Abstract
Mineralized tissue is vital to many characteristic adaptive phenotypes in vertebrates. Three primary tissues, enamel (enameloid), dentin, and bone, are found in the body armor of ancient agnathans and mammalian teeth, suggesting that these two organs are homologous. Mammalian enamel forms on enamel-specific proteins such as amelogenin, whereas dentin and bone form on collagen and many acidic proteins, such as SPP1, coordinately regulate their mineralization. We previously reported that genes for three major enamel matrix proteins, five proteins necessary for dentin and bone formation, and milk caseins and salivary proteins arose from a single ancestor by tandem gene duplications and form the secretory calcium-binding phosphoprotein (SCPP) family. Gene structure and protein characteristics show that SCPP genes arose from the 5′ region of ancestral sparcl1 (SPARC-like 1). Phylogenetic analysis on SPARC and SPARCL1 suggests that the SCPP genes arose after the divergence of cartilaginous fish and bony fish, implying that early vertebrate mineralization did not use SCPPs and that SPARC may be critical for initial mineralization. Consistent with this inference, we identified SPP1 in a teleost genome but failed to find any genes orthologous to mammalian enamel proteins. Based on these observations, we suggest a scenario for the evolution of vertebrate tissue mineralization, in which body armor initially formed on dermal collagen, which acted as a reinforcement of dermis. We also suggest that mammalian enamel is distinct from fish enameloid. Their similar nature as a hard structural overlay on exoskeleton and teeth is because of convergent evolution.
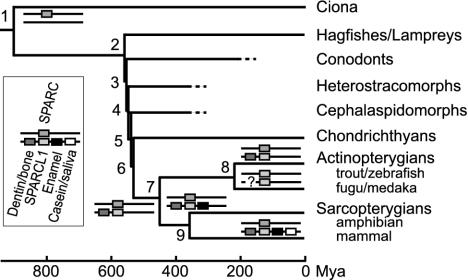
Mineralized tissue is a critical innovation in vertebrate evolution, offering the basis for various adaptive phenotypes: body armor for protection, teeth for predation, and endoskeleton for locomotion. Two distinct types of mineralized tissues emerged in Paleozoic agnathans: tooth-like oral skeleton and dermal skeleton (1–4). The dermal skeleton, which first appeared in the heterostracomorphs (Fig. 1, node 4), consists of surface dentin and basal bone, which are occasionally overlaid by enameloid. Eventually, dermal skeleton developed into simple scales. Based on the histological similarity, these scales have been considered homologous to teeth. Teeth composed of all three tissues first appeared in chondrichthyans (Fig. 1, node 6). Recently, the oral skeleton of conodonts was recognized as the earliest mineralized tissue in vertebrates and proposed to be the likely precursor of all teeth (Fig. 1, node 3) (5). However, there is no phylogenetic support for homology between the oral skeleton and teeth (4). The evolution of mineralized tissues has been enigmatic for more than a century. The investigation of genes involved in tissue mineralization has the potential to provide insight into this long-standing question.
Fig. 1.
Phylogeny and SPARC, SPARCL1, and SCPP gene-duplication history. Symbols for SPARC, SPARCL1, and SCPP genes for dentin/bone, enamel, and casein/saliva are specified in the legend box. The genes for dentin/bone SCPPs, SPARCL1, enamel SCPPs (AMBN and ENAM), and casein/saliva SCPPs are linked on human chromosome 4 in this order (9). No dentin/bone SCPP genes, including SPP1, have been found in fugu or medaka. The scale and phylogeny are based on previous publications (4, 34) and Fig. 4. Dashed lines show extinct agnathan branches.
The mammalian tooth consists of enamel, dentin, cementum, and alveolar bone, each of which is a composite of calcium phosphate, hydroxyapatite (HA). The HA crystallizes on extracellular matrix (ECM) proteins that are secreted by ameloblasts, odontoblasts, cementoblasts, and osteoblasts. These cells develop by reiterative epithelial–mesenchymal interactions. Ameloblasts originate from epithelium and secrete enamel ECM proteins, most of which are virtually specific to enamel (6). In contrast, odontoblasts and osteoblasts derive from mesenchyme and secrete ECM proteins, which are mostly common to these two tissues: ≈90% of type I collagen and other acidic proteins (7). Cementum is a bone-like but distinct collagenous tissue typically surrounding the base of teeth in mammals and certain reptiles (8). We found that genes for three major enamel ECM proteins [amelogenin (AMEL), ameloblastin (AMBN), and enamelin (ENAM)] and five dentin/bone ECM proteins [dentin sialophosphoprotein (DSPP), dentin matrix acidic phosphoprotein 1 (DMP1), integrin-binding sialoprotein (IBSP), matrix extracellular, phosphoglycoprotein (MEPE), and secreted phosphoprotein 1 (SPP1), also called osteopontin] as well as milk caseins and some salivary proteins all arose from a common ancestor by gene duplication to form the secretory calcium-binding phosphoprotein (SCPP) family. With the single exception of AMEL, which is located on chromosomes X and Y, the SCPP genes form a cluster on 4q13-q21 in humans (9).
SCPPs have Ser-Xaa-Glu motifs [SXE, where X denotes any amino acid; phospho-Ser (pSer) or Asp may replace Glu], which associate with Ca2+ after the phosphorylation of Ser residues (10). In addition, dentin/bone SCPPs, except for MEPE, have acidic amino acid (Glu and Asp) clusters, which bind to Ca2+ and HA, and facilitate HA crystal nucleation or modulate its growth (11, 12). These functions are regulated by different degrees of Ser phosphorylation (13). The dentin/bone SCPPs also have an Arg-Gly-Asp motif, which binds to osteoblasts and osteoclasts via the integrin surface receptor, which in turn evokes intracellular signals that regulate bone mineralization or resorption (14).
