Abstract
We show that the mechanism for molecular recognition requires one of the interacting proteins, usually the smaller of the two, to anchor a specific side chain in a structurally constrained binding groove of the other protein, providing a steric constraint that helps to stabilize a native-like bound intermediate. We identify the anchor residues in 39 protein–protein complexes and verify that, even in the absence of their interacting partners, the anchor side chains are found in conformations similar to those observed in the bound complex. These ready-made recognition motifs correspond to surface side chains that bury the largest solvent-accessible surface area after forming the complex (≥100 Å2). The existence of such anchors implies that binding pathways can avoid kinetically costly structural rearrangements at the core of the binding interface, allowing for a relatively smooth recognition process. Once anchors are docked, an induced fit process further contributes to forming the final high-affinity complex. This later stage involves flexible (solvent-exposed) side chains that latch to the encounter complex in the periphery of the binding pocket. Our results suggest that the evolutionary conservation of anchor side chains applies to the actual structure that these residues assume before the encounter complex and not just to their loci. Implications for protein docking are also discussed.
In vivo, proteins encounter many potential binding partners. However, a striking set of specific noncovalent interactions encoded in the three-dimensional structure leads proteins to bind to evolutionarily predetermined unique substrates. The detailed mechanism of how proteins accomplish this difficult task is not yet fully understood. Extensive site-directed mutagenesis experiments can account for the main interactions responsible for the stability of the complex structure. However, thermodynamic experiments are not well suited to distinguish between the interactions that are necessary for recognition from those that only provide the sufficient affinity for the regulation of the protein function. Here our main concern is to provide a theoretical understanding of the origin of the specificity of molecular recognition (1–6).
It is well known (2–11) that protein interactions are critically dependent on just a few residues, or hot spots, at the binding interface. Kinetics analyses of mutagenesis experiments can provide clues regarding the role played by individual residues in protein binding (2, 12, 13). In particular, mutations can change the association or dissociation rate of the interacting proteins. One can identify residues important for recognition when their mutations change the rate of association (on rate). These mutations affect the “specificity” between proteins by enhancing or hindering protein recognition. On the other hand, mutations that only change the rate of dissociation (off rate) do not affect the transition state of protein binding. This dichotomy was explored by Kimura et al. (4), who predicted Lys-15 of bovine pancreatic trypsin inhibitor (BPTI) to be the key side chain in the recognition of BPTI by trypsin on the basis of molecular dynamics (MD) simulations, whereas Arg-17 was predicted to be important only for the stability of the high-affinity complex. Indeed, experiments have shown that a Lys-15 → Ala mutation on BPTI leads to a 200-fold decrease in the association rate of BPTI and trypsin and a 1,000-fold increase in the off rate, whereas the mutation Arg-17 → Ala leads to an almost negligible change in the on rate but a similar 1,000-fold increase in the off rate (13). Thus, it is natural to ask what is different in the molecular interactions of these residues that yield such a different kinetic behavior.
Kimura et al. (4) suggested that specific “key” side chains act as ready-made recognition motifs by acquiring native-like conformations before any physical interaction with the receptor. This hypothesis was found to be consistent with MD simulations of three proteins in explicit solvent as well as by the structural conservation of key residues in some protein families. This behavior is reminiscent of the notion of “anchor residues” in peptide-binding motifs of class I MHC molecules (14). In these systems, the C-terminal side chain of the peptide gets buried in pocket F of the MHC binding groove. Sometimes, one also finds a second anchor residue and even a third one buried at other positions. The occurrence of these well defined anchor residues and anchoring grooves, providing the critical stability required for allele-specific recognition, explains in part why each allelic form of class I molecule binds a broad yet defined range of peptides (15, 16).
The motivation of the present study is to generalize the origin of the specificity of molecular recognition in terms of key anchoring residues. Based on the structure of the binding interface, we identify the anchor residues in 39 protein–protein complexes. We perform MD simulations in explicit solvent in 11 different proteins in this set and show that the selected anchoring side chains frequently visit rotamer conformations similar to those observed in the bound complex. We also show that these anchors interact with structurally constrained pockets matching the anchor structures. Finally, we show that residues on the periphery of the binding pocket are found in positions that are suitable to latch to the encounter complex formed once the anchors are docked. The presence of native-like anchor side chains provides a readily attainable geometrical fit that jams the two interacting surfaces, allowing for the recognition and stabilization of a near-native intermediate. Although molecular recognition benefits from this local “lock-and-key” mechanism, a slower, “induced-fit” process on the periphery of the binding pocket is still necessary for proteins to form the high-affinity complex.
Methods
Protein Complexes. The set of complex structures studied here is listed in Table 1. The structures of complexes and their individual component proteins were obtained from the Protein Data Bank (PDB) (17). MD simulations were performed for 11 individually crystallized (unbound) proteins from this set, including enzyme inhibitors, antigens, and other ligands. These proteins were selected because their complex structures were found to be particularly difficult to predict by using rigid-body docking techniques (see table 1 in ref. 18). Moreover, as detailed in Table 1, the relevant side chains on these systems had a wide range of solvent-accessible surface area (SASA) buried after binding. Thus, by analyzing the role of flexible side chains, we hope to also understand why rigid-body docking fails.
