. Finally, X6 has three stacking interactions and, therefore, should occupy the six subsites between –3 and +3 (Table 1 and Fig. 3). The same arguments hold for the longer substrate xylan, which should also be bound by three stacking interactions. Based on this assignment, each aromatic residue contributes to ΔCp about –100 to –150 cal/mol·K. These results are consistent with theoretical and experimental data (34) and also correlate well with values obtained with the xylan binding module CBM15, where two Trp residues are proposed to mediate the main hydrophobic–stacking interactions with X5, providing ΔCp of about –300 cal/mol·K (35).
Fig. 3.
Substrate positional binding in XT6. (A) Schematic representation of the hydrogen bonding network and stacking interactions in the XT6 binding site. (B) Structure of XT6 in complex with X3 and X2 (cleaved X5), showing only the aromatic amino acids and the sugars. Tyr-203 and Trp-273 form stacking interactions with the sugar rings at subsites +1 and +2, respectively. (C) The suggested positional binding of xylosaccharides to the binding subsites of XT6, obtained by combining the ΔCp with the structural data.
The proposed binding preferences can be challenged by using mutants lacking either the aromatic residues at subsites +1 (Tyr-203) and +2 (Trp-273) or a nonaromatic binding residue, Gln-102 for example. The ΔCp values of the mutants in the aromatic residues were predicted to be less negative by ≈100–150 cal/mol·K, but only for xylosaccharides that occupy these subsites. For the Gln-102 replacement, ΔCp should not change (although the Ka may be reduced). Indeed, the ΔCp values of the mutants were exactly as predicted from the preference of binding suggested above (Fig. 3 and Table 3) with an average ΔCp loss of –105 to –165 cal/mol·K for Y203A and W273A, respectively, and no ΔCp change for Q102A (Table 3). It should be noted that although all of the mutations resulted in reduced Ka, the corresponding ΔCp values support the assigned binding preference.
Table 3. ΔCp of binding for different xylosaccharides with mutants lacking binding residues.
| ΔCp, cal/mol·K
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Xylan
|
|||||||||
| Enzyme (mutant) | X2 | X3 | X4 | X5 | X6 | Birch wood | Beech wood | Stacking in subsites | Ka (X6),* M-1 × 104 |
| XT6(E159Q) | -2 | -94 | -233 | -257 | -380 | -330 | -360 | +1, +2, +3 | 290 |
| XT6(E159QY203A) | 6 | -13 | -154 | -141 | -238 | +2, +3 | 19 | ||
| XT6(E159QW273A) | 0 | -85 | -93 | -89 | -190 | +1, +3 | 54 | ||
| IXT6(E134Q) | -1 | -90 | -253 | -242 | -244 | +1, +2 | 49 | ||
| XT6(E159QQ102A) | -5 | -118 | -239 | -267 | -359 | +1, +2, +3 | 38 | ||
The positional binding scheme deduced from the thermodynamic data of the extracellular xylanase XT6 is also consistent with the results obtained with the intracellular xylanase IXT6. Both the ITC data (Table 2 and Fig. 1) and the crystal structure of IXT6 (PDB ID code 1N82) suggest that the active site of the enzyme consists of five binding subsites compared with the six subsites of XT6. In the + subsites there are two conserved aromatic residues, Tyr-179 at subsite +1 and Phe-249 at subsite +2 (this enzyme lacks an aromatic residue at position +3). Based on the overall arrangements of enzyme–substrate interactions in the active site of family 10 (no stacking interactions at the minus sites), it is expected that the maximum ΔCp value with this enzyme will reflect a contribution of two stacking interactions. Indeed, the ΔCp values observed for X4, X5, and X6 were all similar, about –250 cal/mol·K (Table 3).