We have hypothesized that these SCPP genes arose from the SPARC-like 1 gene (SPARCL1), because SPARCL1 shares the common 5′ gene structure with the SCPP genes and SPARCL1 is located adjacent to the mammalian dentin/bone SCPP genes, whereas the gene for secreted protein, acidic, cysteine-rich (SPARC, also called osteonectin) resides on a different chromosome (Fig. 1) (9). Both SPARC and SPARCL1 are expressed in bone and are composed of three functional domains: acidic calcium-binding domain I, Follistatin-like domain II, and extracellular calcium-binding domain III (15–17). Domain III also binds to type I–V collagens (18). SPARC is the most abundant noncollagenous bone ECM protein and thus has been proposed to be the critical protein for HA crystallization in dentin and bone (15). SPARC has been identified in both protostomes and deuterostomes, whereas SPARCL1 seems to be relatively new and has been found only in amniotes (19–21). This suggests that SPARC and SPARCL1 initially arose by gene duplication, and subsequent tandem gene duplications generated the many SCPP genes.
We have identified SPARC and SPARCL1 genes from various animal taxa. Comparison of these sequences with the dentin/bone SCPP sequences supports our hypothesis. Based on the timing of gene duplications in vertebrate phylogeny and the intimate association between SPARC and fibrillar collagens, we propose a scenario for the origin and subsequent elaboration of vertebrate mineralized tissues.
Methods
Bioinformatic Analysis. By searching expressed sequence tag (EST) and genome sequence databases for homology using human SPARC, we identified a portion of SPARCL1 from fugu (Takifugu rubripes) and zebrafish as well as SPARC from frog (Silurana tropicalis), fugu, zebrafish, medaka (Oryzias latipes), and ascidian (Ciona intestinalis). Based on the sequence, PCR primers for fugu SPARCL1 were designed and used for cloning. These sequences were obtained from the Department of Energy Joint Genome Institute (fugu and Ciona; www.jgi.doe.gov), the Sanger Centre (frog; www.sanger.ac.uk), or GenBank. Previously characterized sequences of SPARC, SPARCL1, and SCPPs were obtained from GenBank, and their accession numbers are shown in Table 1, which is published as supporting information on the PNAS web site. Trout SPP1 is named novel ovarian protein (NOP; GenBank accession nos. AAG35656 and AAG49534). The sea urchin SPARC sequence is available from the Max Planck Institute (cluster 002442.a1.1; www.molgen.mpg.de) but was not used for the phylogenetic analysis, because a portion of domain III was found in only a single EST. All the other sequences were reconstructed from genomic sequence or more than two ESTs. Sequences identified in this study were deposited in the GenBank database (accession nos. AY575071–AY575077 and AY620817).
Multiple-sequence alignments and phylogenetic analyses were conducted as described (9). The molecular clock hypothesis was tested for all available SPARC and SPARCL1 sequences. Genes that evolve significantly faster or slower than the average rate (P < 5%) were eliminated, and a linearized tree was obtained by the two-cluster test by using lintree (22). Divergence time was estimated by using lintree or timer (23). Both Poisson correction (PC) and γ distances were used to estimate amino acid differences (24). The γ parameter was estimated by gamma (25). All of these programs are available from www.bio.psu.edu.
Cloning of Fugu SPARCL1. Fugu SPARCL1 was isolated from cDNA prepared from the whole body of an 18-day-postfertilization embryo. First-strand cDNA was synthesized by using the SMART cDNA construction kit (Clontech). SPARCL1 cDNA fragments were amplified by PCR using specific primers to SPARCL1 (Up2, CTGCTCCTTTTCGGCATCTTAATG; Up3, GTCTTCGGAACTCAGATCCTCGACTT; Up4, GAGTCCGAGATCCCGGCTGACCTC; Up5, CGTGCAAACTCGATGCCGACATAA; Dwn4, CCGGGAATCACTCGGCTGAGCTG; Dwn5, AGCTGGACCAGCACCCATCTGACA; Dwn6, GCGAGCTCCGAGTGCGACAAGAAC; and Dwn7, GCCGCAAAGAGCTGACAGGACGAG) and the SMART cDNAs (5A3, GCAGTGGTATCAACGCAGAGTGGCCA; 5B3, GGTATCAACGCAGAGTGGCCATTACG; 3RA2, AGAGGCCGAGGCGGCCGACAT; and 3RB2, AGGCCGAGGCGGCCGACATGTT). The PCR products were cloned into pCRII-TOPO (Invitrogen). The nucleotide sequences were determined with the primers described above by using a 373-DNA sequencer (Applied Biosystems).
Results
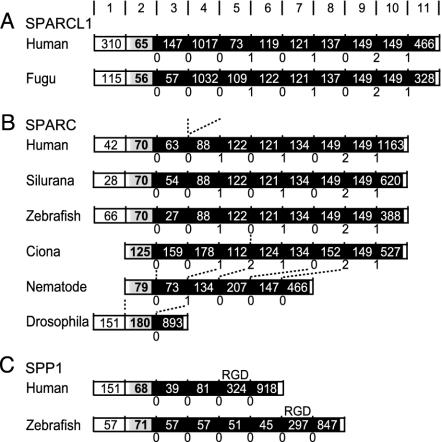
SPARCL1 and SPARC. We identified fugu SPARCL1, indicating that the gene was separated from SPARC before the actinopterygian divergence (Fig. 1, node 7). The analysis of fugu genome sequence revealed the structure of this gene and confirmed features common to the mammalian gene: exon numbers, untranslated first exon, and intron phases (position of intron relative to codons) and locations (Fig. 2A). SPARCL1 has a long exon 4 that is not present in SPARC. This unique exon 4 of SPARCL1 seems to have originated from exon 3 by exon duplication, suggested by their common features: both are extremely rich in Glu, and phase 0 introns (which lie at the boundary between two adjacent codons rather than interrupting a codon) flank both ends (Fig. 2 A). In the fugu genome sequence, we also identified the SPARC gene. Both of these genes have only a single copy in the fugu genome. The amino acid sequence of domain I has experienced considerable evolution, especially in SPARCL1, such that no reliable sequence alignment was found for this domain between fish and mammalian genes. By contrast, domains II and III are highly conserved even between SPARC and SPARCL1 in vertebrates. By genome sequence analysis, we identified domains II and III of zebrafish SPARCL1, which has 81% amino acid sequence identity to fugu SPARCL1. However, no exons coding domain I were identifiable because of the low sequence homology of this domain.
