Abstract
Over the last 9 years, the structures of the various components of the bacterial photosynthetic apparatus or their homologues have been determined by x-ray crystallography to at least 4.8-Å resolution. Despite this wealth of structural information on the individual proteins, there remains an urgent need to examine the architecture of the photosynthetic apparatus in intact photosynthetic membranes. Information on the arrangement of the different complexes in a native system will help us to understand the processes that ensure the remarkably high quantum efficiency of the system. In this work we report images obtained with an atomic force microscope of native photosynthetic membranes from the bacterium Rhodospirillum photometricum. Several proteins can be seen and identified at molecular resolution, allowing the analysis and modeling of the lateral organization of multiple components of the photosynthetic apparatus within a native membrane. Analysis of the distribution of the complexes shows that their arrangement is far from random, with significant clustering both of antenna complexes and core complexes. The functional significance of the observed distribution is discussed.
In photosynthesis, highly efficient multiprotein assemblies convert sunlight into chemical potential energy. This process requires several different membrane proteins that funnel light energy to the primary reaction center (RC) and then ensure a cyclic electron transfer chain that converts this energy into an electrochemical potential (1) and, finally, an ATP synthase that is able to store the energy in the phosphodiester bond of ATP (2). A challenge in structural biology is to analyze the structural basis of this efficiency in native membranes. More precisely, the relationship between the different components of the system that ensure efficient energy and electron transfer needs to be determined (3, 4).
In photosynthetic bacteria, a large amount of structural information about the individual components of the photosynthetic unit (PSU) is available. The PSU is an assembly made up of the RC associated with the light-harvesting proteins LH1 and LH2, containing chlorophylls and carotenoids. All components cooperate in absorbing light effectively and channeling energy to the RC. In particular, high-resolution structures of two LH2s from Rhodopseudomonas acidophila and Rhodospirillum molischianum and of two RCs from Rhodopseudomonas viridis and Rhodobacter sphaeroides are available (5–9). Electron crystallography data have revealed a hexadecameric assembly of LH1 around the RC in Rhodospirillum rubrum (10, 11). More recently, atomic force microscope (AFM) topographs of native membranes of R. viridis could be acquired. The data unambiguously reported an elliptical hexadecameric arrangement of the LH1 around each RC in a noncrystalline native environment (12). A 4.8-Å x-ray structure of the core complex of Rhodopseudomonas palustris was also elliptical, but with 15 LH1 subunits and one unidentifiable peptide subunit surrounding the RC (13). This structural information and functional data were the basis of multiple models proposed for the assembly of the PSU (4, 14, 15), but no direct data could so far be acquired to elucidate the architecture of this in a native system. AFM (16), with its high lateral resolution (17, 18), high signal-to-noise ratio (19), and the possibility of nanodissecting biological samples (20), provides a unique tool for investigating integrated biological systems. By using these advantages, images at molecular and submolecular resolution of single protein-containing native membranes (21, 22) and of multiprotein complexes were acquired (12).
Materials and Methods
Bacterial Culture and Membrane Preparation. Rhodospirillum photometricum [DSM no. 121, DSMZ (German Collection of Microoganisms and Cell Cultures), Braunschweig, Germany] was grown photoheterotrophically on DSM media 27 at low light intensity (20 W/m2) and harvested in late-log phase. Cells were harvested and washed two times with 1 mM Tris·HCl, pH 7.0, before being broken by two passages through a French pressure cell. Lysates were loaded directly onto 5–60% sucrose gradients and centrifuged for 1.5 h. The membranes correspond to the major pigmented band that sedimented to about 40% sucrose and contain the proteins of the photosynthetic apparatus. The membranes were washed with 1 mM Tris·HCl, pH 8.0, in a centrifugal concentrator and kept at 4°C for AFM analysis.