Table 1. Predicted anchor residues in 39 complexes.
| Native-like, %
|
||||||
|---|---|---|---|---|---|---|
| Complex PDB ID code | Receptor/ligand (PDB ID code) | Anchor ResID | ΔSASA, Å2 | ΔGi (rank), kcal/mol | MD ± 7% | Rotamer library |
| Enzyme/inhibitor complexes | ||||||
| 1PPE | Trypsin/CMT-I | Arg-5 | 205.9 | -11.3 (1) | ||
| 1AVW | Trypsin/soybean inhibitor | Arg-563 | 202.7 | -13.2 (1) | ||
| 1BRC | Trypsin/APPI (1AAP) | Arg-15 | 198.8 | -11.9 (1) | 32† | 7.4† |
| 1CGI | α-Chymotrypsinogen/PSTI | Tyr-18 | 186.7 | -8.6 (1) | ||
| 1TGS | Trypsinogen/PSTI | Lys-18 | 169.7 | -11.9 (1) | ||
| 1TAB | Trypsin/BBI | Lys-26 | 167.7 | -10.5 (1) | ||
| 2PTC | β-Trypsin/PTI | Lys-15 | 163.8 | -9.9 (1) | ||
| 2SIC | Subtilisin BPN/Inhibitor | Met-70 | 159.4 | -6.8 (1) | ||
| 1DFJ* | RI/ribonuclease A | Tyr-433 | 159.0 | -2.4 (13) | ||
| 2SNI | Subtilisin novo/CI2 (2CI2) | Ile-56 | 148.4 | -7.7 (1) | 37‡ | 96.6‡ |
| 1UGH* | UDG/UGI | Leu-272 | 146.9 | -5.4 (3) | ||
| 1CHO | α-Chymotrypsin/OMTKY3 | Leu-18 | 133.5 | -8.3 (1) | ||
| 1ACB | α-Chymotrypsin/eglinC | Leu-45 | 132.5 | -8.5 (1) | ||
| 2TEC | Thermitase/eglin C | Leu-45 | 118.5 | -5.3 (1) | ||
| 4HTC | α-Thrombin/hirudin | Ile-1 | 116.5 | -9.25 (1) | ||
| 1CSE | Subtilisin C/eglin C (1ACB) | Leu-45 | 112.1 | -5.7 (1) | 50‡ | 97.4‡ |
| 1MAH | AchE/fasciculin II | Met-33 | 109.1 | -2.6 (8) | ||
| 1FSS | AchE/FasII (1FSC) | Met-33 | 87.9 | -2.6 (6) | 97† | 74.1† |
| 1BRS | Barnase/barstar (1A19) | Asp-39 | 84.9 | -10.2 (1) | 90‡ | 22.8‡ |
| 1DFJ | Ribonuclease inhibitor/ribonuclease A (7RSA) | Asn-67 | 69.1 | -1 (15) | 41‡ | 28.5‡ |
| Antigen/antibody complexes | ||||||
| 1BQL | Hyhel5 Fab/QBL (1DKJ) | Arg-45 | 146.8 | -10.7 (1) | 49† | 38.3† |
| 1MLC | IgG1κ D44.1 Fab/HEL | Arg-68 | 133.4 | -7.2 (1) | ||
| 2VIR* | IgG1-λ Fab/HA | Tyr-102 | 128.4 | -3.0 (5) | ||
| 1AHW | Fab 5G9/tissue factor | Lys-169 | 126.7 | -8.1 (2) | ||
| 1MEL | CAb/lysozyme | Trp-62 | 122.3 | -3.8 (1) | ||
| 1IAI* | IgG1/IgG2A | Tyr-105 | 119.4 | -2.8 (1) | ||
| 2JEL | Jel42 Fab/HPR | Gln-71 | 118.2 | -0.4 (6) | ||
| 1JHL | IgG1 Fv/lysozyme | Lys-116 | 111.1 | -6.2 (1) | ||
| 1DQJ | Hyhel63 Fab/HEL (3LZT) | Arg-21 | 104.6 | -0.5 (7) | 92† | 29.1† |
| 1EO8* | Bh151 Fab/HA | Phe-100 | 100.7 | -3.2 (4) | ||
| 1BVK | Hulys11 Fv/lysozyme | Gln-121 | 100.6 | -0.3 (12) | ||
| 1FBI | IgG1 Fab/lysozyme | Lys-97 | 95.6 | -7.6 (2) | ||
| 1NMB | Fab NC10/neuraminidase | Asn-329 | 90 | -1.7 (4) | ||
| 1NCA | Fab NC41/neuraminidase | Thr-401 | 89.7 | -0.9 (8) | ||
| 1QFU | IgG1-k Fab/HA | Ile-62 | 84 | -2.0 (5) | ||
| 1WEJ | IgG1 Fab/Cyt C (1HRC) | Lys-60 | 83.7 | -7.1 (1) | 93† | 87† |
| Others | ||||||
| 1SPB | Subtilisin/Subtilisin prosegment | Tyr-77 | 161.6 | -3.7 (3) | ||
| 1ATN* | Actin/DNase I | Val-45 | 129.4 | -8.6 (1) | ||
| 1A0O | CheY/CheA (1A0OB) | Phe-214 | 125.1 | -2.3 (4) | 29‡ | 15.1‡ |
| 1AVZ | NEF/SH3 domain (1SHF) | Trp-119 | 100 | -1.4 (3) | 36‡ | 35‡ |
The main anchor is in receptor.