Fig. 2.
Structures of SPARCL1 (A), SPARC (B), and SPP1 (C). Boxes represent the untranslated region (white), signal peptide (localizes the protein in ECM; gray), and the mature protein (black). The length (nucleotide) of each exon is shown in the boxes. Intron phases are described below. Dashed lines show equivalent introns shifted by intron gain, loss, or sliding. (A) Exons 2–5 code domain I, which is separated by phase 0 introns. Exons 6 and 7 code domain II, and exons 8–11 code domain III. (B) Intron 4 in Ciona and intron 5 in nematode slide 1 base upward or downward, respectively. (C) The penultimate exon codes an Arg-Gly-Asp motif.
We also identified the cDNA sequences of Silurana, zebrafish, and Ciona SPARC in EST databases and determined their gene structures by comparison with the corresponding genome sequences (Fig. 2B). All of these SPARC genes show the structures common to the mammalian genes except that the first noncoding exon is missing and intron 4 slid 1 nt upstream into the preceding codon (shifting the intron phase from 0 to 2) in the Ciona gene. In contrast, the protostome SPARC has a quite different structure including the number and phase of exons (Fig. 2B), mainly because of intron loss in protostomes (26).
Teleost SPP1. No SCPP genes have been identified from fish, but trout NOP was suggested previously to have low (≈15%) amino acid sequence identity to tetrapod SPP1 (27). We found zebrafish NOP by searching the EST database with trout NOP. The analysis of the zebrafish genome revealed the common gene structure between zebrafish NOP and mammalian SPP1: in particular, the large penultimate exon of both genes codes an integrin-binding Arg-Gly-Asp motif (Fig. 2C). Thus, we concluded that NOP is orthologous to mammalian SPP1. The amniote SPP1 genes code one or two Asp clusters, whereas teleost SPP1 has no Asp cluster but instead has contiguous SXE motifs (Fig. 3). The Ser residues in these SXE motifs are probably phosphorylated similar to those in mammalian SPP1, in which phosphorylation is required for its function (10, 13). In addition, zebrafish SPP1 has a Glu cluster. Experimental data suggest that the acidic clusters in SCPPs modulate HA formation; the Asp cluster and pSer-rich peptides in SPP1 inhibit HA crystal growth, whereas the Glu clusters in IBSP nucleate HA crystallization (12, 28, 29). Thus, teleost SPP1 probably acts as the modulator of HA crystallization. However, its functions may be distinct from the mammalian proteins because of different amino acid content in its acidic clusters.
Fig. 3.
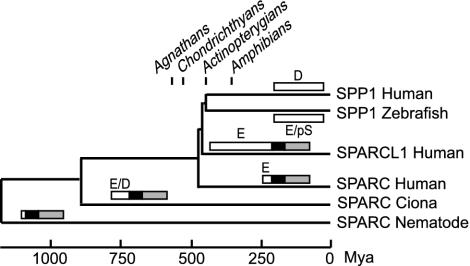
Evolution of SPARC, SPARCL1, and SPP1. Boxes represent domains I (white), II (black), and III (gray) of SPARC and SPARCL1. SPP1 (white box) arose from domain I of SPARCL1. The length of each box is proportionate to that of each domain. Major amino acids appearing in acidic clusters in domain I are represented as E (Glu), D (Asp), or pS (SXE). The scale and the divergence dates of extant animal taxa are based on Fig. 4.
Comparison of Vertebrate SPARC, SPARCL1, and SCPP. The amino acid profiles of domain I are remarkably conserved within SPARC and within SPARCL1 in vertebrates (summarized in Table 1). SPARC has a small (42- to 54-residue) and highly acidic [isoelectric point (pI) 2.7–2.9] domain I: highly abundant acidic residues (33–40% Glu/Asp) and no basic residues. SPARCL1, in contrast, has a large but less acidic domain I (401–427 residues; pI, 4.0–5.2): abundant acidic residues (21–27%) and a considerable number of strongly basic residues (7–14% Lys/Arg). These characteristics of SPARCL1 are common to three of the five dentin/bone SCPPs: DMP1, IBSP, and SPP1 (pI, 3.8–4.2; 24–29% Glu/Asp and 7–9% Lys/Arg). DSPP is cleaved into two proteins, DSP (dentin sialoprotein) and dentin phosphoprotein by a proteinase (30). DSP also has the amino acid contents (pI, 4.5; 21% Glu/Asp and 11% Lys/Arg) common to domain I of SPARCL1. In addition, DSP, DMP1, IBSP, and SPP1 all contain one or two acidic clusters. Phylogenetic conservation of these biochemical characters suggests that the ratio of acidic and basic residues and the cluster of acidic residues rather than the primary sequence are important for HA crystallization.
By contrast, dentin phosphoprotein consists of extensive Ser-Ser-Asp repeats, in which 87% of the Ser residues are phosphorylated in rat, and thus is extremely acidic (pI, 1.1) (7). This extreme acidity is apparently a specialized feature for dentin formation. Among the dentin/bone SCPPs, MEPE is the sole basic protein (pI, 8.5). However, MEPE shows weak sequence homology with DMP1 and is a negative regulator of osteoblasts (31). The analysis of the chicken genome revealed that DMP1, IBSP, and SPP1 reside adjacent to SPARCL1 in the same manner as in mammalian genomes, whereas neither of the two specialized genes, DSPP or MEPE, were identified, suggesting that these two genes arose relatively recently, perhaps in mammals, and rapidly evolved. Interestingly, the duplication of an avian dentin/bone SCPP gene led to a major eggshell matrix protein, ovocleidin-116 (32), which resides between IBSP and SPP1. Based on the pattern of shared characteristics and gene structure, chromosomal location, and calcium-binding ability, we infer that these dentin/bone SCPP genes, and hence all the other SCPP genes, arose from domain I of SPARCL1. The different numbers of exons found in these genes, exclusively flanked by phase 0 introns, suggest that frequent exon duplications facilitated the differentiation of SCPP genes (9).