Partial Purification of Complexes for Protein Composition Analysis. Membranes were solubilized for 8 h with 1% (wt/vol) n-dodecyl-β-maltoside before being layered onto sucrose density gradients (0.2–0.7 M) containing 0.05% n-dodecyl β-maltoside that were centrifuged overnight (16 h at 100,000 × g). The gradients were fractionated and three colored bands were recovered, corresponding to core complexes (near 0.5 M sucrose), LH2 (near 0.35 M sucrose), and cytochrome bc1 (near 0.2 M sucrose). For analysis of membrane protein composition, membranes were solubilized in a buffer (10% glycerol/2% SDS/65 mM Tris·HCl, pH 6.8) at 90°C for 3 min. Denatured proteins were loaded onto 12% denaturing polyacrylamide gels (SDS/PAGE) and developed according to standard laboratory procedures. Gels were either stained for protein with Coomassie blue (Fig. 1) or stained for haem by using tetramethylbenzidine/H2O2.
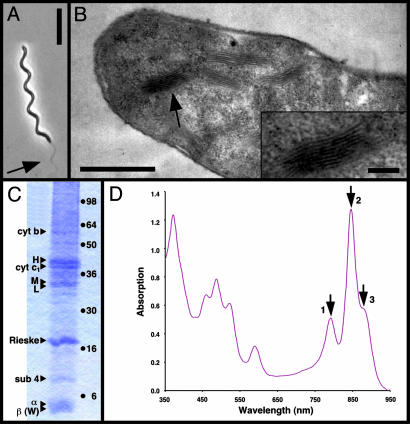
Fig. 1.
Isolation of native membranes from Rsp. photometricum.(A) Phase contrast light microscopy image of Rsp. photometricum. Arrow points at the flagella. (Bar = 10 μm.) (B) Thin section transmission electron microscopy image of Rsp. photometricum. Arrow points at the stacked chromatophore membranes. (Bar = 1 μm.) (Inset) Stack of intracytoplasmic membranes (Bar = 200 nm.) (C) SDS/PAGE analysis of membrane components. Membranes were solubilized and separated on a 12% acrylamide gel before staining with Coomassie blue. Triangles on the left of the gel lane indicate attributions of bands based on molecular weight, haem staining, and partial purification of complexes; points on the right indicate the migration of standard proteins (SeeBlue, Invitrogen). (D) Absorption spectrum of isolated membranes. The three arrows indicate the near-infrared absorption maxima associated with bacteriochlorophyll Qy transitions of the LH2 at 791 nm (1) and 846 nm (2) and LH1 at 882 nm (3). Based on approximate extinction coefficients, the observed absorption spectrum corresponds to about seven LH2 complexes per core complex.
AFM. Mica prepared as described (23) was freshly cleaved and used as a support. The mica was immediately covered by 40 μl of adsorption buffer (10 mM Tris·HCl, pH 7.3/150 mM KCl/20 mM MgCl2). Subsequently, 3 μl of membrane solution was injected into the adsorption buffer drop on the mica surface. After 10 min the sample was rinsed with 10 volumes of recording buffer (10 mM Tris·HCl, pH 7.3/150 mM KCl). Imaging was performed with a commercial Nanoscope-E contact-mode AFM (Digital Instruments, Santa Barbara, CA) equipped with a low-noise laser, and a 160-μm scanner (J-scanner) using oxide-sharpened Si3N4 cantilevers with a length of 100 μm (k = 0.09 N/m; Olympus, Melville, NY). For imaging, minimal loading forces of ≈100 pN were applied; for nanodissection of the top membrane of the vesicles, higher loading forces of ≈600 pN with enhanced scan frequencies (≈6 Hz) were used. High-magnification images (Fig. 3) were recorded at scan frequencies of ≈4 Hz (≈1,200 nm/s) by using optimized feedback parameters. The piezo precision was determined on protein 2D-crystals, at scan ranges between 100 nm and 300 nm, and errors in x- and y-dimensions smaller than 2% were found.
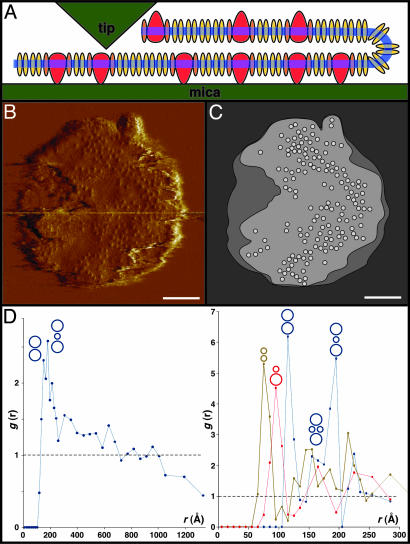
Fig. 3.