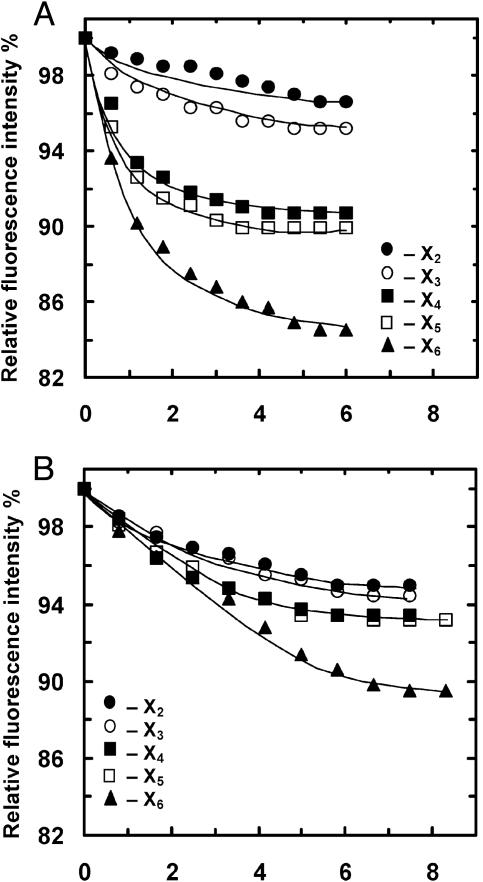
Trp Fluorescence Quenching Analysis Supports the Substrate Binding Preference Indicated by ITC. Because several Trp residues are part of the binding subsites, it is possible to use Trp fluorescence quenching analysis to independently determine the xylosaccharides binding mode for this enzyme. XT6 contains 10 Trp residues, 3 of which are likely to interact with the substrate. Trp-316 at the –2 subsite forms hydrogen bonds (but no stacking interaction), and Trp-273 and Trp-241 form stacking interactions at the +2 and +3 subsites, respectively. In the presence of a molar excess of X6, significant quenching (≈15%) in Trp fluorescence (excitation 295 nm and emission 345 nm) was obtained (data not shown). Titration with various xylosaccharides resulted in quenching curves that are consistent with the binding preference predicted from the ITC analysis (Fig. 4A). Thus, X2 and X3 resulted in quenching of ≈5%, reflecting the interaction with a single Trp residue (probably Trp-316 at –2). X4 and X5 provided additional quenching of up to a total of 10%, suggesting that for both substrates an additional Trp interaction is involved (probably Trp-273 at +2). Finally, X6 resulted in 15% quenching, indicating interaction with a third Trp residue (probably Trp-241, at +3). The results with the W273A mutant are also consistent with the binding assignment. This mutant lacks the Trp residue at the +2 subsite. Thus, it is expected that the degree of quenching will be similar with X2, X3, X4, and X5 that interact only with Trp-316 at the –2 subsite. X6, however, is expected to interact also with Trp-241 at the +3 subsite, thus providing an additional quenching of 5%. As shown in Fig. 4B, these expectations are exactly the results obtained. In addition, the Trp quenching titration experiments allow us to determine the xylosaccharide binding constants (Ka). For X5 and X6 at 25°C, the Ka values were 99 × 104 and 200 × 104 M–1, respectively, well within the range obtained by the ITC analysis. Taking these results together, the Trp fluorescence quenching analysis provided an additional independent confirmation of the substrate binding preference suggested by the ITC analysis.
Fig. 4.
Changes in fluorescence intensities in the presence of different concentrations of xylosaccharides at 25°C. (A) Binding of xylosaccharides to XT6(E159Q). (B) Binding of xylosaccharides to XT6(E159QW273A) (lacking Trp-273 at the +2 subsite).
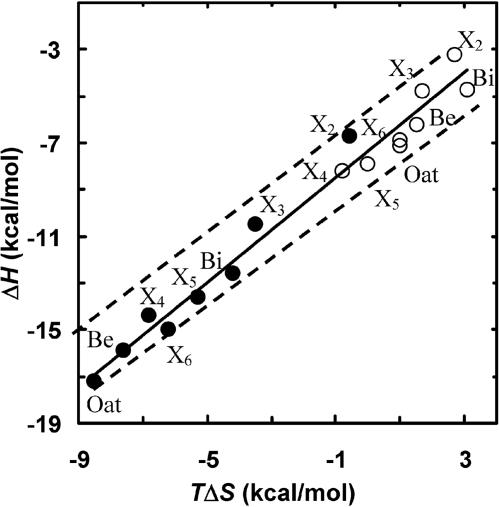
Enthalpy–Entropy Compensation Effect. When ΔHa and TΔSa values for the binding of different xylosaccharides are plotted against each other, a linear relationship is observed with a slope of 1.3 (Fig. 5). Thus, a change in enthalpy is compensated by an opposite change in entropy. This phenomenon, termed enthalpy–entropy compensation, has been described in many systems (36–38), however, the biological relevance of this phenomenon is a matter of debate (39–41). The seemingly linear relationship results, in fact, from the limited range of ΔGa (Ka values) used in the experiments and the Gibbs free energy relationship, ΔGa =ΔHa – TΔSa (see dashed lines in Fig. 5). In experiments where the binding constants do not vary significantly (i.e., by more than three orders of magnitude), the ΔHa to TΔSa relationship must fall within a limited range (within the dashed lines in Fig. 5). However, with both enzymes the binding of X2, X3, and X4 follows a similar trend in which each additional sugar contributes about the same to ΔHa and ΔSa. Thus, the binding mechanism of these sugars is likely to be similar. This trend does not follow for X5, X6, and xylans, which may indicate that somewhat different binding forces are involved. A mechanistic clue for this phenomenon can be derived from the crystal structures of the enzyme–substrate complexes in which the sugar rings at subsites –2 to +2 are held by a larger number of potential hydrogen bonds.