Evolution of SPARC and SPARCL1. Because of its large domain I, at pH 7.0 SPARCL1 has a net charge 1.5–4.5 times more negative than SPARC within any vertebrate species (Table 1). The negative net charge may correlate with the number of Ca2+ bound by this domain. Ciona SPARC shows intermediate characteristics; a relatively large but extremely acidic domain I (112 residues; pI, 3.0) results in a large negative net charge (–51), which is comparable with vertebrate SPARCL1 (–26 to –76). In protostomes, domain I is as small as that of vertebrate SPARC (31–63 residues) but has no acidic cluster and acidity comparable with vertebrate SPARCL1 (pI, 3.8–5.2) and thus has a small negative net charge (–2 to –16). Nevertheless, domain I of nematode SPARC is still capable of binding to Ca2+ (19). Similar to protostome SPARC proteins, sea urchin SPARC has a slight negative charge and no acidic cluster. In contrast, Ciona SPARC has acidic clusters consisting of both Glu and Asp, and vertebrate SPARC and SPARCL1 contain Glu clusters (Fig. 3).
SPARCL1 has a considerable number (11–14 sites) of SXE motifs in domain I, whereas vertebrate SPARC has none and invertebrate SPARC has less than two of these motifs (Table 1). Probably, in developing bone these Ser residues in SPARCL1 are phosphorylated similar to those of other SCPPs. Newly acquired pSer in SPARCL1 may have augmented calcium-binding capacity. The SXE motif seems to have emerged first in SPARCL1 and passed on to SCPPs as they arose.
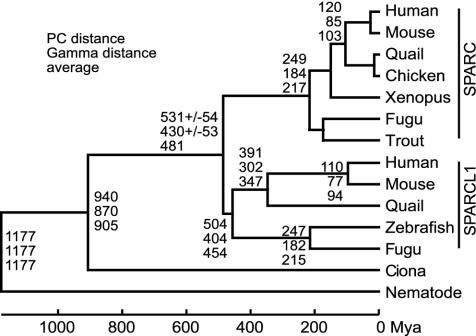
Divergence of SPARC and SPARCL1. Based on the amino acid sequences of domains II and III, we constructed linearized phylogenetic trees for SPARC and SPARCL1 (Fig. 4), consisting of taxa with roughly constant evolutionary rates (24). Both PC and γ distances were used to estimate evolutionary distances. If we set a calibration point 1,177 million years ago (mya) at the divergence of chordates and nematodes (33), the estimated branching dates of vertebrate SPARCL1 correlate well with the previous phylogenetic estimates, although PC distances were above and γ distances below the last estimates (34). Interestingly, the averages of these two distances give divergence dates quite close to the previous estimates of teleost–tetrapod (454 vs. 450 mya), bird–mammal (347 vs. 310 mya), and primate–rodent (94 vs. 91 mya) splits.
Fig. 4.
Linearized phylogenetic tree for SPARC and SPARCL1. Figures at the nodes show divergence time based on PC distance, γ distance, and their average from the top. The γ parameter was estimated: α = 1.20. Standard errors are shown for the divergence of SPARC and SPARCL1. The calibration point was set 1,177 mya at the divergence of nematodes and chordates.
By contrast, the branching dates of vertebrate SPARC are all highly underestimated (Fig. 4), suggesting that the evolutionary rate of vertebrate SPARC was not constant but accelerated after the SPARC–SPARCL1 duplication and slowed after the teleost–tetrapod divergence. The divergence date of Ciona has not been determined, but our estimate, 905 mya on average, is older than the previously estimated cephalochordate–chordate divergence (751 mya), consistent with previous phylogenetic analysis (35).
Based on these considerations, the divergence date of SPARC and SPARCL1 may be estimated to be 531 ± 54 mya by PC distance or 430 ± 53 mya by γ distance with an average of 481 mya. Because the divergence date of chondrichthyans is estimated to be 528 ± 56 mya (34), our estimates raise the possibility that the differentiation of SPARCL1 from SPARC occurred well after the chondrichthyans–teleost divergence (Fig. 3). Although these exact time scales may not reflect actual evolution, the evolutionary distance between vertebrate SPARC and SPARCL1 is only slightly larger than that between tetrapod SPARCL1 and teleost SPARCL1. In fact, fugu SPARCL1 shows higher amino acid sequence identity to quail SPARC (65.8%; 154 of 234) rather than quail SPARCL1 (65.0%; 152 of 234). In contrast, a relatively long evolutionary distance was estimated between teleost and chondrichthyans (34). Because they arose from SPARCL1, the SCPP genes also emerged after the divergence of chondrichthyans (Fig. 1, nodes 6 and 7).
Together these facts make it likely that the developmental mechanism of mammalian tissue mineralization was elaborated during bony fish evolution in actinopterygians or sarcopterygians. Although the genetic tools of tissue mineralization are totally unknown for chondrichthyans, it is quite possible that they have developed their own tools through independent gene duplications and functional selection histories.
Discussion
Many invertebrate metazoans form mineralized skeletons, most of which consist of calcium carbonate, in contrast to the vertebrate HA skeletons (36). However, in vertebrates, both teeth and bone contain considerable amounts of carbonate. Moreover, otolith and avian eggshell consist of calcium carbonate. The zebrafish otolith protein, starmaker, shows extensive sequence convergence with DSPP, and both ovocleidin-116 and SPP1 are present in avian eggshell (32, 37). These facts suggest that ECM proteins do not necessarily specify the types of crystals but instead that environmental ionic condition is more important, leaving the possibility that SPARC might be involved in invertebrate tissue mineralization. However, there is a wide phylogenetic gap between invertebrates and vertebrates having mineralized skeletons (Fig. 1, nodes 1 and 2), inferring their independent origins.