Less than 2 Å rms deviation (rmsd) from bound conformers.
Less than 1 Å rmsd from bound conformers.
MD Simulations. The MD-simulation package gromacs 3.1.4 (19) was used for performing simulations on the selected proteins. The simulations were carried out by using simple point charge water molecules. The systems were minimized by using the steepest-descent method under the gromacs force field. Periodic boundary conditions with a rectangular box were applied. The temperature was coupled to a bath of 300 K with a coupling time constant of 0.1 ps. The pressure was restrained to 1 bar (1 bar = 100 kPa) with a coupling time constant of 0.5 ps. A cut-off radius of 1.1 nm was used to calculate the long-range electrostatic interactions. Initial velocities were generated randomly from a Maxwell distribution at 300 K in accordance with the masses assigned to the atoms. The time step was 2 fs.
Simulations were performed on the independently crystallized (unbound) structure by using as initial condition the unbound x-ray structure of the ligand protein and keeping the heavy atoms of the backbone harmonically restrained. Trajectories were sampled at 2-ps intervals. Initial equilibration was done for 5,000 steps, followed by at least 4-ns production runs. It is important to stress that 4-ns runs are not enough to obtain good thermodynamic averages. However, our goal here is only to show that side chains frequently sample native-like conformations within a nanosecond, i.e., the time scale of an encounter complex (20).
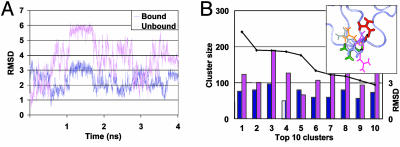
Side-Chain rmsd and Dominant Rotamer Conformation. The side-chain dynamics are analyzed by extracting snapshots from each MD trajectory and overlapping them with the bound protein structure. To avoid systematic errors caused by small differences in the backbone, the bound structure is further translated such that the Cαs of the side chains of interest coincide. Then, for each residue we calculate the rmsd from the corresponding side chain found in the bound structure. The dominant rotamer conformation for a given side chain is determined by calculating the pairwise rmsd for this side chain in the full set of MD snapshots and then clustering the conformations by using a simple greedy algorithm using a clustering radius of 1–2 Å, depending on the residue type. For example, Fig. 1A shows the rmsd of the Arg-39 side chain with respect to the bound (PDB ID code 1DFJ) and unbound (7RSA) conformation along the MD trajectory. Fig. 1B shows the distribution of conformations sampled during the simulation. Fig. 1B Inset shows the cluster center conformation for the largest clusters. The individually crystallized compound is 3.6 Å away from the bound conformation. The center of the dominant cluster, based on the simulation, is 5.9 Å from the unbound conformation and only 2.3 Å from the bound conformation.
Fig. 1.
Side-chain dynamics. (A) The rmsd of Arg-39 of ribonuclease A with respect to the structure found in the complex (1DFJ) and the unbound ligand (7RSA). The rmsd was computed for 2,000 snapshots of a 4-ns MD simulation of 7RSA. (B) Clustering distribution of the conformations of Arg-39 (solid line). The top 10 clusters were derived from a pairwise rmsd analysis of the MD snapshots, by using a clustering radius of 2 Å. Bars indicate the rmsd (left vertical axis) of the side chain in the cluster center with respect to the bound (dark-blue bar) and unbound (pink bar) conformations. (Inset) Cluster centers for the largest clusters as well as the bound (blue), unbound (red), and dominant MD (green) conformations. Note that there is no significant sampling of the unbound rotamer.
SASA. The change in SASA (21) after binding for a given side chain is calculated as ΔSASAα(i) = SASAα(i) – SASAαβ(i), where SASA(i) is the SASA of side chain i, αβ denotes the bound complex, and α denotes only the ligand taken from the bound structure. For each side chain, the percentage of solvent exposure is calculated by using the ratio between its SASA and the standard surface area in a tripeptide segment (22).
Empirical Free-Energy-Based Scoring Function. To estimate the relative importance of the interface residues in binding, we calculate the conformation-dependent portion of the empirical binding free energy by using the expression ΔGi = ΔEelec(i) + ΔGdes(i), where ΔEelec(i) denotes the electrostatic interaction energy between atoms in the ligand residue i and the receptor (calculated by the Coulombic expression with distance-dependent dielectric, ε = 4r), and ΔGdes(i) is an estimate of the desolvation free energy of residue i. The latter is calculated by an empirical atomic contact potential (23) obtained from protein structures by converting frequencies of structural factors. We note that if one further approximates other free-energy contributions such as translational/rotational entropy by a constant [≈5–10 kcal/mol (1 cal = 4.18 J)], then ΔG has been found to be consistent with binding free energies (4, 23).
Results and Discussion
Our goal is to generalize the origin of the specificity of molecular recognition in terms of anchor residues and grooves that stabilize a native-like intermediate.
Identification of Anchor Side Chains from the Geometry of the Binding Interface. Table 1 lists the PDB ID codes and names of 39 complexes considered in this study along with the PDB ID codes of the proteins in which the MD runs were performed. For each complex, we identify the anchor as the residue that both becomes fully buried after binding and results in the largest ΔSASA value among all residues. For each type of protein, ΔSASA values are listed in descending order. For the most part, anchors are on the ligand surface, with only six exceptions marked with an asterisk in Table 1. To give an idea of the energetic importance of these anchors, an empirical estimate of the relative contribution ΔGi of the anchor residue to the binding free energy is shown. In parentheses we note the ranking of this free energy relative to that of the other residues. Then, we show the percentage of the simulation time during which the residue remains native-like or close to its bound conformation, where “close” means within 2 Å rmsd for long side chains (Arg, Lys, and Met) and 1 Å rmsd for others. For comparison, the last column shows the expected probability of the bound rotamer on the unbound structure as estimated by using Dunbrack's rotamer library (24).