Molecular organization of the photosynthetic apparatus. (A Top) Raw-data AFM topograph of a native chromatophore membrane (full-color scale 3 nm). (A Middle) Fitting of the LH2 and core-complex averages corresponding to the relative positions and orientations in the topograph. (A Bottom) Fitting of the LH2 structure, a core-complex model, and a cytochrome bc1 dimer model, corresponding to the relative positions and orientations in the topograph. (B Left) Raw-data AFM topograph of an example of the organization of multiple LH2 around a core complex (full-color scale 3 nm). (B Center) Fitting of the LH2 and core-complex averages corresponding to the relative positions and orientations in the topograph. (B Right) Fitting of the LH2 structure, a core-complex model corresponding to the relative positions, and orientations in the topograph. (Bars = 5 nm.)
Image Analysis. Averages of LH2 and the core complexes were calculated from trace and retrace topographs by using the xmipp single particle analysis program package (24). For this, particles were extracted and were then aligned by using reference-free alignment procedures. For LH2, rotational power spectrum analysis resulted in a peak corresponding to ninefold symmetry (data not shown). Particle selection in the xmipp program package gave access to the x and y coordinates of the particle position within an image, and then the averages were rotationally docked to generate Fig. 3 A Middle and B Center.
Model Building. For the LH2 we used the nonameric Rps. acidophila LH2 structure (PDB ID code 1KZU) (8). For the core complex, a model was built by using the Rhodobacter sphaeroides RC structure (PDB ID code 1M3X) (25) and an elliptical assembly of 16 Phaeospirillum molischianum LH2 subunits (PDB ID code 1LGH) (9). For the bc1 we used a dimer of four subunits of the yeast bc1 structure (PDB ID code 1KB9) (26). The transmembrane helices and the pigment molecules are displayed.
Two-Dimensional Pair Correlation Function. A program was written into which the x and y coordinates of all complexes within a membrane image are entered. From this, the separation vector, r, between all complexes can be calculated. Hence, the probability g(r) of finding a complex (B) in a distance interval from r1 to r2 from a given complex (A) can be normalized, taking into account the surface area covered within the interval and the total number of complexes (B) within the membrane. If the distribution is random, g(r) is always close to 1 (27). For each complex, the distance to the closest membrane border was set as the maximum search radius, rmax.
Results
One of the requirements for AFM of native membranes is the presence of relatively large (>200 nm × 200 nm), flat membrane fragments (12, 22). To obtain fragments of a complex photosynthetic membrane, we used the relatively unknown bacterium Rsp. photometricum (28), closely related to the better-known species Rsp. rubrum. This purple photosynthetic bacterium is particularly large, frequently growing to lengths of >50 μm, as shown in Fig. 1A. The photosynthetic apparatus is associated with stacked, disk-like vesicles of intracytoplasmic membranes (see Fig. 1B, arrow), strongly reminiscent of grana in chloroplasts or the Golgi body in eukaryotic cells. Within these membranes is found a complex mixture of different proteins, including peripheral and core light-harvesting complexes (LH2 and LH1), the RC, and the cytochrome bc1 electron transfer complex, the polypeptides of which are identified on SDS gels, as shown in Fig. 1C. The attribution of bands was based on the molecular weight, haem staining, and partial purification of the complexes (see Materials and Methods). Purified LH2 contained the α and β bands; purified core complexes contained, in addition, the L, M, and H bands. Partially purified RCs were enriched in the L, M, and H bands, whereas partially purified cytochrome bc1 was enriched in the cytochrome b, c1 Rieske, and subunit 4 bands. The cytochrome c1 band was stained for haem (data not shown). The absorption spectrum, which reflects the average light-harvesting distribution, is illustrated in Fig. 1D. The relatively low, broad absorption at 795 nm attributed to the B800 bacteriochlorophyll is a characteristic of the in vivo absorption in this species (28). We have observed this particularity in Rsp. photometricum cells grown under different conditions.