Fig. 5.
Enthalpy–entropy compensation plot for the binding of X2, X3, X4, X5, X6, beech wood xylan (Be), birch wood xylan (Bi), and oat spelts xylan (Oat) to IXT6 (○) and to XT6 (•) at 303 K. The dashed lines are the theoretical range of ΔGa, where the binding constants vary by three orders of magnitude.
Conclusions. This study demonstrates that ΔCp values can be used as a reliable and accurate tool to evaluate the contribution of stacking–hydrophobic interactions to the binding mechanisms. If structural data are available, it may also provide the binding preference of the substrate where more than one binding alternative exists. Because ITC measures enthalpy directly, there is very little error involved in the ΔCp values regardless of the binding mechanisms and constants. This approach may be applicable for many other systems in which the binding is characterized with measurable changes in ΔCp.
Acknowledgments
This study was supported by the German–Israeli Foundation for Scientific Research and Development (to Y.S. and G.S.), the French–Israeli Association for Scientific and Technological Research (to Y.S.), the Israel Science Foundation (to G.S. and Y.S.), and the Otto Meyerhof Center for Biotechnology, Technion, established by the Minerva Foundation (Munich).
Abbreviations: ITC, isothermal titration calorimetry; IXT6, intracellular xylanase from Geobacillus stearothermophilus T-6; XT6, extracellular xylanase from G. stearothermophilus T-6; X2, xylobiose; X3, xylotriose; X4, xylotetraose; X5, xylopentaose; X6, xylohexaose.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1R85 and 1R87).
References
- 1.Fersht, A. (1999) Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding (Freeman, New York).
- 2.Wolfenden, R. & Snider, M. J. (2001) Acc. Chem. Res. 34, 938–945. [DOI] [PubMed] [Google Scholar]
- 3.Jencks, W. P. (1975) Adv. Enzymol. Relat. Areas Mol. Biol. 43, 219–410. [DOI] [PubMed] [Google Scholar]
- 4.Wiseman, T., Williston, S., Brandts, J. F. & Lin, L. N. (1989) Anal. Biochem. 179, 131–137. [DOI] [PubMed] [Google Scholar]
- 5.Livingstone, J. R., Spolar, R. S. & Record, M. T., Jr. (1991) Biochemistry 30, 4273–4244. [DOI] [PubMed] [Google Scholar]
- 6.Lemieux, R. U. (1996) Acc. Chem. Res. 29, 373–380. [Google Scholar]
- 7.Spolar, R. S. & Record, M. T., Jr. (1994) Science 263, 777–784. [DOI] [PubMed] [Google Scholar]
- 8.Tomme, P., Creagh, A. L., Kilburn, D. G. & Haynes, C. A. (1996) Biochemistry 35, 13885–13894. [DOI] [PubMed] [Google Scholar]
- 9.Cooper, A. (2000) Biophys. Chem. 85, 25–39. [DOI] [PubMed] [Google Scholar]
- 10.Quiocho, F. A. (1989) Pure Appl. Chem. 61, 1293–1306. [Google Scholar]
- 11.Quiocho, F. A., Vyas, N. K. & Spurlino, J. C. (1989) Trans. Am. Crystallogr. Assoc. 25, 23–35. [Google Scholar]
- 12.Hu, G., Oguro, A., Li, C., Gershon, P. D. & Quiocho, F. A. (2002) Biochemistry 41, 7677–7687. [DOI] [PubMed] [Google Scholar]
- 13.Nishio, M., Hirota, M. & Umezwa, Y. (1998) The CH/p Interaction: Evidence, Nature, and Consequences (Wiley, New York).