For several complexes, the largest ΔSASA buried by a single residue is comparable with the second or even third largest value (<100 Å2). This is the case for many of the antigen/antibody systems listed in Table 1. In these cases, the secondary residues are also identified as anchors that should act cooperatively for the recognition to take place. These residues are not shown in Table 1 but are mentioned in the discussion of Fig. 2.
Fig. 2.
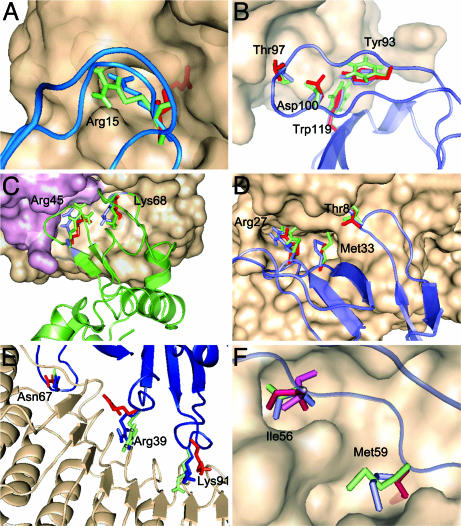
Anchor residues in six complexes. Simulated proteins are shown in cartoon form, and the receptor is shown as surface except for the 1DFJ complex in E. Each anchor side chain is shown in stick conformations that represent the crystal structure of the complex (blue), the individually crystallized ligand (red), and the dominant conformation from the MD simulation (green). (A) Trypsin/APPI complex (1BRC). (B) HIV-1 NEF/FYN tyrosine kinase SH3 domain complex (1AVZ). (C) Hyhel-5 Fab/lysozyme complex (1BQL). (D) Complex of acetylcholinesterase and fasciculin II (1FSS). (E) Ribonuclease inhibitor/ribonuclease A complex (1DFJ). (F) Subtilisin novo/chymotrypsin inhibitor 2 complex (2SNI), the two most dominant rotamers (in green and magenta), are shown for Ile-56.
Considering only the main anchor, we found that Arg and Lys side chains tend to exhibit the largest ΔSASA values; for these anchors, the interface is at least partially polar. Indeed, anchors are polar or charged in 17 of the cases studied. This major role of polar/charged residues is in good agreement with the results of Fernandez and Scheraga (5), who showed that the exclusion of water from the polar regions of the protein surface is an important factor in defining protein–protein associations. Aromatic residues are also prevalent as anchors at the binding interface. These results are supported by the data of Ma et al. (6), who observed a number of conserved polar and aromatic residues close to the middle of the contact region. Finally, in ≈20% of the complexes, aliphatic residues play the role of anchor side chains. Only one complex in the set, barnase/barstar, has an acidic residue as its anchor.
Anchor Side Chains Are Native-Like. According to our MD simulations (Table 1), the anchor side chains spend substantial fractions of the simulation time (30–90%) in rotameric states that are close to the conformation of the side chain in the complex. More interestingly, the preference for the bound state occurs in the absence of the binding partner. Fig. 2 shows six examples of anchor side chains, each in the unbound and bound structure and at the center of the largest rotamer cluster found in the simulation.
Fig. 2A shows the trypsin/amyloid β-protein inhibitor domain (APPI) complex (PDB ID code 1BRC), representing the case in which a single anchor residue dominates the recognition process. Residue Arg-15 of the ligand, APPI, has a ΔSASA value of almost 200 Å2, and its interactions are a major contributor to the total binding free energy. Fig. 2A shows the dramatic difference between the position of Arg-15 in the x-ray structure of the unbound structure and its dominant conformation from the MD simulation. We argue that, because of steric clashes, an attempt to dock trypsin to APPI while Arg-15 is on its unbound rotamer would not be very productive. However, the dominant rotamer observed in the MD is much closer to the bound-state conformation, suggesting a much easier encounter of the two molecules. During the full extent of our simulation, Arg-15 never returned (within 2 Å) to the initial unbound conformation.
The lead anchor of HIV-1 NEF/FYN tyrosine kinase SH3 domain complex (PDB ID code 1AVZ) Trp-119 buries a relatively small area (100 Å2) after binding. Fig. 2B shows Trp-119, which stays within 1 and 2 Å of the bound conformation for 36% and 96% of the MD, respectively. Trp-119 is stabilized in this native-like conformation by Tyr-93 that is almost fully buried (and therefore also native-like) in the free state. Thr-97 buries the second largest ΔSASA (70 Å2) and also resembles the rotamer conformation of the bound state. The energetically important “hot-spot” residue Asp-100 forms a salt bridge with a flexible residue in the receptor Arg-77. However, Asp-100 is already 83% buried (i.e., fully constrained) in the free ligand, thus it cannot be considered as an anchor residue. Overall, the SH3 interface that is found buried in the complex is not very flexible and resembles that of the unbound structure. The latter is not true for side chains in the periphery of the binding interface (Fig. 3B).