We isolated and deposited the vesicular photosynthetic membranes onto mica and obtained AFM topographs of the surface of these structures (Fig. 2B) that expose their cytoplasmic surface. At this resolution, the relief is provided by the membrane thickness and the proteins that project most from the membrane. The lower mica-attached bilayer exposes a thickness of 7.5 ± 0.3 nm (n = 30), in good agreement with the total thickness of the RC, whereas the top bilayer exposed to the liquid reveals an uneven height profile with an average thickness of 9.5 ± 0.5 nm (n = 30), as expected for a membrane resting on trapped periplasmic components. On these membranes, the main topography observed can be attributed to the photosynthetic RC, the H subunit of which extends 2.8 ± 0.3 nm (n = 24) into the cytoplasm. The assignment of the inside-out orientation of the membranes and of the major protruding domains (Fig. 2B) goes hand in hand. The biochemical and biophysical properties of the purified vesicular membranes can be compared with vesicular intracytoplasmic membranes from other species. Photo-oxidized RCs are still reduced by soluble electron carrier proteins. Furthermore, the protrusion height over the membrane of the putative H subunit compares favorably with topographs of the cytoplasmic surface of core complexes of Rsp. rubrum (29). Finally, the two-dimensional pair correlation function between core complexes calculated on the overview image is identical to the short-range pair correlation between the core complexes in the high magnification images (Fig. 2D).
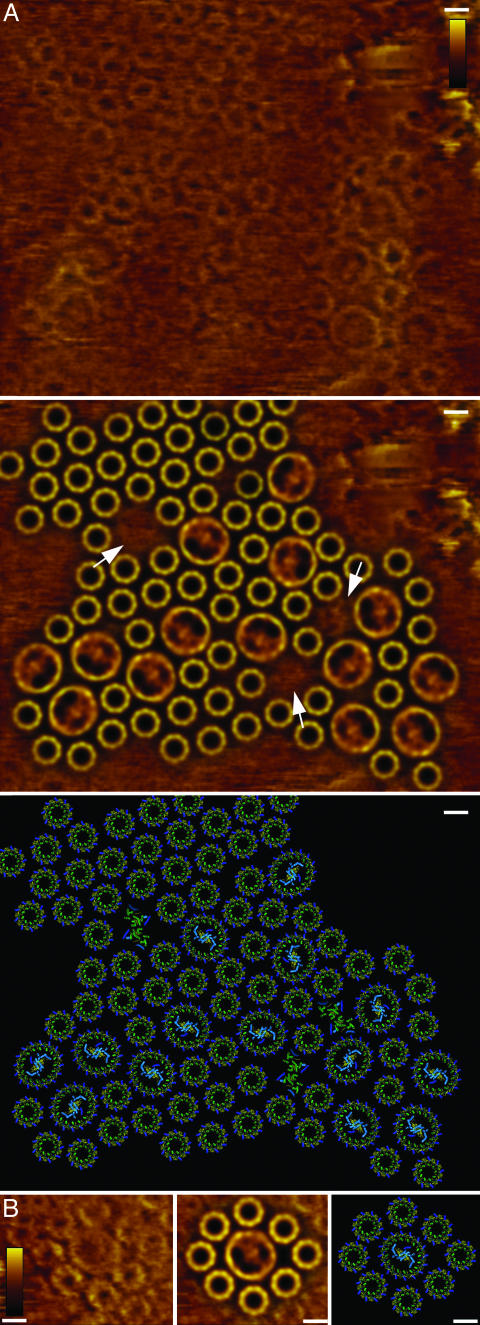
Fig. 2.
