Abstract
The plant circadian clock allows the anticipation of daily changes to the environment. This anticipation aids the responses to temporally predictable biotic and abiotic stress. Conversely, disruption of circadian timekeeping severely compromises plant health and reduces agricultural crop yields. It is therefore imperative that we understand the intricate regulation of circadian rhythms in plants, including the factors that affect motion of the transcriptional clockwork itself.
Testing circadian defects in the model plant Arabidopsis thaliana (Arabidopsis) traditionally involves crossing specific mutant lines to a line rhythmically expressing firefly luciferase from a circadian clock gene promoter. This approach is laborious, time-consuming, and could be fruitless if a mutant has no circadian phenotype. The methodology presented here allows a rapid initial assessment of circadian phenotypes. Protoplasts derived from mutant and wild-type Arabidopsis are isolated, transfected with a rhythmically expressed luminescent reporter, and imaged under constant light conditions for 5 days. Luminescent traces will directly reveal whether the free-running period of mutant plants is different from wild-type plants. The advantage of the method is that any Arabidopsis line can efficiently be screened, without the need for generating a stably transgenic luminescent clock marker line in that mutant background.
Keywords: Plant Biology, Issue 115, Circadian clocks, Chronobiology, Arabidopsis thaliana, Protoplasts, Luminescent imaging, Transfection, Clock genes, Phenotyping
Introduction
Most living organisms possess an endogenous timekeeper to aid efficient negotiation of daily changes to the environment. This timekeeper, the circadian clock, regulates many aspects of the metabolism and physiology of an organism to anticipate predictable changes in the external environment. In plants for example, anticipation of temporally predictable pathogen or herbivore attacks strongly reduces overall susceptibility1-4. Starch metabolism is tightly regulated by the circadian clock to ensure that starch reserves last until dawn whether grown at long or short day conditions5. Indeed, plants with circadian clocks matching the 24 hr environment show increased growth rates, carbon fixation rates and survival rates compared to plants running a clock of mismatched period6. The extensive influence of circadian rhythms in all these processes arises to a large extent from the rhythmic regulation of up to a third of the total transcriptome in Arabidopsis7. These rhythmic gene products are involved in a broad range of cellular processes including metabolic pathways, hormone signaling, and stress responses7. On top of that, direct circadian regulation of protein function is established by circadian regulation of phosphorylation8 and most probably other post-translational modifications (PTMs).
At the center of the circadian clock that drives this daily transcriptional reprogramming lies a network of transcriptional/translational feedback loops (TTFLs), including the morning-expressed genes CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) that repress the expression of the evening genes TIMING OF CAB 1 (TOC1), GIGANTEA (GI) and the members of the evening complex (EC); LUX ARRHYTHMO (LUX), EARLY FLOWERING 3 (ELF3), and ELF49-11. Together with the PSEUDO RESPONSE REGULATOR-genes (PRR3, 5, 7, and 9), TOC1 represses CCA1/LHY expression whereas the EC acts as a positive regulator12-14. Additional to feedback mechanisms, post-transcriptional and post-translational regulation of the TTFL network components plays an important role in tuning circadian rhythms. To date, the most abundant PTM identified on circadian clock proteins is phosphorylation15. The phosphorylation of CCA1 by Casein Kinase 2 (CK2) affects its binding to target promoters16 and is important for temperature compensation of the circadian clock17. The PRR proteins are phosphorylated differentially over the circadian cycle, and phosphorylation of TOC1 affects interaction with its negative regulator, the evening-phased clock component ZEITLUPE (ZTL)18. These examples illustrate how PTMs and protein-protein interactions tune the TTFL network into a 24-hr transcriptional oscillator. For a more detailed introduction to the transcriptional clock network and its regulation, excellent recent reviews are available (e.g. reference15).
However, what remains less clear is the extent to which rhythmically regulated processes and other aspects of cellular metabolism feed back into the timekeeping mechanism itself. Extensive screening of Arabidopsis mutants for clock defects could gain further understanding of which mechanisms or signaling pathways could be involved in the generation of 24 hr rhythms from a TTFL network. To this end, Kim and Somers previously published a break-through method for the efficient and rapid analysis of clock function in any Arabidopsis line19. Based on the use of transient expression in protoplasts, this methodology surpasses the need for stable transgenic lines or crosses to luminescent clock marker lines. Here, we visualize a high yielding protoplast isolation method20 combined with an optimized protocol to screen circadian defects by luminescent imaging of transiently expressed clock markers in a 96-well plate reader.
