Abstract
Although community-acquired pneumonia remains a major public health problem, murine models of bacterial pneumonia have recently facilitated significant preclinical advances in our understanding of the underlying cellular and molecular pathogenesis. In vivo mouse models capture the integrated physiology and resilience of the host defense response in a manner not revealed by alternative, simplified ex vivo approaches. Several methods have been described in the literature for intrapulmonary inoculation of bacteria in mice, including aerosolization, intranasal delivery, peroral endotracheal cannulation under 'blind' and visualized conditions, and transcutaneous endotracheal cannulation. All methods have relative merits and limitations. Herein, we describe in detail a non-invasive, technically non-intensive, inexpensive, and rapid method for intratracheal delivery of bacteria that involves aspiration (i.e., inhalation) by the mouse of an infectious inoculum pipetted into the oropharynx while under general anesthesia. This method can be used for pulmonary delivery of a wide variety of non-caustic biological and chemical agents, and is relatively easy to learn, even for laboratories with minimal prior experience with pulmonary procedures. In addition to describing the aspiration pneumonia method, we also provide step-by-step procedures for assaying the subsequent in vivo pulmonary innate immune response of the mouse, in particular, methods for quantifying bacterial clearance and the cellular immune response of the infected airway. This integrated and simple approach to pneumonia assessment allows for rapid and robust evaluation of the effect of genetic and environmental manipulations upon pulmonary innate immunity.
Keywords: This Month in JoVE, Issue 115, Klebsiella pneumoniae, intratracheal, bacterial pneumonia, host defense, bacteremia, BSL2, mouse
Introduction
Community-acquired pneumonia remains the leading cause of death from infection in the U.S., with little overall change in mortality rates over the past 40 years despite improvements in vaccination and antibiotic strategies1,2. Despite the lack of perceptible progress at the public health level, in recent years dramatic advances have been made in our understanding of the molecular and cellular pathogenesis of pneumonia, with many of these steps forward made possible by the use of mouse models of lung infection. The genetic tractability of the mouse, the similarity of the murine and human immune systems, and the vast array of murine-targeted immunologic reagents that have become commercially available have together facilitated rapid progress of the field.
Mouse models of bacterial pneumonia described in the literature have generally relied upon one of four technical routes for pathogen inoculation: i) aerosolization; ii) intranasal delivery; iii) peroral delivery; and iv) surgical intratracheal injection (i.e., tracheotomy)3. All routes of infection have advantages and disadvantages3. In particular, relative exposure of the upper airway, potential for admixture of oronasal flora, requirements for general anesthesia, variability of the inoculum delivered to the distal lung, lobar distribution of the delivered pathogens, technical expertise requirements, and procedural morbidity vary widely across these approaches.
Commonly used peroral infection techniques include endotracheal (translaryngeal) cannulation via either a 'blind' (non-visualized) approach, or under direct laryngeal visualization3-5. Both methods, while robust, require substantial training and also carry risk of trauma to the upper airway. In the present report, we describe a technically non-intensive, non-invasive, inexpensive, and rapid method of peroral infection, whereby bacteria (Klebsiella pneumoniae in the example provided) pipetted into the oropharynx of an anesthetized mouse are delivered to the lungs via aspiration (i.e., inhalation). We and others have used the aspiration pneumonia technique successfully6-9. This versatile and easily learned lung-delivery method can be extended to the delivery of many additional non-caustic agents to the lungs, including cytokines and other proteins, pathogen-associated molecules (e.g., lipopolysaccharide), cells (i.e., adoptive transfer), and toxins (e.g., bleomycin). In addition to discussing important technical considerations, we also describe an integrated, quantitative approach for assessing the subsequent host response to pneumonia, including downstream measurement of bacterial clearance (i.e., colony forming unit [CFU] quantitation in the lung and peripheral organs) and leukocyte accumulation in the airspace.
Protocol
All experiments were performed in accordance with the Animal Welfare Act and the U.S. Public Health Service Policy on Humane Care and Use of Laboratory Animals after review by the Animal Care and Use Committee of the NIEHS.
1. Preparation of K. pneumoniae Culture
Caution: Perform all steps in a biosafety level 2 (BSL2) hood or other BSL2 designated area and discard waste per institute BSL2 guidelines.
For suspension growth of K. pneumoniae, thaw a glycerol stock of bacteria and inoculate a working culture by transferring 1 ml of K. pneumoniae stock to 50 ml of tryptic soy broth (TSB) in a 500 ml or larger flask.