Sea urchin SPARC has small domain I with low calcium-binding capacity, suggesting that this species did not develop a SPARC-based mineralization system. In fact, SPARC has not been identified in the spicule matrix (36). In contrast, Ciona SPARC has the large acidic domain I including acidic clusters, which implies the possibility of the involvement of SPARC in its mineralized tissue (38). Alternatively, the large calcium-binding capacity may facilitate stabilization of structural proteins and enable more diverse and complex cell–ECM protein and cell–cell interactions. Indeed, this is one possible origin of physiological processes that were subsequently co-opted for mineralization and the evolution of structural and protective hard tissue in vertebrates.
In addition to SCPPs, two γ-carboxyglutamic acid (GLA)-containing proteins, matrix GLA (MGP) and bone GLA (BGP; osteocalcin), are important for mammalian tissue mineralization. Teleost MGP is expressed in cartilage but not in bone (39). BGP is expressed not in cartilage but in dentin and bone and has never been found in chondrichthyans (40). These facts suggest that neither MGP nor BGP were involved in early dentin/bone formation. By contrast, SPARC remains as a candidate gene for involvement in the earliest tissue mineralization (Fig. 1, nodes 3 and 4), a possibility consistent with the observation that SPARC is expressed in teleost bone and scales (41). Shark MGP has three SXE motifs, two of which are partially phosphorylated (42). In SPP1, the inhibitory effect on mineralization depends on the extent of Ser phosphorylation (13). Thus, the inhibition of mineralization regulated by pSer might have developed early in chondrichthyans, the process later being used by SCPPs.
An experimentally constructed chimeric protein consisting of a Glu cluster of IBSP and the collagen-binding site of decorin induces HA crystals that bind with collagen (43), suggesting that a collagen-binding site and an acidic cluster are enough for HA crystallization on collagen. Both of these elements are retained in vertebrate SPARC and SPARCL1 (15, 44). Thus, SPARC seems to be a key molecule for the initiation of vertebrate tissue mineralization. Later, SPARCL1 might also have become involved in tissue mineralization, as suggested by the expression of this gene in bone (16). However, in mammals, the functions of these two proteins, especially SPARCL1, may be reduced or derived, because many SCPPs, MGP, and BGP overlap or replaced their original functions.
The dermal skeleton of agnathans mineralizes at the epithelium–mesenchymal interface, which is delimited by a basal lamina. SPARC is distributed in the basal lamina both in protostomes and deuterostomes (18, 21), suggesting that the involvement of SPARC in mineralized tissue began when the first dermal skeleton appeared in vertebrates (Fig. 1, node 4). In vertebrate dermis, SPARC associates with fibrillar collagens, which are abundant even in lampreys, and facilitates the maturation of type I collagen bundles, reinforcing their mechanical strength (45, 46). Thus, we propose that the initial dermal skeleton formed on dermal fibrillar collagen underlying a basal lamina and that the primitive calcareous deposits primarily acted as the reinforcement of dermis; these deposits tightly bundle collagen fibrils, as do chemical crosslinks. Indeed, at least some ancient dentin- and bone-related tissues are assumed to be collagenous (47). Although there is no direct fossil evidence to date, the most primitive body armor may not have covered the entire head or body, but even incompletely mineralized stiff dermis seems to be effective in protecting and supporting elaborated anterior brain, peripheral sense organs, and paired special sense organs and also to be efficient in transmitting muscular force for propulsion and resisting hydrostatic pressure for faster swimming (2, 4, 48). Mineralized tissue was probably highly adaptive in many ways, and the amount of mineral was augmented rapidly. As some taxa evolved to specialize in predation, the dermal skeleton became more important as defensive armor.
Type I [α1(I)2α2(I)] collagen is the most abundant protein in dentin and bone, whereas type II [α1(II)3] is the main collagen in cartilage (49). One of these three collagen genes [α1(I), α2(I), and α1(II)] or the type III [α1(III)3] collagen gene is linked to each of the four Hox gene clusters in the mammalian genome; thus, the differentiation of these fibrillar collagens may have been enabled by chromosomal (or segmental) duplications that produced Hox clusters during vertebrate evolution. Only a single Hox cluster seems to have existed in the last common ancestor of lampreys and gnathostomes (Fig. 1, node 2) (50). The initial duplication separated a primordial type I collagen gene from an undifferentiated precursor gene for types II and III collagens. This duplication occurred before the divergence of chondrichthyans (Fig. 1, node 6), which have at least two Hox clusters (51), and hence may be related to the initial vertebrate tissue mineralization. The differential expression of these duplicated collagen genes may have been the basis for the subsequent differentiation of pluripotent mesenchymal cells into novel types of cells that later specialized into odontoblasts and osteoblasts. During this experiment, these novel cells, still under specialization or derived into distinct cell types, may have produced various mineralized tissues found only in extinct agnathans such as aspidin and diverse forms of dentin (1, 47). Integrin genes and the six Dlx genes are also linked to and have been duplicated along with the Hox clusters (52). Among these genes, αIIb, α3, α4, α5, αV, and β3 integrins are expressed in bone cells; Dlx3 and Dlx5 regulate cartilage, bone, or tooth growth; and all six Dlx genes coordinately specify jaw and perhaps also dental patterning (14, 53, 54). These chromosomal duplications seem to have been important in vertebrate tissue mineralization and connect transcription factors active during jaw and tooth patterning with subsequent mineralization.