Protein distribution within the intracytoplasmic membrane. (A) Schematic presentation of the nanodissection of the membrane vesicles with the AFM tip (red, RC; orange, LH1; yellow, LH2). Before nanodissection of the upper membrane layer the vesicles exposed their cytoplasmic surface, both to the mica and to the tip. (B) AFM deflection image of an intra-cytoplasmic membrane vesicle. The separation of the top membrane from the bottom membrane by nanodissection is visible on the left edge of the vesicle. (Bar = 100 nm.) (C) Outline of the vesicle and position map of the RCs. (Bar = 100 nm.) (D) Graphs of the pair correlation function analysis calculated from overview data (Left) (core–core; n = 3,750 distances) and from the high-resolution data (Right) (LH2–LH2, green line, n = 485; core–LH2, red line, n = 162; and core–core, blue line, n = 49). This function is a measure of the probability of finding a particular type of complex at a certain distance from another complex; for a random distribution this gives a constant value of 1. For LH2–LH2, core–LH2, and core–core, discrete peaks corresponding to favored complex–complex interactions, as indicated by the sketches above the peaks, were found. The curves in Right were calculated from the image in Fig. 3A.
Low-resolution topographs thus allow us to describe the overall distribution of the RCs (Fig. 2C). The RCs are not randomly distributed and, indeed, appear to be significantly clustered. To better analyze this distribution we have calculated the two-dimensional pair correlation function. For a random distribution this function gives a constant value of 1. However, when calculated for the RCs, we obtain a high probability of finding another RC at a short distance (up to ≈25 nm) from the first and a value significantly lower than unity at long distances (larger than 100 nm) (Fig. 2D Left). These probabilities confirm that the distribution is far from random and suggest relatively strong attractive interactions, either direct or indirect, between the RCs as well as significant large-scale clustering. Other relatively large regions of membrane appear to be devoid of RCs. Closer examination of these regions showed that they contained hexagonally packed LH2.
Because of tip geometry and the scanning mechanism, only relatively flat and firmly attached samples can be imaged at high lateral and vertical resolution with the AFM. On the membrane vesicles, the upper membrane layer could not be imaged at high resolution because of rugosity and weak attachment. To alleviate these problems we removed the upper membrane layer by nanodissection with the AFM tip (Fig. 2 A).
This protocol allowed us to obtain high-resolution topographs of the firmly attached lower bilayer exposing the periplasmic membrane surface. On this, we could study the precise organization of the photosynthetic apparatus. The largest membrane patch, representative for the sample, that we can describe at molecular resolution (Fig. 3A Top) covers a surface area of 12,000 nm2 and contains 14 RCs surrounded by elliptical LH1 in assembly with 72 LH2 (Fig. 3A Middle).
The LH2 forms circular assemblies with a top-ring diameter of 50 Å and ninefold symmetry (30). In some areas the LH2 are organized in a quasicrystalline hexagonal packing with dimensions of a = b = 77 ± 3 Å; γ = 60 ± 2° (n = 18). The LH2 in Rps. acidophila, Rhodovulum sulfidophilum, Rhodobacter sphaeroides, and Rubrivivax gelatinosus possess a nonameric organization (8, 30–34), but the closely related bacterium Phaeospirillum (formerly Rsp.) molischianum has an octameric LH2 (9). The core complexes observed are formed of a closed ellipse of LH1 around the RC. In agreement with previous studies (11, 12), the LH1, with major and minor axis lengths of 100 Å and 90 Å, is large enough to house 16 subunits. However, our resolution is insufficient to distinguish a closed hexadecameric structure from a core complex containing 15 LH1 subunits and a W subunit, as observed in the core complex of Rps. palustris (13). The resolution is sufficient, though, to exclude an LH1 arrangement with a large gap, as observed in the dimeric core complexes of Rhodobacter sphaeroides (35–37). The RC reveals an asymmetric topography within the ellipse, because of the one-sided topography contribution of the periplasmic surface of the H subunit, in addition to the quasisymmetric L and M subunits (12, 13, 29).