We will compare wild-type Col-0 Arabidopsis to well-characterized clock mutants to confirm the methodology described here successfully detects altered circadian rhythms. Protoplasts derived from these lines are transfected with a reporter construct that expresses firefly luciferase from the circadian CCA1 promoter; CCA1pro:LUC19. Luciferase catalyzes the multistep reaction in which luciferin is converted to electronically excited oxyluciferin that emits light21, which is imaged over time to assess the period, amplitude and phase of transcriptional rhythms in these protoplasts.
The protocol consists of three major parts; isolating the protoplasts, transfecting the protoplasts with the reporter plasmid, and luminescent imaging. These three parts should always be performed on a single day, using freshly prepared buffers and reagents. Plant growth and purification of sufficient amounts of reporter plasmid should be performed in advance and are not visualized in this protocol.
Protocol
NOTE: In this protocol, reagents and volumes described are expressed per Arabidopsis line that is tested, and will result in six replicate wells for that genotype. Multiply the materials with the number of lines to be analyzed when analyzing more than one line. In the video, wild-type Arabidopsis will be compared to the circadian clock mutant line ztl (although representative results from additional lines will be provided subsequently).
| Enzyme solution | |
| Cellulase | 0.5% (w/v) |
| Pectinase R10 | 0.25% (w/v) |
| D-mannitol | 400 mM |
| CaCl2 | 10 mM |
| KCl | 20 mM |
| Bovine Serum Albumin | 0.1% (w/v) |
| MES, pH 5.7 | 20 mM |
| W5 solution | |
| NaCl | 150 mM |
| CaCl2 | 125 mM |
| KCl | 5 mM |
| MES, pH 5.7 | 2 mM |
| Glucose | 5 mM |
| MMg solution | |
| MES, pH 5.7 | 4 mM |
| D-mannitol | 400 mM |
| MgCl2 | 15 mM |
| PEG solution | |
| PEG4000 | 40% (w/v) |
| D-mannitol | 200 mM |
| CaCl2 | 100 mM |
| Imaging solution | |
| W5 solution | to final volume |
| Fetal Bovine Serum | 5% (v/v) |
| Luciferin | 1.2 mM |
| Ampicillin | 50 mg/ml |
Table 1: List of solutions.
1. Preparation of Materials and Buffers
- Plant Materials
- Keep Arabidopsis Col-0 seeds in water in a 1.5 ml tube at 4° C in the dark for four days. Pipette the seeds onto soil in a pot and transfer to long day conditions (16 hr light 8 hr dark) at 21 °C for 7 days. Leave a lid on the pot for the first three days to ensure high humidity. Transplant six seedlings to a new 8 cm x 13 cm x 5 cm pot and continue growing the plants under the same conditions for another 21 days. Make sure the soil is moist throughout.
- Reporter Plasmid
- Purify the CCA1pro:LUC reporter plasmid19 from bacterial culture in advance, using a DNA isolation kit according to manufacturer's instructions. NOTE: 20 µg reporter plasmid is required (10 µl at 2 µg/µl in dH2O) for a transfection.
- Solutions NOTE: For this protocol five solutions are needed: Enzyme solution, Wash 5 (W5) solution, Mannitol-Magnesium (MMg) solution, polyethylene glycol (PEG) solution, and imaging solution (Table 1). Prepare these before continuing to the protoplast isolation step.
- Prepare 10 ml of enzyme solution as per Table 1, adding the enzymes last, and filter sterilize the solution through a 0.22 µm syringe filter.
- Prepare the remaining solutions in the following volumes: 15 ml W5 solution, 10 ml MMg solution, 1 ml PEG solution and 1.5 ml imaging solution as per Table 1. NOTE: It is important that the PEG solution is prepared no less than an hr before the transfection to ensure that the PEG has fully dissolved. Filter sterilize the imaging solution through a 0.22 µm syringe filter.