Fig. 3.
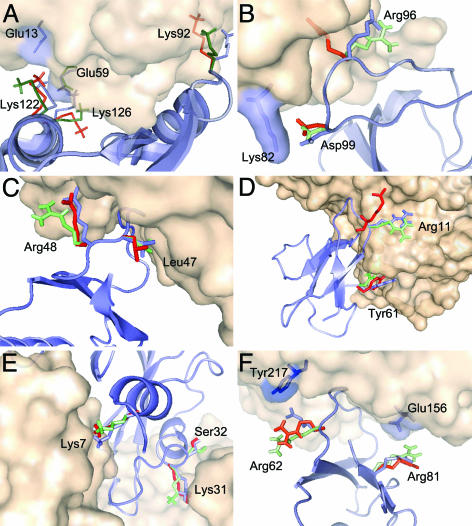
Latch residues in six complexes. Details are as described for Fig. 2. (A) CheA/CheY complex (1A0O). (B) HIV-1 NEF/FYN tyrosine kinase SH3 domain complex (1AVZ). (C) Subtilisin Carlsberg/Eglin C (1CSE). (D) Acetylcholinesterase/fasciculin II complex (1FSS). (E) Ribonuclease inhibitor/ribonuclease A complex (1DFJ). (F) Subtilisin novo/chymotrypsin inhibitor 2 complex (2SNI). In some cases in which the clarity of the picture is not compromised, the interacting residue on the other side of the interface is also shown as sticks inside the surface representation.
The Hyhel-5 Fab/lysozyme complex (PDB ID code 1BQL) is shown in Fig. 2C. The main anchor residue, Arg-45, has a ΔSASA value of 147 Å2; a second anchor residue, Lys-68, is found buried with a ΔSASA = 93 Å2. Both side chains show native-like properties, sampling during 50% and 97% of the time conformations that were less than 2 Å rmsd from their corresponding bound rotamer. It is interesting to recall that lysozyme with the mutation Lys-68 → Arg also was simulated in ref. 4. As expected, despite the mutation, residue 68 remains in an equivalent conformer in both systems. The latter further confirms the importance of the structure of this side chain for recognition.
The complex of acetylcholinesterase with fasciculin II (PDB ID code 1FSS), shown in Fig. 2D, has a large interface that includes several anchor residues with relatively small ΔSASA values. The main anchor Met-33 is in a native-like conformation during most of the simulation. The ΔSASA encompassed by Met-33 is comparable with the next largest ΔSASA of 78 Å2 resulting from the burial of Arg-27; this anchor is in a native-like conformer during 95% of the MD simulation. Similarly, the bound-like residue Thr-8 buries 60 Å2 after binding. We note that anchors in 1FSS are more constrained by the surrounding residues in the free ligand than in other systems; similarly, the grooves into which these residues dock are not as deep. However, as for the SH3 domain protein, peripheral residues are more flexible (see below).
Our analysis focuses on the largest ΔSASA regardless of whether they occur in the ligand, receptor, or both. Our evidence indicates that anchors are more likely to be in the smaller (ligand) protein. The complex of ribonuclease A and ribonuclease inhibitor (1DFJ) (Fig. 2E) has anchor side chains in both receptor and ligand proteins. This protein has a very unique shape and differs from a regular protein–protein interface. In 1DFJ, the ligand (ribonuclease A) sits on top of a horseshoe-shaped receptor (inhibitor), essentially “plugging a hole.” On the receptor side, the anchor side chain Tyr-433 has a large buried area, but we did not study its dynamics by simulations. On the ligand side, our simulations confirm that the lead anchor Asn-67 is native-like (Table 1).
The most striking aspect of ribonuclease A is the dynamics of Arg-39. Despite having a major contribution to the binding free energy and a ΔSASA of >130 Å2, Arg-39 is not a well defined anchor residue, because it is not found fully buried in the complex. Nevertheless, it is interesting to analyze its dynamic behavior, because a straightforward analysis indicates that if after plugging the hole Arg-39 is as in 7RSA, then this side chain would never be able to rearrange to its bound conformation without fully unbinding and trying again (Fig. 2E). However, MD simulations (Fig. 1) unambiguously show that a solvated Arg-39 rapidly turns around from its unbound state and points in the direction of the bound conformation (toward the hole). Similarly, the solvent-exposed Lys-91 is also found in a bound-like conformation during the whole simulation.
Finally, Fig. 2F shows the subtilisin novo with chymotrypsin inhibitor 2 complex (PDB ID code 2SNI), in which the main anchors are Ile-56 and Met-59 (nonpolar residues). In this case, Ile-56 provides a considerable fraction of the binding free energy and remains within 1 Å of the bound conformation for >36% of the simulation time. Interestingly, MD shows that Ile-56 moves back and forth between two states, one being the dominant bound-like state. As shown in Fig. 2F, both states fit nicely in the binding groove. The P1 site for this ligand is Met-59, which buries 119 Å2 after binding. Although the unbound rotamer (Fig. 2F) is found blocking the binding interface, MD shows that, in solution, this side chain should turn around to a native-like position at which it stays for ≈43% of the (simulation) time.
In summary, the interactions of anchor residues with their environment (including solvent) lead their flexible conformations to a state similar to that found in the complex structure. This dynamic rearrangement is not trivial, because a direct comparison between the residence times of anchors in bound-like structures with the probability of finding these same structures based on a rotamer (24) library (Table 1) are quite different.