Based on the raw-data images (Fig. 3 A Top and B Left), we can propose a model of multiprotein assemblies in native photosynthetic membranes. The complexes can be fitted first by their averages (Fig. 3 A Middle and B Center) and second by the atomic models (Fig. 3 A Bottom and B Right), taking their position and rotational orientation into account. Some regions in the AFM topograph (Fig. 3, arrows) cannot be easily assigned to previously imaged complexes (12, 30). We can speculate that these regions house cytochrome bc1 dimers (38), because (i) these photosynthetic membranes contain, besides the LH and the RC components, the cytochrome bc1 complex (Fig. 1C), (ii) there is no topography resembling LH2 in these regions, and (iii) these regions are too small to house a core complex but large enough to contain a bc1 dimer (Fig. 3A Bottom). It is possible that the more protruding domains of these proteins were damaged during nanodissection of the top layer of the chromatophore vesicles or show high flexibility.
To analyze the lateral organization at different scales and to better understand the relationship between the various components in the membrane we calculated pair correlation functions (Fig. 2D, ref. 27) from the high-resolution (Fig. 3A) and the overview (Fig. 2B) data. From the high-resolution topographs we were able to calculate, in addition to the core–core correlation function, the LH2–LH2 and core–LH2 correlation functions. In each case we observed several strong, sharp, short distance peaks, which we can interpret in terms of particular favored molecular architectures, illustrated in Fig. 2D Right. In the LH2–LH2 curve the first peak is at 75 Å, in good agreement with the 77-Å hexagonal packing parameter. In the core–LH2 curve the first peak is at 95 Å, corresponding to an LH2 in direct contact with a core (Fig. 3B). In the core–core curve the first peak, at 115 Å, corresponds to two cores in direct contact; this is slightly less than the 133-Å distance between two cores in contact determined from electron microscopy of Blastochloris viridis membranes (39). The second peak is at 195 Å, about twice the 95-Å peak corresponding to the core–LH2 distance, which we therefore assign to an LH2 sandwiched between two cores. This peak has a strong shoulder near 160 Å, corresponding to a structure with two cores bridged by a pair of LH2 complexes. Importantly, we observe the same organization for core–core correlations in data from both the high-resolution (Fig. 2D Right) and overview (Fig. 2D Left) topographs.
Discussion
The lateral organization that we observe is important for understanding the light-harvesting and electron transfer processes. Although there is no fixed photosynthetic unit structure (4, 14), the assembly is highly organized. The RCs are clustered, as is already visible from the low-resolution topographs. This observation is important because it has already been suggested by functional studies in a number of different organisms that the RCs are grouped together (40) to explain the efficiency and saturation behavior of the RC. This clustering of RCs was originally suggested on the basis of triplet fluorescence quenching (41). Functionally, it increases the possibility of an excitation being trapped by an RC under conditions where the rate of electron transport and photon capture are approximately balanced. Under these circumstances, an excitation as it diffuses through the antenna system will find, with a reasonable probability, a first RC that is already occupied in performing the initial photochemical reactions. The clustering of several core complexes means that the excitation can rapidly continue its diffusion, thus increasing the chances of the excitation finding an RC ready to perform primary photochemistry.
Furthermore, the LH2 complexes are themselves strongly clustered. This clustering does not cause a separation of core complexes and LH2, ensuring the connectivity between the two, as is shown in Fig. 3B, where a core complex is imaged surrounded by eight LH2 complexes (14). However, the arrangement does ensure the absence of single isolated LH2 complexes unconnected from the light-harvesting system. An LH2 complex lacking physical contact to other light-harvesting components is nonfunctional, i.e., its excitation energy will be lost upon photon capture. The observation of a partial segregation of LH2 and core complexes is intriguing because, on the one hand, it increases RC connectivity, but on the other it raises the average distance between an LH2 and RC. However, it should be noted that on the basis of the expected hopping rate between LH2 complexes (≈4 ps) and the LH2 fluorescence lifetime (≈1 ns), the excitation will be able to travel a significant distance, 250 hops of a random walk on the antenna system, to find an RC.
In this article, we have been able to observe and analyze the heterogeneous lateral organization of a complex native membrane. Our AFM topographs yield information about the organization of the PSU, i.e., how LH2 arrange around the core complex. Surprisingly, a well confined PSU does not exist: whereas core complexes can be found surrounded by a ring of eight LH2 complexes (14), core complexes in contact with other core complexes, without intercalating LH2, are found with high probability. This heterogeneity was not previously suspected in these photosynthetic membranes and suggests that biological membranes might contain many local domains specialized for particular functions, beyond the well documented lateral heterogeneity of, for example, lipid rafts in eukaryotic membranes.