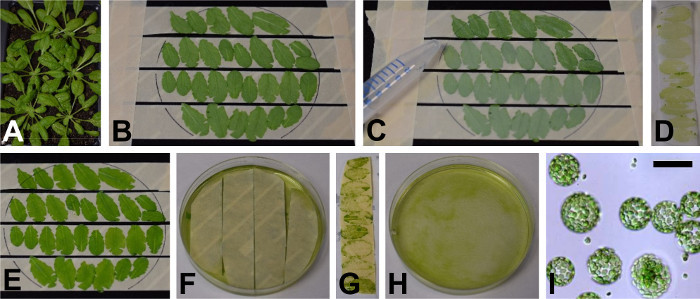
Figure 1: Protoplast isolation. The leaves of adult Arabidopsis plants (A) are fixed on autoclave tape with the lower epidermal layer facing up (B). (C) Magic tape is gently pressed onto the lower epidermal layer with the tip of a 15 ml conical tube. (D) The magic tape is removed to expose the mesophyll cells (E). (F) Strips of autoclave tape with leaves attached are floated on enzyme solution containing cellulase and pectinase, releasing protoplasts into the solution. (G) The leaf skeletons remain on autoclave tape after enzyme digestion and are discarded, leaving the protoplasts in enzyme solution (H). (I) Final resulting mesophyll protoplasts (Scale bar = 50 µm). Please click here to view a larger version of this figure.
2. Protoplast Isolation
Label a petri dish and add 10 ml filter-sterilized Enzyme solution.
Fix four strips of autoclave tape with the sticky side up on a clean lab surface, and mark the size of the petri dish on the strips.
Cut leaves of 4-week old plants (Figure 1a), and press the upper epidermis onto the autoclave tape (i.e. the lower epidermal surface facing up (Figure 1b)).
Gently press strips of magic tape onto the lower epidermal surface using a 15 ml conical tube. Take care not to crush the leaf tissue (Figure 1c). Carefully pull the magic tape off to strip the lower epidermis (Figure 1d) and expose the mesophyll cells (Figure 1e).
Cut the autoclave tape to fit the petri dish, use tweezers to move the tape into the petri dish and float on the enzyme solution (Figure 1f), leaves facing down. Rotate at 60 rpm on a platform shaker for 60 min, during which the protoplasts will be released into the solution.
Use tweezers to remove and discard the strips of autoclave tape (Figure 1g) and pipette the solution containing the protoplasts (Figure 1h) into a 50 ml conical tube. Use a wide-bore pipette tip (such as a P5000 tip or a P1000 tip with the end cut off).
Centrifuge the protoplasts at 100 x g for 3 min at 4 °C and remove the supernatant with a pipette. Take care, as the pellet is very loose.
Add 10 ml W5 solution to the protoplasts and resuspend by gently swirling the tube.
Rest the protoplasts on ice for 30-60 min. During this step, assess yield by counting protoplasts in a hemocytometer (Figure 1i).
Collect the protoplasts by centrifugation at 100 x g for 3 min at 4 °C and remove the supernatant with a pipette. Take care, as the pellet is very loose.
Resuspend the protoplasts to a concentration of 5 x 105 protoplasts/ml in MMg solution.
3. Protoplast Transfection
Add 10 µl reporter plasmid (2 µg/µl) to a 15 ml conical tube.
Add 400 µl protoplasts to the tube.
Add one volume (410 µl) PEG solution and mix by inverting the tube gently 12 times to transfect the protoplasts.
After 8-15 min incubation at room temperature, dilute the protoplast-DNA-PEG mixture with four volumes (1640 µl) W5 solution and mix by inverting gently six times.
Collect the transfected protoplasts by centrifugation at 100 x g for 2 min at room temperature and remove the supernatant with a pipette. Once again, take care, as the pellet is very loose.
Resuspend the protoplasts in 1,250 µl imaging solution.
Aliquot 200 µl into six replicate wells of a 96-well plate, fill any empty wells with W5 solution, and seal the plate with an adhesive clear lid suitable for luminescent imaging.
4. Imaging
Transfer the plate to any luminescent imaging setup suitable for plant imaging. Change the adhesive lids on the plates 10-15 min after transfer to avoid condensation and pressure differences among the wells, then start imaging. Read the plate every ~45 min (3 sec per well) for five days at room temperature. NOTE: The frequency of plate reads and the read time per well might vary depending on the imaging setup.
5. Data Analysis
Analyze the luminescence results using any analysis software (for example, Biological Rhythms Analysis Software System (BRASS)22). Compare the mutant line to wild-type controls to reveal circadian defects.