Make glycerol stocks by adding 900 µl of cultured log-phase K. pneumoniae to 100 µl of sterile glycerol and freezing at -20 °C or -80 °C. For long term storage, -80 °C is recommended.
Culture bacteria from step 1.1 by shaking flask overnight (14 - 18 hr) at 37 °C at 200-225 rpm.In order to promote log phase growth, dilute 1-5 ml of this overnight culture into a flask containing 50 ml TSB in the morning and place back at 37 °C with shaking for ~2.5 hr.
- Transfer 1 ml of K. pneumoniae culture from the last step into a 1.5 ml tube and spin at 7,500 x g for 2 min. A white pellet should be visible. Remove supernatant without disturbing the pellet, gently resuspend bacteria with pipette in 1 ml of sterile saline, and spin again at 7,500 x g for 2 min.
- Perform this wash step 2x, bringing up the final, washed pellet in 1 ml of sterile saline.
Perform log-wise serial dilutions of this neat inoculum. For example, add 400 µl of the inoculum into 3.6 ml of sterile saline serially to make a 10-1-10-9 dilution series.
- Determine the OD600 of washed K. pneumoniae by placing 1 ml of the 1:10 and 1:100 dilutions into disposable cuvettes and measuring the OD600, blanking first with sterile saline. Measure duplicates to ensure accuracy. An OD600 of 1.0 is roughly equivalent to 4-7 x 108 CFU/ml. Note that the relationship between OD and CFU/ml may not be linear.
- Use the OD600 from the dilutions to calculate concentration of K. pneumoniae, applying the OD:CFU/ml conversion above, and factoring in the dilution.
Use the K. pneumoniae concentration to prepare the desired inoculum CFU dose in a <100 µl volume for delivery to mice (see below).
Also plate 100 µl of 10-6-10-9 dilutions and 100 µl of sterile saline onto individual tryptic soy agar (TSA) plates at RT. Incubate at 37 °C overnight and enumerate bacterial colonies in order to calculate exact concentration that was used in experiment and to control for contamination. NOTE: Actual bacterial concentration of final inoculum may be ± 500 CFU/ml that predicted by absorbance. Experimenters should ideally confirm the OD600:CFU/ml relationship in pilot studies prior to use in vivo in mice, empirically readjusting the OD600:CFU/ml conversion based on their personal experience.
2. Murine Intratracheal (i.t.) Aspiration of K. pneumoniae from Oropharynx
Ensure that mice are uniquely identified by tail mark, tattoo, or other approved identifier.
Draw up dosing inoculum of bacteria using P200 pipette prior to removing mice from isoflurane in order to expedite procedure. For best results with i.t. aspiration, use a volume of <100 µl to prevent overflow. Herein, use 2000 CFU/50 µl of K. pneumoniae.
Anesthetize mice in a clear chamber with isoflurane gas (e.g., 2% isoflurane at flow rate of 1 L/min) or as per institutional guidelines. The number of mice to anesthetize together is determined by comfort level of the experimenter, typically 1-2 at a time. Observe breathing and confirm level of anesthesia once deep breaths are visible and 2-3 sec can be counted between breaths. NOTE: If there is concern about possible confounding effects of volatile (inhaled) anesthetics, anesthesia can be achieved instead with intraperitoneal injection of ketamine/xylazine. With the method described above, deep anesthesia only lasts a few min. If more prolonged anesthesia is induced, eye ointment should be used to prevent ocular desiccation.
Once mouse is anesthetized, position it in a semirecumbent supine position, suspended by the maxillary incisors from a rubber band stretched between pegs on a slanted Plexiglas board (Figure 1).
Gently pull the mouse tongue to the side with a pair of blunt non-ridged forceps and deposit dose into the oral cavity with a P200 pipette. Exercise great care to avoid inducing trauma to either the tongue or the oropharynx.
While keeping the tongue retracted, gently occlude the nose with a gloved finger until the mouse inhales. Continue to cover the nose until the mouse has taken two or more inhalations, and no liquid is visible in the oral cavity. Covering the nose helps ensure that the mouse will inhale the bacteria into the lungs, as mice are obligate nose breathers.
Remove the mouse from the inoculation board and return to its cage, placing the mouse on its back to prevent bedding or debris from blocking the nares while the mouse is recovering from anesthesia.