Acknowledgments
We thank Dr. Daniel Picot for help with model assembly. This work was supported by the Institut Curie, the Commissariat à l'Energie Atomique, the Centre National de la Recherche Scientifique, and European Economic Community Grant HPRM-CT-2002-00269 (to S.S.).
Abbreviations: AFM, atomic force microscope/microscopy; LH, light-harvesting protein; PSU, photosynthetic unit; RC, reaction center.
References
- 1.Bibby, T., Nield, J., Partensky, F. & Barber, J. (2001) Nature 413, 590. [DOI] [PubMed] [Google Scholar]
- 2.Abrahams, J. P., Leslie, A. G. W., Lutter, R. & Walker, J. E. (1994) Nature 370, 621–628. [DOI] [PubMed] [Google Scholar]
- 3.Kühlbrandt, W. (1995) Nature 374, 497. [Google Scholar]
- 4.Hu, X., Ritz, T., Damjanovic, A., Autenrieth, F. & Schulten, K. (2002) Q. Rev. Biophys. 35, 1–62. [DOI] [PubMed] [Google Scholar]
- 5.Deisenhofer, J., Epp, O., Miki, K., Huber, R. & Michel, H. (1984) J. Mol. Biol. 180, 385–398. [DOI] [PubMed] [Google Scholar]
- 6.Deisenhofer, J., Epp, O., Miki, K., Huber, R. & Michel, H. (1985) Nature 318, 618–624. [DOI] [PubMed] [Google Scholar]
- 7.Allen, J. P., Feher, G., Yeates, T. O., Komiya, H. & Rees, D. C. (1987) Proc. Natl. Acad. Sci. USA 84, 6162–6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDermott, G., Prince, S. M., Freer, A. A., Hawthornthwaite-Lawless, A. M., Papiz, M. Z., Cogdell, R. J. & Isaacs, N. W. (1995) Nature 374, 517–521. [DOI] [PubMed] [Google Scholar]
- 9.Koepke, J., Hu, X., Muenke, C., Schulten, K. & Michel, H. (1996) Structure (London) 4, 581–597. [DOI] [PubMed] [Google Scholar]
- 10.Karrasch, S., Bullough, P. A. & Ghosh, R. (1995) EMBO J. 14, 631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jamieson, S. J., Wang, P., Qian, P., Kirkland, J. Y., Conroy, M. J., Hunter, C. N. & Bullough, P. A. (2002) EMBO J. 21, 3927–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheuring, S., Seguin, J., Marco, S., Lévy, D., Robert, B. & Rigaud, J.-L. (2003) Proc. Natl. Acad. Sci. USA 100, 1690–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roszak, A. W., Howard, T. D., Southall, J., Gardiner, A. T., Law, C. J., Isaacs, N. W. & Cogdell, R. J. (2003) Science 302, 1969–1972. [DOI] [PubMed] [Google Scholar]
- 14.Papiz, M. Z., Prince, S. M., Hawthornthwaite-Lawless, A. M., McDermott, G., Freer, A. A., Isaacs, N. W. & Cogdell, R. J. (1996) Trends Plant Sci. 1, 198–206. [DOI] [PubMed] [Google Scholar]
- 15.Hu, X., Damjanovic, A., Ritz, T. & Schulten, K. (1998) Proc. Natl. Acad. Sci. USA 95, 5935–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binnig, G., Quate, C. F. & Gerber, C. (1986) Phys. Rev. Lett. 56, 930–933. [DOI] [PubMed] [Google Scholar]
- 17.Engel, A., Schoenenberger, C.-A. & Müller, D. J. (1997) Curr. Opin. Struct. Biol. 7, 279–284. [DOI] [PubMed] [Google Scholar]
- 18.Müller, D. J., Fotiadis, D., Scheuring, S., Müller, S. A. & Engel, A. (1999) Biophys. J. 76, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engel, A., Gaub, H. & Müller, D. J. (1999) Curr. Biol. 9, R133–R136. [DOI] [PubMed] [Google Scholar]