Representative Results
Protoplasts of wild-type Arabidopsis were compared to the known clock mutants cca1/lhy (short period mutant2), ztl (long period mutant23), and the overexpression line CCA1-OX (arrhythmic24). All were transiently transfected with the CCA1pro:LUC reporter and imaged in constant light on a plate reader over 5 days.
Here we show that in the cca1/lhy mutant, the free-running period of circadian-controlled gene expression is shortened (Figure 2a), in line with the published period analyzed by stable transgenic expression of luminescent clock markers2,9. The period of circadian oscillations in protoplasts of the ztl mutant is longer than in wild type (Figure 2b), in accordance with previously published results from seedlings, cell suspension cultures and protoplasts19,23,25.
Overexpression of CCA1 locks the clock in morning-phase and global transcription becomes arrhythmic24. Consistently, we observed no oscillations in CCA1pro:LUC expression in CCA1-OXprotoplasts (Figure 2c).
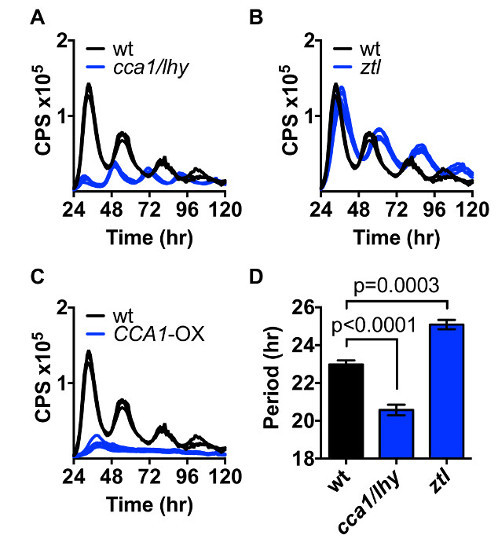 Figure 2:Protoplast Circadian Rhythms Protoplasts of Col-0, cca1/lhy, ztl, and CCA1-OXwere transiently transformed with the CCA1pro:LUC construct and circadian rhythms in luminescence were imaged over 5 days (A-C). (D) Circadian period estimates based on luminescence traces in Col-0, cca1/lhy and ztl (A-B). Mean ±SEM, n = 3-5, t-test p-value as indicated. Please click here to view a larger version of this figure.
Figure 2:Protoplast Circadian Rhythms Protoplasts of Col-0, cca1/lhy, ztl, and CCA1-OXwere transiently transformed with the CCA1pro:LUC construct and circadian rhythms in luminescence were imaged over 5 days (A-C). (D) Circadian period estimates based on luminescence traces in Col-0, cca1/lhy and ztl (A-B). Mean ±SEM, n = 3-5, t-test p-value as indicated. Please click here to view a larger version of this figure.
| Genotype | Period (protoplasts) | Period (transgenic) | Reported by: | References |
| Col-0 | 23.0 ±0.21 | 24.4 ±0.09 | pCCA1:LUC in Col-0 | (2) |
| cca1/lhy | 20.6 ±0.13 | 19.9 ±0.11 | pCCA1:LUC in Col-0 | (2) |
| ~18 | RNA in Ler background | (9) | ||
| ztl | 25.1 ±0.12 | 27.4 ±0.5 | CAB2:LUC in C24 ecotype | (22) |
| CCA1-OX | arrhythmic | arrhythmic | pCCA1:LUC (LD data only) | (2) |
| arrhythmic | RNA and protein in Col-0 background | (23) |
Table 2: Circadian Rhythms in Protoplasts Compared to Transgenic Seedlings. The circadian period of pCCA1:LUC expression in protoplasts compared with the period lengths first reported for the mutation and, if available, with pCCA1:LUC expression in Col-0.
Discussion
Here, we present a rapid assay to screen circadian period defects in Arabidopsis lines, including protoplast isolation, transfection, and luminecent imaging. The circadian period in wild-type Arabidopsis and the clock mutants cca1/lhy, ztl, and CCA1-OX was calculated from measurements of luminescence from a CCA1pro:LUC reporter and found to be consistent with published data generated from whole plants using more time-consuming transgenic approaches (Table 2). Using this assay to screen Arabidopsis mutants avoids the initial requirement to generate transgenic luminescent lines, which is time consuming and might only reveal that a particular mutant exhibits wild-type rhythms. A clear advantage of this protocol is that any Arabidopsis line can be screened in short time for altered circadian transcriptional rhythms, which will help the identification of more genes affecting timekeeping in plants.