Once all mice have been dosed and have awoken from anesthesia, house mice in a cubicle/room containing only mice that have been dosed with K. pneumoniae. Monitor mice daily, including body weights if going beyond 48 hr post-infection.
Use daily mash and/or supplemental heat if allowing the mice to go beyond 48 hr.
3. Bronchoalveolar Lavage Fluid (BALF) Collection and Analysis
Euthanize mice per institutional guidelines. Here, use a lethal injection of 150 mg/kg sodium pentobarbital or equivalent dose of commercial euthanasia solution followed by exsanguination, as this avoids possible complications to the lungs associated with CO2 inhalation and cervical dislocation.
Position mouse on back and sterilize mouse by spraying fur with 70% ethanol particularly over the chest and neck area.
Make a longitudinal cut just below the sternum, then holding the sternum with forceps, nick the diaphragm, allowing the lung to fall back into the chest cavity. Cut up through the chest cavity on either side of the vasculature and then up through the neck, exposing the trachea.
As the trachea is surrounded by longitudinal muscles on either side, carefully cut through the middle and push tissue to the sides, as this avoids cutting the surrounding vasculature, which could introduce blood into the trachea. If the vasculature is nicked, clean the surrounding area with gauze prior to accessing the tracheal lumen.
Use surgical scissors or a needle to nick the trachea about ¼ of the way down from the head and insert a cannula attached to a 1 ml syringe preloaded with 1 ml of 1x phosphate-buffered saline (PBS) caudally into the trachea. If using mice <8 weeks of age or females, reduce the lavaging volume to 600-800 µl.
Push the volume into the lung slowly, allowing all lobes to inflate and then pull the volume back out with the syringe, repeating 3x. If PBS is coming out of the nostrils the cannula has not been pushed far enough into the trachea or inflation is occurring too quickly. Alternatively, if the lungs are not inflating well, the cannula has been pushed in too far, so withdraw slightly.
Collect pooled washes in a 15 ml tube on ice.
Spin the airspace lavage fluid at 1,200 x g for 5 min, collect supernatant into a 1.5 ml tube and freeze at -80 °C. Subsequently, use the supernatant (i.e., BALF) for measurement of protein, cytokines, and for various other biochemical assays. The pellet represents cells from the bronchoalveolar space.
If red blood cells (RBCs) are evident in the cell pellet based on color, lyse with 1 ml of ACK lysis buffer for 1 min followed by addition of 5 ml of 1x PBS to stop the lysis reaction and dilute the buffer. Handle all specimens identically, either treated or not with ACK lysis buffer.
Spin cells at 1,200 x g for 5 min, decant the supernatant, and bring the cells up in 1 ml of 1x PBS.
Vortex and count cells on a hemocytometer for enumeration of total airspace cells.
Based on density of cells add approximately 80-150 µl (~80 µl for an infected mouse and ~150 for a naive mouse) to a slide chamber on a cytospin centrifuge. Spin cells onto microscope slides for subsequent staining to determine cell differential populations.
Allow cells to dry on slides overnight. Stain cells with tri-stain (e.g., Hema 3 [see table]) by dipping the slides 7 times in fixative, 9 times in stain 1, and 7 times in stain 2 (no rinses between stains), and allow to dry. After stain has dried, coverslip slides and manually count differentials by microscope. Generally, neutrophils and monocyte/macrophages are easily distinguished by their size and nuclear morphology.
4. Determining Bacterial Load in Lung and Peripheral Tissues
Before beginning: prepare dilution tubes with 900 µl of 1x PBS for lung and spleen and 90 µl of 1x PBS for blood. Label plates for culturing bacteria and set at room temperature; therefore, once tissue has been collected time will be minimized between homogenization and plating for bacterial growth. Note: Due to the heterogeneous deposition of bacteria in the lungs that can occur with aspiration, it is recommended that individual mice be used for analysis of either total airway cellularity or total (bilateral) lung bacterial burden.
Euthanize the mouse as per step 3.1 and subsequently collect blood from the left ventricle, placing into a heparinized tube to avoid clotting. Remove all lung lobes and place in a 15 ml tube containing 5 ml of 1x PBS, followed by removal of the spleen and placement into 2 ml of 1x PBS. Place tubes on ice. Take care to clean instruments with ethanol between each tissue/mouse.
Homogenize the tissue on ice, preferably with disposable homogenizers that can be changed between each sample. Disposable homogenizer tips can be autoclaved for reuse between experiments.