Structure of the Binding Grooves That Accommodate Anchor Side Chains. As in peptide–MHC interactions, anchor residues are complemented with a well defined native-like groove into which to dock. MD does not significantly change the shape of the pockets, because residues forming them are already almost 80% buried in the free state, and the complexes considered here do not undergo major backbone rearrangements after binding.
For example, the grooves for the anchors of enzymes 1PPE, 1BRC, 1TGS, and 2PTC are made of the Asp and Ser residues of the triad, both with only 5- to 12-Å2 SASA in their corresponding free receptor; the anchoring groove of Trp-62 on the antibody of 1MEL is made up by Ile-102, Tyr-32, and Gly-54, with the first two residues been 62% and 84% buried in the free state, respectively. Several antibody/antigen complexes have more than one anchor that often bury <100 Å2 each. The grooves associated with some of these anchors are broader and less deep than for enzymes. For instance, anchor Lys-68 rests on residues Trp-33 and Glu-50 of the antibody in 1BQL, 60% and 84% buried in the free state, respectively. The second anchor, Arg-45, is constrained by residues Trp-90, Glu-50, and Trp-47, found 63%, 84%, and 97% buried in the free antibody, respectively. For anchors Lys-99 and Lys-60 in 1WEJ, they rest on Tyr-32L–Trp-92L (65% and 55% buried, respectively) and Tyr-33H–Asp-100H–Asp-52H (82%, 66%, and 80% buried, respectively).
These straightforward observations from the complex structures in Table 1 show that both anchor residues and their grooves are native-like. Although anchors are native-like because of nonbonded interactions, grooves are relatively constrained by their own folded structure, providing well defined recognition pockets for the anchor residues to dock.
Kinetic/Energetic Implications of Multiple Anchor Residues. As shown in Table 1, ΔSASA > 150 Å2 for anchors in most enzyme/inhibitor complexes. Anchors are usually functionally important sites. For instance, they correspond to the P1 site for the first eight enzymes in Table 1; also, for 2SIC and 2SNI, P1 sites are secondary anchors. Furthermore, these side chains have the largest contribution to the binding free energy. Most of the anchor residues contribute >6 kcal/mol to the binding free energy, which is consistent with the estimated free energy required for the formation of the native-like intermediate (2). The above notwithstanding, the mechanism emerging from our analysis suggests that the kinetic benefits of having two interacting surfaces jammed together by a relatively large needle-like side chain is critical for recognition.
For systems in which ΔSASA < 100 Å2, the energetic/kinetic contribution of the main anchor is not enough to stabilize the intermediate state. In these cases, we found that a second and (less frequently) third anchor act cooperatively to provide a stronger foothold for the binding to proceed, each anchor having only a limited contribution to the binding free energy.
Partially Solvent Exposed Bonds, or “Latches,” Lock the Native-Like Encounter Complex. Once the encounter complex is formed, the remaining free energy arises from induced interactions of flexible side chains that latch the two proteins. Contrary to anchor residues, latches are relatively free to adjust, because they interact in the periphery of the binding interface and remain 30–70% solvent-exposed even in the complex. Our MD simulations indicate that latch side chains do not necessarily take native-like conformations before encountering their receptor. More interestingly, the MD simulations consistently show that, in solution, latches do not block the binding interface even in cases in which the unbound ligand structure suggests otherwise.
Regardless of whether the main anchor is on the receptor or ligand, latches can be in any or both of the molecules. In most cases, we simulated only the protein with the anchor; therefore, we will mostly describe the dynamics of latches present in these proteins. There are two types of latches. One type corresponds to side chains in one protein that rearrange to form a bond with a relatively rigid residue in the other. A second type is pairs of flexible residues (often forming salt bridges) that simultaneously induce their optimal configuration. In this case, both residues are latches, and both remain with a significant amount of SASA in the complex. Fig. 3 shows examples of these two classes of latches for six systems.
Fig. 3A shows the CheY/CheA complex (PDB ID code 1A0O). Although the anchor in this complex is Phe-56 in CheA, the main latches (Lys-126, Lys-122, and Lys-92) are in CheY, and they are a good example of the types of interactions that lock a complex structure. For this case, we have solvated the unbound crystal structure of CheY (PDB ID code 1CHN). The dominant MD conformations for these large side chains show that, overall, none interfere with the approach of CheY. Indeed, the MD simulations indicate that Lys-92, which in the unbound structure is found somewhat blocking the binding site, moves away from the interface toward a position more amenable to latch once CheA is in a native-like position. Lys-126 moves closer to its bound rotamer and eventually will form a salt bridge with Glu-59, a flexible latch in CheA that is found only 50% buried in the complex. Finally, Lys-122 forms a salt bridge with Glu-13 (80% buried in CheA).
The main latch in the tyrosine kinase SH3 domain (1SHF) of the 1AVZ complex, Asp-99 (59% buried in the complex), is shown in Fig. 3B. Asp-99 buries 41 Å2 after binding, the largest amount of any residue that is not an anchor, and forms a salt bridge with latch Lys-82 in the receptor. The notion that these two side chains would undergo an induced-fit rearrangement is quite apparent in Fig. 3B. The residue Arg-96 is not fully buried and strongly interacts with the receptor. However, it does not qualify as a latch, because its interactions are very unfavorable. It is interesting that the MD simulations move the Arg-96 side chain from a rotamer that blocks the binding interface in the unbound structure to one in which it no longer interferes with the approach of the receptor.