Isolation of protoplasts can be laborious, especially if the mutant line displays a dwarfed phenotype. Previously reported methods for generating protoplasts involve cutting leaves or seedlings into thin strips and vacuum infiltrating the strips with enzyme solution to allow the enzymes to reach the cell walls26,27. The subsequent digestion requires at least 3 hr incubation in the dark to release the protoplasts into the enzyme solution. When vacuum infiltration is not applied, incubation time up to 18 hr is advised. In the protocol presented here, vacuum infiltration is unnecessary because the epidermal cell layer impenetrable to the enzymes is removed by tape. When generating protoplasts by cutting plant material it is necessary to separate protoplasts from cell wall debris after digestion; this is typically done by filtering the solution or purifying it on a sugar gradient27,26. Here, any undigested tissue will remain on the autoclave tape after digestion, so filtration and sugar gradient purification of the protoplasts can be omitted. There is a chance that the stress resulting from prolonged protoplasting procedures affects cell viability and the circadian clock. The tape-based protoplasting method20 visualized here yields a high number of protoplasts; and given the viability of cells over a circadian time series and the matching period lengths of protoplasts and whole seedlings (Table 2), the methodology described here is preferred over alternative methods to generate protoplasts.
The transfection step of the protocol is a critical step that impacts on the viability of the protoplasts. The PEG solution enables the DNA to be delivered into the cell, and both incubation time in this solution (step 3.4) and the percentage of PEG should be determined empirically. The standard concentration is 20%, with lower concentrations reducing the transformation efficiency and higher concentrations reducing viability28. Incubation time as short as five minutes could yield efficient transfection of the protoplasts27.
The protoplasts can be imaged on any luminescent imaging platform. In this video, we have used a luminescence plate reader equipped with external lights (red and blue LEDs, 630 and 470 nm, respectively, at a combined intensity of ~20 µE), which allows for high-throughput screening and frequent measurements. Other imaging equipment that detects luminescence, such as alternative plate readers or setups based around a charge-coupled device (CCD) camera, could be applied equally well as long as illumination is available for photosynthesis. The advantage of using a CCD camera mounted on a controllable cabinet is that light and temperature can be programmed. A disadvantage is the longer capture time, introducing prolonged periods of darkness that can cause stress to the protoplasts in addition to perturbing endogenous timekeeping. Regardless of which experimental platform is chosen, adjustment of settings is likely to be necessary compared to settings for seedlings or mature plants. In our experience, increasing the concentrations of reporter plasmid during transfection and/or luciferin in the imaging buffer might resolve any erratic or quickly dampening rhythms that might occur in initial experiments.
In future experiments, the methodology described here can be applied to study the circadian phenotype of any mutant Arabidopsis line. With minor modifications, the effect of e.g. various drugs on timekeeping could be studied in this setup. Furthermore, the transfection can be modified to include artificial microRNA for gene silencing19, further expanding the versatility of this protocol. Protocols to generate rice protoplasts are well-established29 and the imaging protocol presented here could equally be applied to further our understanding of the clock in economically important monocot crop plants.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We would like to thank Prof. David Somers and Dr. Jeongsik Kim for the luciferase reporter plasmid and initial method development. Prof. Karen Halliday kindly provided all the circadian mutant lines used in this study. This research was supported by Royal Society research grants RS120372 and RS140275. GvO is a Royal Society University Research Fellow (UF110173).