The cartilage of cephalochordates, hagfishes, and lampreys consists not of collagens but of unique proteins (55). Based on histological observation, the mineralized cartilage of extinct agnathans also seems to be noncollagenous (Fig. 1, node 5); hence, their mineralization was assumed to represent a parallel evolutionary move toward the mineralized endoskeleton found in modern vertebrates (56). This is consistent with the observation that SPARC is not expressed in lamprey gill pouches, which consist of noncollagenous cartilage (57), suggesting that SPARC associates only with collagenous skeleton. Thus, ancient noncollagenous cartilage and dermal skeleton may have used different mineralization systems.
SPARC expression is intimately associated with hypertrophy of chondrocytes and mineralized cartilage in mammals (58), which suggests that the fundamental machinery of cartilage mineralization in endoskeleton was derived from that of the evolutionarily preceding dermal skeleton; both share the same tools, SPARC and fibrillar collagen. Later, the common machinery facilitated mineralized cartilage to become the model of endochondral bone that first appeared in actinopterygians (Fig. 1, node 7) (4). The remodeling from cartilage to bone must have been regulated strictly by both chondrocytes and osteoblasts. These two types of cells are distinguished by type I and II collagens that were fully established after the final Hox-related chromosomal duplication. The date of this duplication seems to be slightly earlier than the emergence of endochondral bone (51), suggesting that the differentiation of type II collagen might have been important for the innovation of endochondral bone.
SPP1 was found in the zebrafish EST database as five distinct entries, showing substantial expression level of this gene, whereas no SPP1 was found in the databases for medaka ESTs or the fugu genome sequence (Takifugu and Tetraodon nigro-viridis; Fig. 1, node 8). The absence may be attributed to the small sizes of these databases and SPP1 not having yet been sequenced, although 95% of Takifugu and 70% of Tetraodon genomes have been sequenced. Alternatively, SPP1 may truly be missing; these fish have a specialized bone-resorbing mechanism. Both fugu and medaka have acellular (osteocyte-deprived) bone and no mammalian-like multinucleated osteoclasts but morphologically distinguishable bone-resorbing cells (59). Furthermore, the activity of bone-resorbing cells may be limited in normal conditions (60). SPP1 anchors osteoclasts to resorbing bone surface (61), but this function may be degenerate in the specialized bone-resorbing cells, and fugu and medaka may have secondarily lost SPP1.
Three major mammalian enamel protein genes, AMEL, AMBN, and ENAM, arose from SPARCL1, most likely after the chondrichthyan divergence. The N terminus of both Xenopus AMEL and AMBN shows relatively high sequence identity to mammalian proteins: ≈60% for 62 residues and ≈50% for 96 residues, respectively (62, 63). Thus, we expected to find the fish orthologs in genome sequences or EST databases, but we found none of these genes. This suggests that actinopterygians have no mammalian-type enamel proteins (Fig. 1, node 7), an inference that is also supported by what is known of the developmental processes: mammalian enamel is epithelial in origin, whereas fish enameloid is mainly of mesenchymal origin (64), and teleost enameloid forms on collagen, whereas mammalian enamel does not involve collagen but rather enamel SCPPs. In addition, the Ordovician agnathan Pycnaspis had cap enameloid that was penetrated by fine canals extending out toward its external surface (1). Thus, in many respects, enameloid is closer to dentin than to mammalian enamel (3, 47). Immunohistochemical analysis has long been interpreted as suggesting that fish have mammalian-like enamel ECM proteins (65, 66). However, these genes have never been isolated. Two distinct enamel proteinases are involved in mammalian enamel mineralization: matrix metalloproteinase-20 (MMP20) and Kallikrein-4 (KLK4) (67). The human genes coding these proteins form distinct gene clusters: nine MMP genes on 11q22 and 15 KLK genes on 19q13. Our phylogenetic analysis showed that both of these clusters formed after the teleost–mammal divergence, corroborating the recent origin of mammalian enamel (data not shown). Moreover, enamel or enameloid tissues found in conodont oral skeleton and dermal skeleton of the heterostracomorphs are not linked phylogenetically to modern enameloid in gnathostomes (Fig. 1, nodes 3, 4, and 6) (4). These observations lead us to the conclusion that enamel and enameloid have multiple origins and that mammalian enamel is a unique tissue probably developed in sarcopterygians (Fig. 1, nodes 7–9). If so, their similar hard structural nature as an overlay on exoskeleton and teeth is a convergent evolution.
Mammalian ameloblasts secrete proteinases that digest enamel matrix proteins and remove digested proteins from mineralizing matrix by endocytosis (6); highly mineralized enamel thus is formed by the removal of the protein content. In chondrichthyans, odontoblasts but not the inner dental epithelium (IDE; epithelium overlying enameloid) deposit enameloid matrix proteins. Although no enameloid proteinases have been identified, the IDE removes enameloid proteins, suggesting that the initial function of IDE may have been the removal of protein matrix to form highly mineralized enameloid (64). Later, in sarcopterygians IDE began to secrete a dentin/bone SCPP above basal lamina. Collagen was probably the mineralization matrix of this unique tissue. Afterward, this SCPP specialized into the enamel matrix protein. In this tissue, collagen is no longer required as a protein matrix. The differentiation from a dentin/bone SCPP to an enamel SCPP required a shift of major expression domains from mesenchyme to epithelium. This shift facilitated the generation of casein and salivary proteins from an enamel SCPP in the mammalian lineage (Fig. 1, node 9); both mammary glands and salivary glands are of epithelial origin (9). The same shift independently occurred in birds, which secrete an eggshell-specific SCPP from the eggshell gland. Thus, SCPPs developed various parallel specializations that facilitated adaptive evolution in vertebrates.
We hypothesized that the primitive mechanism of mammalian tissue mineralization initially developed in the dermal skeleton of early agnathans or perhaps in the conodont oral skeleton. However, this fundamental mechanism was considerably modified in actinopterygians and sarcopterygians in particular. Genetic analysis of agnathans and critical gnathostomes will further enhance the interpretation of early vertebrate fossil records, and elucidate the evolution of mineralized tissues that is pivotal to the sustained success of vertebrates.