- Once tissue has been homogenized, perform serial dilutions and plate. Plate samples of a given tissue/dilution in duplicate on a single TSA plate by drawing a dividing line on the plastic. Plating and dilution suggestions are shown below (in all cases, 10 µl of sample is spread onto plate) and are based on a 24 hr post-infection necropsy. If analyzing 48-72 hr samples, a larger number of dilutions may need to be plated due to increased bacterial burden at later time points.
- For Blood - dilute 1:10 by adding 10 µl of blood into 90 µl of sterile 1x PBS. Plate neat and 1:10 diluted samples in duplicate. Once blood has been plated, spin residual blood at 9,300 x g for 10 min to extract plasma for measurement of cytokines and chemokines.
- For Lung - dilute 1:10 - 1:100 by adding 100 µl of homogenized lung into 900 µl of sterile 1x PBS serially. Plate neat homogenate and all dilutions in duplicate.
- For Spleen - dilute 1:10-1:100 by adding 100 µl of homogenized spleen into 900 µl of sterile 1x PBS serially. Plate neat homogenate and both dilutions in duplicate.
Allow spread samples to dry briefly.
Invert TSA plates and place in a static 37 °C incubator overnight.
Determine the number of colonizing bacteria by averaging the bacterial colonies from both sides of the plate and from each dilution. Factor in the initial dilutions of 5 ml for lung and 2 ml for spleen to determine total tissue CFUs.
Representative Results
C57BL/6 mice were infected with 2000 CFU of K. pneumoniae 43816 (serotype 2) via oropharyngeal aspiration into the lungs. At this dose, mice typically begin to show clinical symptoms 12-24 hr post-infection including lethargy, ruffled fur, and weight loss of 5-10% (Figure 2A). Within 48-72 hr post-infection, many of the mice show symptoms of illness and morbidity that is typically preceded by an average of 20% weight loss and results in hunched postures with decreased activity and decreased responsiveness to stimulation. Studies of a longer duration may require a lower dose of K. pneumoniae. Infection with 500 CFU of K. pneumoniae results in milder illness peaking 3-5 d post-infection that is characterized by a weight loss of 10-15% with recovery and weight gain beginning by day 5-6 (Figure 2B). Body weight is often predictive of survival, with a weight loss of 20% rarely associated with recovery.
The ultimate success of the host response may be best indicated by survival studies, wherein mice are monitored for 10-14 days, and euthanized (per institutionally approved guidelines) for signs of moribundity, such as minimal responsiveness to tactile stimulation and/or weight loss >20%. Survival studies are often most effective if one targets an infecting inoculum that approximates 50% lethality (i.e., LD50 dose) in the control group (Figure 2C). An LD50 dose allows for detection of both relatively increased and decreased survival in the experimental group. Differential survival may be inoculum-dependent, so several infection doses may sometimes be necessary to detect an altered host defense phenotype.
Lower respiratory tract infection with K. pneumoniae is characterized by a robust influx of leukocytes into the airway. Immune cell enumeration and differentiation by staining of the BAL cell pellet demonstrates an increase in airway immune cells within 6h post-infection (Figure 3A). Cellularity peaks by 24 hr, with neutrophils being the predominant immune cell in the airway (Figure 3A-B). Both genders of C57BL/6 mice demonstrate equivalent airway leukocytosis in response to K. pneumoniae at 24 hr and 48 hr post-infection (Figures 3C-D).
Bacterial pneumonia induces local (intrapulmonary) and systemic pro-inflammatory mediators including cytokines and chemokines. Local cytokine/chemokine levels can be detected in the BALF with a conventional ELISA or by using a multiplex cytokine system. Cytokines including IL-6, RANTES, TNFα, G-CSF, and others are detectible at both 24 hr and 48 hr post-infection with K. pneumoniae and provide insight into the lung microenvironment and its recruitment of immune cells (Figure 4).
Collection of lung, blood, and peripheral tissue(s) such as spleen allows for quantitation of tissue bacterial load, providing insight into both local pulmonary clearance of microbes as well as extrapulmonary dissemination of bacteria. Serial dilution and culturing of the lung demonstrates expansion of microbial burden between 24 hr and 48 hr post-infection (Figure 5A). Similar findings are noted in the blood and spleen (Figure 5B-C). Of note, a bimodal bacterial burden can be noted in the blood and spleen at 24 hr post-infection, with some mice having high-grade bacterial dissemination, and others, even in the face of high-grade bacterial burden in lung, displaying no detectable bacteria in the blood or spleen. K. pneumoniae bacterial load in C57BL/6 mice is not gender-specific, as males and females demonstrate equivalent burden in various tissues post-inoculation (Figure 5D-F).