References
- Goodspeed D, Chehab EW, Min-Venditti A, Braam J, Covington MF. Arabidopsis synchronizes jasmonate-mediated defense with insect circadian behavior. Proc. Natl. Acad. Sci. U. S. A. 2012;109(12):4674–4677. doi: 10.1073/pnas.1116368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, et al. Crosstalk between the circadian clock and innate immunity in Arabidopsis. PLoS Pathog. 2013;9(6):1003370. doi: 10.1371/journal.ppat.1003370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevia MA, Canessa P, Müller-Esparza H, Larrondo LF. A circadian oscillator in the fungus Botrytis cinerea regulates virulence when infecting Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 2015;112(28):8744–8749. doi: 10.1073/pnas.1508432112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V, Meier S, Petersen LN, Ingle RA, Roden LC. Defence responses of Arabidopsis thaliana to infection by Pseudomonas syringae are regulated by the circadian clock. PLoS One. 2011;6(10):26968. doi: 10.1371/journal.pone.0026968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A, Schlereth A, Stitt M, Smith AM. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. 2010;107(20):9458–9463. doi: 10.1073/pnas.0914299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, et al. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309(5734):630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9(8):130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary MK, Nomura Y, Wang L, Nakagami H, Somers DE. Quantitative Circadian Phosphoproteomic Analysis of Arabidopsis Reveals Extensive Clock Control of Key Components in Physiological, Metabolic, and Signaling Pathways. Mol. Cell. Proteomics. 2015;14(8):2243–2260. doi: 10.1074/mcp.M114.047183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T, et al. LHY and CCA1 Are Partially Redundant Genes Required to Maintain Circadian Rhythms in Arabidopsis. Dev. Cell. 2002;2(5):629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- Kikis EA, Khanna R, Quail PH. ELF4 is a phytochrome-regulated component of a negative-feedback loop involving the central oscillator components CCA1 and LHY. Plant J. 2005;44(2):300–313. doi: 10.1111/j.1365-313X.2005.02531.x. [DOI] [PubMed] [Google Scholar]
- Lu SX, et al. CCA1 and ELF3 Interact in the Control of Hypocotyl Length and Flowering Time in Arabidopsis. Plant Physiol. 2012;158(2):1079–1088. doi: 10.1104/pp.111.189670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, et al. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell. 2010;22(3):594–605. doi: 10.1105/tpc.109.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon LE, et al. Temporal repression of core circadian genes is mediated through EARLY FLOWERING 3 in Arabidopsis. Curr. Biol. 2011;21(2):120–125. doi: 10.1016/j.cub.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron JM, et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U. S. A. 2012;109(8):3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Harmer SL. Wheels within wheels: the plant circadian system. Trends Plant Sci. 2013;1:1–10. doi: 10.1016/j.tplants.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM. CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 2004;101(9):3292–3297. doi: 10.1073/pnas.0400163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portolés S, Más P. The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 2010;6(11):e1001201. doi: 10.1371/journal.pgen.1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, et al. Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 2008;283(34):23073–23083. doi: 10.1074/jbc.M803471200. [DOI] [PubMed] [Google Scholar]
- Kim J, Somers DE. Rapid assessment of gene function in the circadian clock using artificial microRNA in Arabidopsis mesophyll protoplasts. Plant Physiol. 2010;154(2):611–621. doi: 10.1104/pp.110.162271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F-H, et al. Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods. 2009;5:16. doi: 10.1186/1746-4811-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin TO. Firefly luciferase: the structure is known, but the mystery remains. Structure. 1996;4(3):223–228. doi: 10.1016/s0969-2126(96)00026-3. [DOI] [PubMed] [Google Scholar]
- Edwards KD, et al. Quantitative analysis of regulatory flexibility under changing environmental conditions. Mol. Syst. Biol. 2010;6(424):424. doi: 10.1038/msb.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE Encodes a Novel Clock-Associated PAS Protein from Arabidopsis. Cell. 2000;101(3):319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Tobin EM. Constitutive Expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) Gene Disrupts Circadian Rhythms and Suppresses Its Own Expression. Cell. 1998;93(7):1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Kim W-Y, Geng R, Somers DE. Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. U. S. A. 2003;100(8):4933–4938. doi: 10.1073/pnas.0736949100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Jung H-I, Vatamaniuk OK. Isolation of protoplasts from tissues of 14-day-old seedlings of Arabidopsis thaliana. J. Vis. Exp. 2009. pp. e15–e17. [DOI] [PMC free article] [PubMed]
- Yoo S-D, Cho Y-H, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2(7):1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Damm B, Schmidt R, Willmitzer L. Efficient transformation of Arabidopsis thaliana using direct gene transfer to protoplasts. Mol. Gen. Genet. 1989;217(1):6–12. doi: 10.1007/BF00330935. [DOI] [PubMed] [Google Scholar]
- Zhang Y, et al. A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. 2011;7(1):30. doi: 10.1186/1746-4811-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]