- 14.Shallom, D. & Shoham, Y. (2003) Curr. Opin. Microbiol. 6, 219–228. [DOI] [PubMed] [Google Scholar]
- 15.Mackenzine, L. F., Wong, Q., Warren, R. A. & Withers, S. G. (1998) J. Am. Chem. Soc. 120, 5583–5585. [Google Scholar]
- 16.Kulkarni, N., Shendye, A. & Rao, M. (1999) FEMS Microbiol. Rev. 23, 411–456. [DOI] [PubMed] [Google Scholar]
- 17.Hazlewood, G. P. & Gilbert, H. J. (1998) Prog. Nucleic Acid Res. Mol. Biol. 61, 211–241. [DOI] [PubMed] [Google Scholar]
- 18.Sunna, A. & Antranikian, G. (1997) Crit. Rev. Biotechnol. 17, 39–67. [DOI] [PubMed] [Google Scholar]
- 19.Thomson, J. A. (1993) FEMS Microbiol. Rev. 104, 65–82. [DOI] [PubMed] [Google Scholar]
- 20.Davies, G. & Henrissat, B. (1995) Structure (London) 3, 853–859. [DOI] [PubMed] [Google Scholar]
- 21.Bravman, T., Zolotnitsky, G., Shulami, S., Belakhov, V., Solomon, D., Baasov, T., Shoham, G. & Shoham, Y. (2001) FEBS Lett. 495, 39–43. [DOI] [PubMed] [Google Scholar]
- 22.Faergeman, N. J., Sigurskjold, B. W., Kragelund, B. B., Andersen, K. V. & Knudsen, J. (1996) Biochemistry 35, 14118–14126. [DOI] [PubMed] [Google Scholar]
- 23.Leatherbarrow, J. R. (2001) grafit (Erithacus Software, Horley, U.K.), Version 5.0.
- 24.Teplitsky, A., Mechaly, A., Stojanoff, V., Sainz, G., Golan, G., Feinberg, H., Gilboa, R., Reiland, V., Zolotnitsky, G., Shallom, D., et al. (2004) Acta Crystallogr. D 60, 836–848. [DOI] [PubMed] [Google Scholar]
- 25.Davies, G., Wilson, K. S. & Henrissat, B. (1997) Biochem. J. 321, 557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White, A., Withers, S. G., Gilces, N. R. & Rose, D. R. (1994) Biochemistry 33, 12546–12552. [DOI] [PubMed] [Google Scholar]
- 27.Dominguez, R., Souchon, H., Spinelli, S., Dauter, Z., Wilson, K. S., Chauvaux, S., Beguin, P. & Alzari, P. M. (1995) Nat. Struct. Biol. 2, 569–576. [DOI] [PubMed] [Google Scholar]
- 28.Derewenda, U., Swenson, L., Green, R., Wei, Y., Morosoli, R., Shareck, F., Kluepfel, D. & Derewenda, Z. S. (1994) J. Biol. Chem. 269, 20811–20814. [PubMed] [Google Scholar]
- 29.Harris, G. W., Jenkins, J. A., Connerton, I. & Pickersgill, R. W. (1996) Acta Crystallogr. D 52, 393–401. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt, A., Schlacher, A., Schwab, W. & Kratky, C. (1998) Protein Sci. 7, 2081–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natesh, R., Bhanumoorthy, P., Vithayathil, P. J., Sekar, K., Ramakumar, S. & Viswamitra, M. A. (1999) J. Mol. Biol. 288, 999–1012. [DOI] [PubMed] [Google Scholar]
- 32.Fujimoto, Z., Kuno, A., Kaneko, S., Yoshida, S., Kobayashi, H., Kusakabe, I. & Mizuno, H. (2000) J. Mol. Biol. 300, 575–585. [DOI] [PubMed] [Google Scholar]
- 33.Pell, G., Szabo, L., Charnock, S. J., Xie, H., Gloster, T. M., Davies, G. J. & Gilbert, H. J. (2004) J. Biol. Chem. 279, 11777–11788. [DOI] [PubMed] [Google Scholar]
- 34.Makhatadze, G. I. & Privalov, P. L. (1990) J. Mol. Biol. 213, 375–384. [DOI] [PubMed] [Google Scholar]
- 35.Pell, G., Williamson, M. P., Walters, C., Du, H., Gilbert, H. J. & Bolam, D. N. (2003) Biochemistry 42, 9316–9323. [DOI] [PubMed] [Google Scholar]
- 36.Chervenak, M. C. & Toone, E. J. (1995) Biochemistry 34, 5685–5695. [DOI] [PubMed] [Google Scholar]
- 37.Srimal, S., Surolia, N., Balasubramanian, S. & Surolia, A. (1996) Biochem. J. 315, 679–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie, H., Bolam, D. N., Nagy, T., Szabo, L., Cooper, A., Simpson, P. J., Lakey, J. H., Williamson, M. P. & Gilbert, H. J. (2001) Biochemistry 40, 5700–5707. [DOI] [PubMed] [Google Scholar]
- 39.Cornish-Bowden, A. (2002) J. Biosci. (Bangalore, India) 27, 121–126. [DOI] [PubMed] [Google Scholar]
- 40.Sharp, K. (2001) Protein Sci. 10, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper, A., Johnson, C. M., Lakey, J. H. & Nollmann, M. (2001) Biophys. Chem. 93, 215–230. [DOI] [PubMed] [Google Scholar]