 Figure 1:Procedure Board for Aspiration Pneumonia in Mice. Study mice are positioned, head up and back to board (i.e., semirecumbent), suspended by the incisors from a rubber band stretched between two pegs mounted to a Plexiglas or similar board.
Figure 1:Procedure Board for Aspiration Pneumonia in Mice. Study mice are positioned, head up and back to board (i.e., semirecumbent), suspended by the incisors from a rubber band stretched between two pegs mounted to a Plexiglas or similar board.
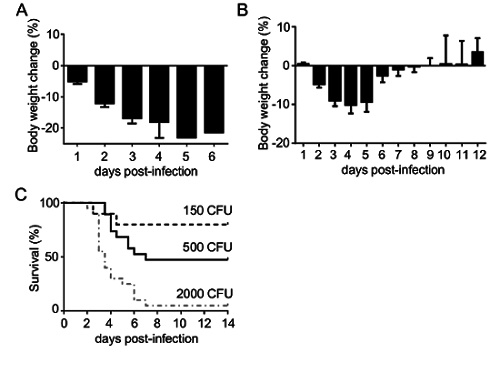 Figure 2:Assessment of Weight Loss and Morbidity Following Pulmonary Aspiration of K. pneumoniae. C57BL/6 mice (N = 10-20 per condition) were infected by aspiration of K. pneumoniae 43816 (serotype 2). (A) Daily weights, indexed to pre-infection weight, from mice infected with 2000 CFU. In this study, no mice survived past day 6. (B) Infection with 500 CFU results in daily weight loss through day 4 post-infection with a transition towards weight gain beginning by day 5. (C) Survival curves from 150 CFU, 500 CFU, and 2000 colony forming unit (CFU) infections, indicating that 500 CFU approximates the LD50. Data in panels A and B are mean ± SEM. Please click here to view a larger version of this figure.
Figure 2:Assessment of Weight Loss and Morbidity Following Pulmonary Aspiration of K. pneumoniae. C57BL/6 mice (N = 10-20 per condition) were infected by aspiration of K. pneumoniae 43816 (serotype 2). (A) Daily weights, indexed to pre-infection weight, from mice infected with 2000 CFU. In this study, no mice survived past day 6. (B) Infection with 500 CFU results in daily weight loss through day 4 post-infection with a transition towards weight gain beginning by day 5. (C) Survival curves from 150 CFU, 500 CFU, and 2000 colony forming unit (CFU) infections, indicating that 500 CFU approximates the LD50. Data in panels A and B are mean ± SEM. Please click here to view a larger version of this figure.
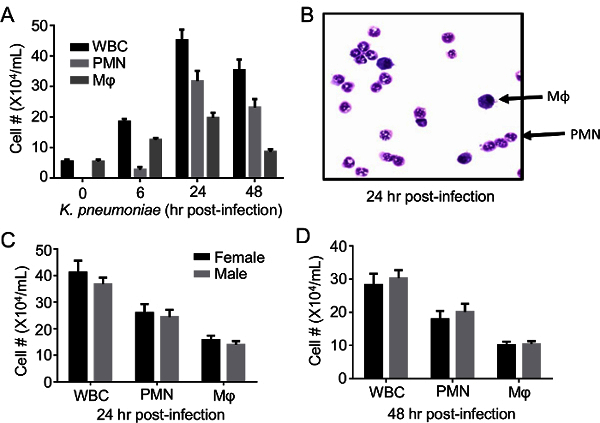 Figure 3: Infection withK. pneumoniae Results in Time-dependent Airway Leukocytosis. C57BL/6 mice were given intrapulmonary infection with 2000 CFU of K. pneumoniae. (A) Enumeration of total leukocytes (WBC), neutrophils (PMN), and macrophages (Mφ) in airspace fluid at various time points post-infection. (B) Upon staining, PMNs and macrophages can be easily identified by nuclear morphology and size in cytospin fields and counted. (C-D) Infection in males and females results in similar numbers of recruited inflammatory cells at 24 hr and 48 hr post-infection. Data derive from 6-25/mice per condition and are mean ± SEM. Please click here to view a larger version of this figure.