Supplementary Material
Acknowledgments
We thank Dr. Alan Walker, Dr. Anne Buchanan, Mr. Jongmin Nam, and Mr. Samuel J. Sholtis for critical discussion. This work was supported by the financial support of U.S. National Science Foundation Awards SBR 9804907 and BCS-0343442 and research funds from Pennsylvania State University.
Abbreviations: HA, hydroxyapatite; ECM, extracellular matrix; SCPP, secretory calcium-binding phosphoprotein; AMEL, amelogenin; AMBN, ameloblastin; ENAM, enamelin; DSPP, dentin sialophosphoprotein; DMP1, dentin matrix acidic phosphoprotein 1; IBSP, integrin-binding sialoprotein; MEPE, matrix extracellular, phosphoglycoprotein; SPP1, secreted phosphoprotein 1; SXE, Ser-Xaa-Glu; pSer, phospho-Ser; SPARC, secreted protein, acidic, cysteine-rich; SPARCL1, SPARC-like 1; NOP, novel ovarian protein; PC, Poisson correction; mya, million years ago; MGP, matrix γ-carboxyglutamic acid; BGP, bone γ-carboxyglutamic acid.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY575071–AY575077 and AY620817).
References
- 1.Ørvig, T. (1967) in Structure and Chemical Organization of Teeth, ed. Miles, A. E. W. (Academic, New York), Vol. I, pp. 45–110. [Google Scholar]
- 2.Reif, W.-E. (1982) Evol. Biol. 15, 287–368. [Google Scholar]
- 3.Reif, W.-E. (2001) Neues Jahrb. Geol. Palaontol. Abh. 219, 285–304. [DOI] [PubMed] [Google Scholar]
- 4.Donoghue, P. C. & Sansom, I. J. (2002) Microsc. Res. Tech. 59, 352–372. [DOI] [PubMed] [Google Scholar]
- 5.Smith, M. M. & Coates, M. I. (2001) in Major Events in Early Vertebrate Evolution, ed. Ahlberg, P. E. (Taylor and Francis, London), pp. 223–240.
- 6.Zeichner-David, M., Diekwisch, T., Fincham, A., Lau, E., MacDougall, M., Moradian-Oldak, J., Simmer, J., Snead, M. & Slavkin, H. C. (1995) Int. J. Dev. Biol. 39, 69–92. [PubMed] [Google Scholar]
- 7.Linde, A. & Goldberg, M. (1993) Crit. Rev. Oral Biol. Med. 4, 679–728. [DOI] [PubMed] [Google Scholar]
- 8.Peyer, B. (1968) Comparative Odontology (Univ. of Chicago Press, Chicago).
- 9.Kawasaki, K. & Weiss, K. M. (2003) Proc. Natl. Acad. Sci. USA 100, 4060–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sørensen, E. S., Højrup, P. & Petersen, T. E. (1995) Protein Sci. 4, 2040–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunter, G. K., Hauschka, P. V., Poole, A. R., Rosenberg, L. C. & Goldberg, H. A. (1996) Biochem. J. 317, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pampena, D. A., Robertson, K. A., Litvinova, O., Lajoie, G., Goldberg, H. A. & Hunter, G. K. (2004) Biochem. J. 378, 1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jono, S., Peinado, C. & Giachelli, C. M. (2000) J. Biol. Chem. 275, 20197–20203. [DOI] [PubMed] [Google Scholar]
- 14.Schaffner, P. & Dard, M. M. (2003) Cell Mol. Life Sci. 60, 119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Termine, J. D., Kleinman, H. K., Whitson, S. W., Conn, K. M., McGarvey, M. L. & Martin, G. R. (1981) Cell 26, 99–105. [DOI] [PubMed] [Google Scholar]
- 16.Soderling, J. A., Reed, M. J., Corsa, A. & Sage, E. H. (1997) J. Histochem. Cytochem. 45, 823–835. [DOI] [PubMed] [Google Scholar]
- 17.Brekken, R. A. & Sage, E. H. (2000) Matrix Biol. 19, 569–580. [DOI] [PubMed] [Google Scholar]
- 18.Bradshaw, A. D. & Sage, E. H. (2001) J. Clin. Invest. 107, 1049–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarzbauer, J. E. & Spencer, C. S. (1993) Mol. Biol. Cell 4, 941–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Girard, J. P. & Springer, T. A. (1995) Immunity 2, 113–123. [DOI] [PubMed] [Google Scholar]
- 21.Martinek, N., Zou, R., Berg, M., Sodek, J. & Ringuette, M. (2002) Dev. Genes Evol. 212, 124–133. [DOI] [PubMed] [Google Scholar]
- 22.Takezaki, N., Rzhetsky, A. & Nei, M. (1995) Mol. Biol. Evol. 12, 823–833. [DOI] [PubMed] [Google Scholar]
- 23.Glazko, G. V. & Nei, M. (2003) Mol. Biol. Evol. 20, 424–434. [DOI] [PubMed] [Google Scholar]
- 24.Nei, M. & Kumar, S. (2000) Molecular Evolution and Phylogenetics (Oxford Univ. Press, New York).
- 25.Gu, X. & Zhang, J. (1997) Mol. Biol. Evol. 14, 1106–1113. [DOI] [PubMed] [Google Scholar]
- 26.Patthy, L. (1999) Gene 238, 103–114. [DOI] [PubMed] [Google Scholar]
- 27.Bobe, J. & Goetz, F. W. (2001) FEBS Lett. 489, 119–124. [DOI] [PubMed] [Google Scholar]
- 28.Hunter, G. K., Kyle, C. L. & Goldberg, H. A. (1994) Biochem. J. 300, 723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter, G. K. & Goldberg, H. A. (1994) Biochem. J. 302, 175–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDougall, M., Simmons, D., Luan, X., Nydegger, J., Feng, J. & Gu, T. T. (1997) J. Biol. Chem. 272, 835–842. [DOI] [PubMed] [Google Scholar]
- 31.Gowen, L. C., Petersen, D. N., Mansolf, A. L., Qi, H., Stock, J. L., Tkalcevic, G. T., Simmons, H. A., Crawford, D. T., Chidsey-Frink, K. L., Ke, H. Z., et al. (2003) J. Biol. Chem. 278, 1998–2007. [DOI] [PubMed] [Google Scholar]
- 32.Mann, K., Hincke, M. T. & Nys, Y. (2002) Matrix Biol. 21, 383–387. [DOI] [PubMed] [Google Scholar]
- 33.Wang, D. Y., Kumar, S. & Hedges, S. B. (1999) Proc. R. Soc. London Ser. B 266, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar, S. & Hedges, S. B. (1998) Nature 392, 917–920. [DOI] [PubMed] [Google Scholar]
- 35.Hedges, S. B. (2001) in Major Events in Early Vertebrate Evolution, ed. Ahlberg, P. E. (Taylor and Francis, London).