For subtilisin Carlsberg/Eglin C (PDB ID code 1CSE), the main latches are Arg-48 and Leu-47 in Eglin C (Fig. 3C). The MD suggests that Arg-48 moves away from the interface before its encounter with the receptor, whereas Leu-47 stays close to a native-like position.
Fig. 3D shows the acetylcholinesterase/fasciculin II complex (1FSS), in which the main latches on the ligand are Arg-11 (53% buried in the complex) and Tyr-61 (63% buried). Arg-11 moves away from the interface and from the unbound rotamer to a conformer close to the bound structure from where it can easily reach the rigid Glu-79 (84% buried) in the receptor. The C-terminal Tyr-61 interacts with Lys-338 (80% buried in the free state), and it stays in a close-to-native position.
Fig. 3E shows three latches of the ribonuclease inhibitor/ribonuclease A complex (1DFJ); two more latch-like residues, Arg-39 and Lys-91, were discussed earlier (Fig. 2E). As shown in Fig. 3E, the dynamics of Lys-31 and Lys-7 are dominated by native-like conformers. Finally, the subtilisin novo/chymotrypsin inhibitor 2 complex (2SNI) is shown in Fig. 3F. Our MD simulation of the solvated ligand (2CI2) shows the latch side chains Arg-43 and Arg-62 in close-to-native conformations. We note that Arg-43 nicely moves away from the interface before docking.
Conclusions
The analysis of any protein–protein complex at the atomic length scale reveals that the interface, rather than being smooth and flat, includes side chains deeply protruding into well defined cavities on the other protein. It is not well understood yet how two flexible protein surfaces that generally do not have perfect shape complementarity when isolated can associate so rapidly and with moderate energetic costs. The results presented here show that interacting surfaces have evolved by developing very specific anchoring interactions that early on in the binding process bury a ligand (or receptor) side chain deep in a well defined binding groove on the receptor (or ligand), forming a native-like encounter complex.
In all complexes we have studied thus far, the anchor is the side chain whose burial after complex formation yields the largest possible decrease in SASA. These anchors are functionally important residues (P1 site in enzymes), hot spots that have a significant contribution to the free energy, or kinetically important residues with a role in binding that has not yet been assessed experimentally.
Generally, the larger the buried surface of the main anchor residue, the fewer secondary anchors are required to bring about complex formation. If ΔSASA > 100 Å2, the anchoring interaction generally involves a single side chain. However, for ΔSASA < 100 Å2, one anchor is not enough to stabilize the native intermediate, and a second or (less frequently) third native-like side chain is observed at the binding interface. These secondary anchors still bury a large SASA, and the individual interaction of each anchor is not as dominant as for one-anchor proteins.
Once the anchor side chains have docked, the encounter complex is constrained both energetically and kinetically in a weakly bound native-like conformation. This intermediate allows for additional intermolecular interactions to take place. In particular, we showed that, in all cases we tested, latch side chains are found in conformations conducive to a relatively straightforward clamping of the intermediate into the high-affinity complex.
From a kinetic point of view, the greatest benefit from having a native-like motif on the protein surface is a fast recognition process, in which Brownian motion and the partial affinity triggered by anchoring interactions are enough to form the binding intermediate. Thus, we expect these interactions to dominate the on rate of the reaction (4). Once the intermediate is formed, we expect that the likelihood of latches cementing the high-affinity complex would be larger than that of the intermediate detaching (25). Latch bonds are then expected to control the off rate (4). The fact that native-like anchors are fairly stable on a time scale of nanoseconds suggests that recognition of the native-like encounter complex can take place in a few nanoseconds. This estimate is consistent with Brownian dynamics simulations of protein association (26).
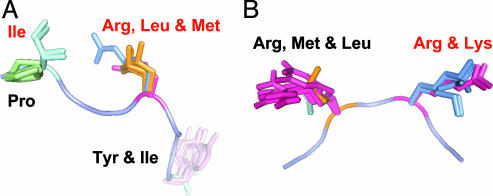
The description of protein association in terms of anchor residues and anchoring grooves generalizes the known mechanism of peptide binding to MHC. Interestingly, the notion that sequence-specific anchor residues have been critical for the evolution of a broad range of peptides might well apply for protein–protein interactions. For instance, Fig. 4 shows two sets of homolog proteins that inhibit seven or more different receptors. In both cases, regardless of the residue type, the conformations of the anchors in the complexes overlap. The overwhelming conservation of the actual structure of anchor residues at the level of the detailed molecular structure of their side chain gives strong support to the notion that anchors play a critical role in binding.
Fig. 4.
Structurally conserved anchor residues for different homologs. Anchor residues are written in red letters. The active site loop from the reference structure is shown with overlap residues from all of the homologs; residues of the same type have the same color. (A) Chymotrypsin inhibitor 2 (from 2SNI) is compared with ligands in PDB ID entries 1LW6, 1SBN, 1MEE, 2SEC, 1ACB, and 3TEC. (B) APPI (from 1BRC) is compared with ligands in PDB ID entries 1BTH, 1BZX, 1CBW, 1EAW, 1F5R, 1FAK, 2KAI, and 1AN1. Here, a small turn in the Lys anchor is observed in the two complexes that inhibit α-chymotrypsin, PDB ID entries 1MTN and 1ACB.