Figure 3: Infection withK. pneumoniae Results in Time-dependent Airway Leukocytosis. C57BL/6 mice were given intrapulmonary infection with 2000 CFU of K. pneumoniae. (A) Enumeration of total leukocytes (WBC), neutrophils (PMN), and macrophages (Mφ) in airspace fluid at various time points post-infection. (B) Upon staining, PMNs and macrophages can be easily identified by nuclear morphology and size in cytospin fields and counted. (C-D) Infection in males and females results in similar numbers of recruited inflammatory cells at 24 hr and 48 hr post-infection. Data derive from 6-25/mice per condition and are mean ± SEM. Please click here to view a larger version of this figure.
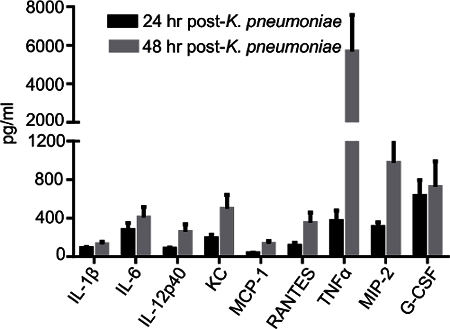 Figure 4:Cytokines and Chemokines are Induced in the Airspace in Response to Bacterial Infection. C57BL/6 mice were given intrapulmonary infection with 2000 CFU of K. pneumoniae. BALF cytokines and chemokines were quantified by multiplex assay at 24 hr and 48 hr post-infection. Data derive from 5-15 mice/condition and are mean ± SEM. Please click here to view a larger version of this figure.
Figure 4:Cytokines and Chemokines are Induced in the Airspace in Response to Bacterial Infection. C57BL/6 mice were given intrapulmonary infection with 2000 CFU of K. pneumoniae. BALF cytokines and chemokines were quantified by multiplex assay at 24 hr and 48 hr post-infection. Data derive from 5-15 mice/condition and are mean ± SEM. Please click here to view a larger version of this figure.
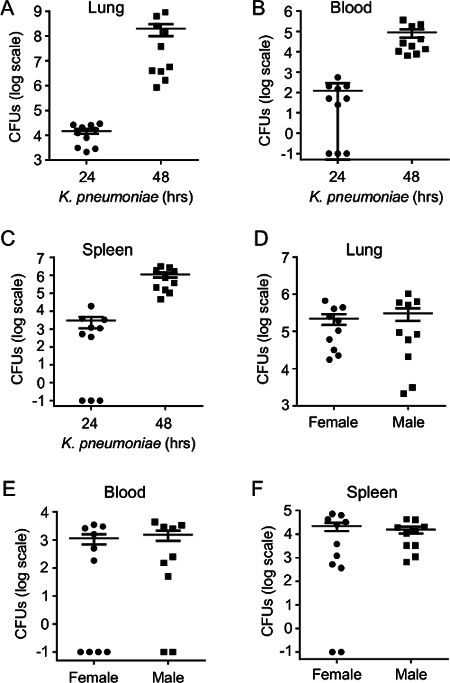 Figure 5:Time-dependent Expansion of Bacterial Burden in Lung and Peripheral Tissues during Pneumonia. C57BL/6 mice were given intrapulmonary infection with 2000 CFU of K. pneumoniae. (A) Lung parenchymal bacterial burden was measured by plating serial dilutions of lung homogenate on TSA 24 hr and 48 hr post-infection. (B) Bloodstream and (C) splenic bacterial burden were similarly quantified. (D-F) Bacterial burden in various tissues is equivalent in male and female mice after induction of pneumonia. Data derive from 10 mice/condition and are mean ± SEM. Zero values in panels B, C, E, and F were assigned a value of 0.1 to permit graphing on a log scale. Please click here to view a larger version of this figure.
Figure 5:Time-dependent Expansion of Bacterial Burden in Lung and Peripheral Tissues during Pneumonia. C57BL/6 mice were given intrapulmonary infection with 2000 CFU of K. pneumoniae. (A) Lung parenchymal bacterial burden was measured by plating serial dilutions of lung homogenate on TSA 24 hr and 48 hr post-infection. (B) Bloodstream and (C) splenic bacterial burden were similarly quantified. (D-F) Bacterial burden in various tissues is equivalent in male and female mice after induction of pneumonia. Data derive from 10 mice/condition and are mean ± SEM. Zero values in panels B, C, E, and F were assigned a value of 0.1 to permit graphing on a log scale. Please click here to view a larger version of this figure.