- 20.Hoh, J. H., Lal, R., John, S. A., Revel, J.-P. & Arnsdorf, M. F. (1991) Science 253, 1405–1408. [DOI] [PubMed] [Google Scholar]
- 21.Müller, D. J., Schabert, F. A., Büldt, G. & Engel, A. (1995) Biophys. J. 68, 1681–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fotiadis, D., Liang, Y., Filipek, S., Saperstein, D. A., Engel, A. & Palczewski, K. (2003) Nature 421, 127–128. [DOI] [PubMed] [Google Scholar]
- 23.Schabert, F. A. & Engel, A. (1994) Biophys. J. 67, 2394–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marabini, R., Masegosa, I. M., San Martin, C., Marco, S., Fernandez, J. J., de la Fraga, C. G. & Carazo, J. M. (1996) J. Struct. Biol. 116, 237–240. [DOI] [PubMed] [Google Scholar]
- 25.Camara-Artigas, A., Brune, D. & Allen, J. P. (2002) Proc. Natl. Acad. Sci. USA 99, 11055–11060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lange, C., Nett, J. H., Trumpower, B. L. & Hunter, C. N. (2001) EMBO J. 20, 6591–6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baksh, M. M., Jaros, M. & Groves, J. T. (2004) Nature 427, 139–141. [DOI] [PubMed] [Google Scholar]
- 28.Giesberger, G. (1947) Antoine van Leeuwenhoek J. Microbiol. Ser. 13, 135–148. [Google Scholar]
- 29.Fotiadis, D., Qian, P., Philippsen, A., Bullough, P. A., Engel, A. & Hunter, C. N. (2004) J. Biol. Chem. 279, 2063–2068. [DOI] [PubMed] [Google Scholar]
- 30.Scheuring, S., Reiss-Husson, F., Engel, A., Rigaud, J.-L. & Ranck, J.-L. (2001) EMBO J. 20, 3029–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montoya, G., Cyrklaff, M. & Sinning, I. (1995) J. Mol. Biol. 250, 1–10. [DOI] [PubMed] [Google Scholar]
- 32.Walz, T., Jamieson, S. J., Bowers, C. M., Bullough, P. A. & Hunter, C. N. (1998) J. Mol. Biol. 282, 833–845. [DOI] [PubMed] [Google Scholar]
- 33.Ranck, J.-L., Ruiz, T., Pehau-Arnaudet, G., Arnoux, B. & Reiss-Husson, F. (2001) Biochim. Biophys. Acta 1506, 67–78. [DOI] [PubMed] [Google Scholar]
- 34.Scheuring, S., Seguin, J., Marco, S., Lévy, D., Breyton, C., Robert, B. & Rigaud, J.-L. (2003) J. Mol. Biol. 325, 569–580. [DOI] [PubMed] [Google Scholar]
- 35.Jungas, C., Ranck, J.-L., Rigaud, J.-L., Joliot, P. & Vermeglio, A. (1999) EMBO J. 18, 534–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scheuring, S., Francia, F., Busselez, J., Melandri, B., Rigaud, J.-L. & Lévy, D. (2004) J. Biol. Chem. 279, 3620–3626. [DOI] [PubMed] [Google Scholar]
- 37.Siebert, C. A., Qian, P., Fotiadis, D., Engel, A., Hunter, C. N. & Bullough, P. A. (2004) EMBO J. 23, 690–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia, D., Yu, C. A., Kim, H., Xia, J. Z., Kachurin, A. M., Zhang, L., Yu, L. & Deisenhofer, J. (1997) Science 277, 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ikeda-Yamasaki, I., Odahara, T., Mitsuoka, K., Fujiyoshi, Y. & Murata, K. (1998) FEBS Lett. 425, 505–508. [DOI] [PubMed] [Google Scholar]
- 40.Drews, G. (1985) Microbiol. Rev. 49, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monger, T. G. & Parson, W. W. (1977) Biochim. Biophys. Acta 460, 393–407. [DOI] [PubMed] [Google Scholar]