It is well known that one difficulty in using rigid-body techniques to dock proteins is that unbound side chains are often found blocking the binding interface (Figs. 2 and 3). Not surprisingly, these side chains often have large B-factors (≥50) or missing electron densities. At the same time, we cannot rule out that crystallization conditions (including solvent) and crystal packing also might affect the resolution. For example, Arg-39 in ribonuclease A (Fig. 1), a side chain with a crystal conformation that is not preserved by MD, has an intermediate B-factor of ≈25 but also has many crystal water molecules within 5 Å of the residue.
MD simulations indicate that interface side chains tend to be in positions that do not interfere with the binding process. It is tempting to speculate that this dynamical behavior is the consequence of the evolution of specificity in protein interactions. Our results suggest that protein–protein docking can be improved substantially by replacing the unbound x-ray conformation of an anchor side chain by its dominant conformation from MD, because they would generally provide better surface and energy complementarity (D.R., S.V., and C.J.C., unpublished data).
In summary, the generality of our findings provide a compelling scenario in which details of the three-dimensional structure of individual proteins encode the necessary information for them to bind to their unique substrates. The mechanism emerging from the dynamics of solvated proteins indicates that anchor residues provide most of the specificity necessary for protein–protein recognition, whereas latch residues regulate the stability for protein function.
Acknowledgments
This work was supported by National Institutes of Health Grants GM061867 and GM064700.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MD, molecular dynamics; SASA, solvent-accessible surface area; rmsd, rms deviation; APPI, amyloid β-protein inhibitor domain.
References
- 1.Ringe, D. (1995) Curr. Opin. Struct. Biol. 5, 825–829. [DOI] [PubMed] [Google Scholar]
- 2.Schreiber, G. & Fersht, A. R. (1996) Nat. Struct. Biol. 3, 427–431. [DOI] [PubMed] [Google Scholar]
- 3.Camacho, C. J., Weng, Z., Vajda, S. & DeLisi, C. (1999) Biophys. J. 76, 1166–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimura, S., Brower, R., Vajda, S. & Camacho, C. J. (2001) Biophys. J. 80, 635–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez, A. & Scheraga, H. A. (2003) Proc. Natl. Acad. Sci. USA 100, 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma, B., Elkayam, T., Wolfson, H. & Nussinov, R. (2003) Proc. Natl. Acad. Sci. USA 100, 5772–5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clackson, T. & Wells, J. A. (1995) Science 267, 383–386. [DOI] [PubMed] [Google Scholar]
- 8.Novotny, J., Bruccoleri, R. E. & Saul, F. A. (1989) Biochemistry 28, 4735–4749. [DOI] [PubMed] [Google Scholar]
- 9.Bogan, A. A. & Thorn, K. S. (1998) J. Mol. Biol. 280, 1–9. [DOI] [PubMed] [Google Scholar]
- 10.DeLano, W. (2002) Curr. Opin. Struct. Biol. 12, 14–20. [DOI] [PubMed] [Google Scholar]
- 11.Kortemme, T. & Baker, D. (2002) Proc. Natl. Acad. Sci. USA 99, 14116–14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selzer, T., Albeck, S. & Schreiber, G. (2000) Nat. Struct. Biol. 7, 537–541. [DOI] [PubMed] [Google Scholar]
- 13.Castro, M. & Anderson, S. (1996) Biochemistry 35, 11435–11446. [DOI] [PubMed] [Google Scholar]
- 14.Falk, K., Rotzschke, O., Stevanovic, S., Jung, G. & Rammensee, H. (1991) Nature 351, 290–296. [DOI] [PubMed] [Google Scholar]
- 15.Madden, D. R. (1995) Annu. Rev. Immunol. 13, 587–622. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, C., Anderson, A. & DeLisi, C. (1998) J. Mol. Biol. 281, 929–947. [DOI] [PubMed] [Google Scholar]
- 17.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000) Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comeau, S. R., Gatchell, D., Vajda, S. & Camacho, C. J. (2004) Bioinformatics 20, 45–50. [DOI] [PubMed] [Google Scholar]
- 19.Lindahl, E., Hess, B. & van der Spoel, D. (2001) J. Mol. Modeling 7, 302–317. [Google Scholar]
- 20.Northrup, S. & Erickson, H. (1992) Proc. Natl. Acad. Sci. USA 89, 3338–3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, B. & Richards, F. M. (1971) J. Mol. Biol. 55, 379–400. [DOI] [PubMed] [Google Scholar]
- 22.Fraczkiewicz, R. & Braun, W. (1998) J. Comput. Chem. 19, 319–333. [Google Scholar]
- 23.Zhang, C., Vasmatzis, G., Cornette, J. L. & DeLisi, C. (1997) J. Mol. Biol. 267, 707–726. [DOI] [PubMed] [Google Scholar]
- 24.Dunbrack, R. L., Jr. (2002) Curr. Opin. Struct. Biol. 12, 431–440. [DOI] [PubMed] [Google Scholar]
- 25.Camacho, C. J. & Vajda, S. (2001) Proc. Natl. Acad. Sci. USA 98, 10636–10641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camacho, C. J., Kimura, S. R., DeLisi, C. & Vajda, S. (2000) Biophys. J. 78, 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]