Discussion
Murine models of bacterial pneumonia, partnered with gene targeting and in vivo biological and pharmacological interventions, have provided critical insights into the pulmonary host defense response. Great advances have been made in particular in our understanding of the chemokines and adhesion molecules that govern recruitment of neutrophils to the infected airspace10,11. In vivo models of pneumonia, unlike cell-based or alternative approaches, have also provided key insights into endocrine communications that occur between the infected lung and other organs, such as the liver (acute phase response)12 and adrenal glands (stress glucocorticoids)13. Finally, a critical and clinically highly relevant point that is revealed by in vivo pneumonia models is that successful pathogen clearance is not tantamount to success of the host. In many cases, an overexuberant immune response may lead to death of the host due to excessive bystander lung injury, even in the face of successful pathogen clearance8,14. Given this, parallel measurements of pathogen clearance and of the immune response are generally most informative, and survival studies may ultimately be required to reveal the integrated resilience of the host.
Herein, we have described a simple and non-invasive method for delivery of bacteria to the murine lung via aspiration. Compared to aerosolization, this method carries lower potential risk for respiratory exposure of study personnel, provides for higher concentrated inoculum delivery to the deep lung, and avoids potential confounding by ocular and upper airway immune responses. Intranasal inoculation also carries the potentially confounding concern of upper airway infection, including the possibility of locally invasive infection of the central nervous system4, and has also been reported to result in highly variable inocula in the lung5. Compared to endotracheal intubation via either the peroral or transcutaneous route, this method requires minimal training, is more rapid (~1 min per mouse), and, at least compared to the latter method, has lower morbidity. Potential disadvantages of the oropharyngeal aspiration method – some of which are shared by other methods – include the requirement for general anesthesia, the probability of patchy (i.e., asymmetric and heterogeneous) pathogen delivery, predominant infection of the lower lobes15, and inability to direct the pathogen unilaterally to a single lung. Due to patchy distribution in the lungs after aspiration (data not shown) – an outcome that is encountered with all lung infection methods other than aerosolization15 – we generally do not perform unilateral lung assays on infected mice. Both lungs are either lavaged together to assess the immune response, or necropsied together for pooled bacterial quantitation and/or molecular analysis (e.g., gene expression, myeloperoxidase assay, cytokine ELISA). Particularly critical steps of the protocol are step 1.8 -confirming the OD600: CFU/mL relationship prior to in vivo infection and also confirming the inoculum concentration through plating in every experiment; as well as step 3.6 – ensuring a non-traumatic lavage of the airway with good volume returns into the syringe.
Although co-infection with oral flora (i.e., polymicrobial pneumonia) is a theoretical concern, we have not encountered polymorphic bacterial colonies in lung homogenates of mice infected with K. pneumoniae or S. pneumoniae. For investigators concerned about this possibility, however, control mice can be exposed to aspirated vehicle (i.e., buffer), if desired. Although some contamination of the stomach lumen (through swallowing of residual unaspirated bacteria) is possible with the method we describe, we have never encountered gastrointestinal clinical signs or changes on gross pathology. Moreover, this possibility is common to the aerosolization and intranasal methods, and can even occur following coughing in the intratracheal inoculation method.
Some methodological concerns are universal to all pneumonia models. Control and experimental mouse groups should be age-matched ideally within 2-3 weeks of each other. Although we have not found overt gender differences in pathogen clearance or airway inflammation in the K. pneumoniae model, mice should be gender-matched, as significant differences between genders in the innate immune response have been reported16. Careful consideration to the origin of control mice should be given. Littermate controls are generally preferable over commercially purchased controls as incomplete backcrossing, microbiome-related differences in immune cell populations, and other epigenetic differences are all possible confounders that may affect the immune response17,18. Careful attention should be given to the specific serotype and culturing history of the infecting bacterium as substantial differences in virulence may be seen. Based on our experience with K. pneumoniae, we recommend performing initial pilot studies using a range of infecting inocula (e.g., 500-2,000 CFU) as the morbidity/mortality of specific bacterial doses reported in the literature - even with the same bacterial serotype and mouse strain - may not translate well to one's own laboratory, likely due to a variety of technical discrepancies. Inter-group differences between control and experimental mice in inflammatory and host defense outcomes may be inoculum-dependent. Due to the logistical realities of animal breeding and staffing, we often conduct experiments with 5-6 mice per experimental group. Although this occasionally provides adequate statistical power for CFU and cell count outcomes, we find that we often need to repeat a study of this size one or more times in order to achieve adequate power.
For some mice in a given experiment, no detectable bacterial growth in blood and spleen may be encountered 24 hr post-infection (Figure 5). We interpret this to indicate that 24 hr may be near the kinetic threshold for detectable extrapulmonary dissemination. If low/undetectable growth in blood/spleen is observed, lung CFUs from the same mouse should always be examined to address the possibility of a technical error (i.e., very low/undetectable lung infection possibly indicating an error in dosing of the mouse).
In closing, the oropharyngeal aspiration delivery method for bacterial pneumonia in mice is a robust technique that is relatively easy to acquire from a cost, training, and equipment standpoint, and is realistic even for laboratories without extensive expertise in pulmonary procedures. The method is easily coupled to downstream microbiological and immunological assays of the host defense response, and can moreover be used as a lung delivery method for a wide range of non-caustic exposures.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This work was supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences (Z01 ES102005).
References
- Mizgerd JP. Acute lower respiratory tract infection. N Engl J Med. 2008;358(7):716–727. doi: 10.1056/NEJMra074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterer GW, Rello J, Wunderink RG. Management of community-acquired pneumonia in adults. Am J Respir Crit Care Med. 2011;183(2):157–164. doi: 10.1164/rccm.201002-0272CI. [DOI] [PubMed] [Google Scholar]
- Mizgerd JP, Skerrett SJ. Animal models of human pneumonia. Am J Physiol Lung Cell Mol Physiol. 2008;294(3):L387–L398. doi: 10.1152/ajplung.00330.2007. [DOI] [PubMed] [Google Scholar]
- Revelli DA, Boylan JA, Gherardini FC. A non-invasive intratracheal inoculation method for the study of pulmonary melioidosis. Front Cell Infect Microbiol. 2012;2:164. doi: 10.3389/fcimb.2012.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Nebreda L, et al. Intratracheal administration of influenza virus is superior to intranasal administration as a model of acute lung injury. J Virol Methods. 2014;209:116–120. doi: 10.1016/j.jviromet.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aujla SJ, et al. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med. 2008;14(3):275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, et al. Th17 cells mediate clade-specific, serotype-independent mucosal immunity. Immunity. 2011;35(6):997–1009. doi: 10.1016/j.immuni.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper DW, et al. ATP-binding cassette transporter G1 deficiency dysregulates host defense in the lung. Am J Respir Crit Care Med. 2010;182(3):404–412. doi: 10.1164/rccm.200910-1580OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KM, et al. Influenza A exacerbates Staphylococcus aureus pneumonia by attenuating IL-1beta production in mice. J Immunol. 2013;191(10):5153–5159. doi: 10.4049/jimmunol.1301237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizgerd JP. Molecular mechanisms of neutrophil recruitment elicited by bacteria in the lungs. Semin Immunol. 2002;14(2):123–132. doi: 10.1006/smim.2001.0349. [DOI] [PubMed] [Google Scholar]
- Balamayooran G, Batra S, Fessler MB, Happel KI, Jeyaseelan S. Mechanisms of neutrophil accumulation in the lungs against bacteria. Am J Respir Cell Mol Biol. 2010;43(1):5–16. doi: 10.1165/rcmb.2009-0047TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinton LJ, et al. Hepatocyte-specific mutation of both NF-kappaB RelA and STAT3 abrogates the acute phase response in mice. J Clin Invest. 2012;122(5):1758–1763. doi: 10.1172/JCI59408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowdy KM, et al. Key role for scavenger receptor B-I in the integrative physiology of host defense during bacterial pneumonia. Mucosal Immunol. 2015;8(3):559–571. doi: 10.1038/mi.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madenspacher JH, et al. p53 Integrates host defense and cell fate during bacterial pneumonia. J Exp Med. 2013;210(5):891–904. doi: 10.1084/jem.20121674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brain JD, Knudson DE, Sorokin SP, Davis MA. Pulmonary distribution of particles given by intratracheal instillation or by aerosol inhalation. Environ Res. 1976;11(1):13–33. doi: 10.1016/0013-9351(76)90107-9. [DOI] [PubMed] [Google Scholar]
- Card JW, et al. Gender differences in murine airway responsiveness and lipopolysaccharide-induced inflammation. J Immunol. 2006;177(1):621–630. doi: 10.4049/jimmunol.177.1.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]