- 36.Wilt, F. H., Killian, C. E. & Livingston, B. T. (2003) Differentiation (Berlin) 71, 237–250. [DOI] [PubMed] [Google Scholar]
- 37.Söllner, C., Burghammer, M., Busch-Nentwich, E., Berger, J., Schwarz, H., Riekel, C. & Nicolson, T. (2003) Science 302, 282–286. [DOI] [PubMed] [Google Scholar]
- 38.Lambert, G. & Lambert, C. C. (1990) in Skeletal Biomineralization: Patterns, Precesses, and Evolutionary Trends, ed. Joseph, G. C. (Van Nostrand Reinhold, New York), Vol. I, pp. 461–469. [Google Scholar]
- 39.Simes, D. C., Williamson, M. K., Ortiz-Delgado, J. B., Viegas, C. S., Price, P. A. & Cancela, M. L. (2003) J. Bone Miner. Res. 18, 244–259. [DOI] [PubMed] [Google Scholar]
- 40.Pinto, J. P., Ohresser, M. C. & Cancela, M. L. (2001) Gene 270, 77–91. [DOI] [PubMed] [Google Scholar]
- 41.Lehane, D. B., McKie, N., Russell, R. G. & Henderson, I. W. (1999) Gen. Comp. Endocrinol. 114, 80–87. [DOI] [PubMed] [Google Scholar]
- 42.Price, P. A., Rice, J. S. & Williamson, M. K. (1994) Protein Sci. 3, 822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hunter, G. K., Poitras, M. S., Underhill, T. M., Grynpas, M. D. & Goldberg, H. A. (2001) J. Biomed. Mater. Res. 55, 496–502. [DOI] [PubMed] [Google Scholar]
- 44.Hambrock, H. O., Nitsche, D. P., Hansen, U., Bruckner, P., Paulsson, M., Maurer, P. & Hartmann, U. (2003) J. Biol. Chem. 278, 11351–11358. [DOI] [PubMed] [Google Scholar]
- 45.Johnels, A. G. (1950) Acta Zool. (Stockholm) 31, 177–185. [Google Scholar]
- 46.Bradshaw, A. D., Puolakkainen, P., Dasgupta, J., Davidson, J. M., Wight, T. N. & Helene Sage, E. (2003) J. Invest. Dermatol. 120, 949–955. [DOI] [PubMed] [Google Scholar]
- 47.Halstead, B. L. (1987) in Developmental and Evolutionary Aspects of the Neural Crest, ed. Maderson, P. F. (Wiley, New York), pp. 339–358.
- 48.Gans, C. & Northcutt, R. G. (1983) Science 220, 268–274. [DOI] [PubMed] [Google Scholar]
- 49.Boot-Handford, R. P. & Tuckwell, D. S. (2003) BioEssays 25, 142–151. [DOI] [PubMed] [Google Scholar]
- 50.Fried, C., Prohaska, S. J. & Stadler, P. F. (2003) J. Exp. Zool. 299B, 18–25. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, G. P., Amemiya, C. & Ruddle, F. (2003) Proc. Natl. Acad. Sci. USA 100, 14603–14606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larhammar, D., Lundin, L. G. & Hallbook, F. (2002) Genome Res. 12, 1910–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Merlo, G. R., Zerega, B., Paleari, L., Trombino, S., Mantero, S. & Levi, G. (2000) Int. J. Dev. Biol. 44, 619–626. [PubMed] [Google Scholar]
- 54.Depew, M. J., Lufkin, T. & Rubenstein, J. L. (2002) Science 298, 381–385. [DOI] [PubMed] [Google Scholar]
- 55.Wright, G. M., Keeley, F. W. & Robson, P. (2001) Cell Tissue Res. 304, 165–174. [DOI] [PubMed] [Google Scholar]
- 56.Janvier, P. & Arsenault, M. (2002) Nature 417, 609. [DOI] [PubMed] [Google Scholar]
- 57.Ringuette, M., Damjanovski, S. & Wheeler, D. (1991) Biochem. Cell Biol. 69, 245–250. [DOI] [PubMed] [Google Scholar]
- 58.Aeschlimann, D., Wetterwald, A., Fleisch, H. & Paulsson, M. (1993) J. Cell Biol. 120, 1461–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Witten, P. E. (1997) Cell Tissue Res. 287, 591–599. [DOI] [PubMed] [Google Scholar]
- 60.Glowacki, J., Cox, K. A., O'Sullivan, J., Wilkie, D. & Deftos, L. J. (1986) Proc. Natl. Acad. Sci. USA 83, 4104–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reinholt, F. P., Hultenby, K., Oldberg, A. & Heinegard, D. (1990) Proc. Natl. Acad. Sci. USA 87, 4473–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Toyosawa, S., O'HUigin, C., Figueroa, F., Tichy, H. & Klein, J. (1998) Proc. Natl. Acad. Sci. USA 95, 13056–13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shintani, S., Kobata, M., Toyosawa, S. & Ooshima, T. (2003) Gene 318, 125–136. [DOI] [PubMed] [Google Scholar]
- 64.Sasagawa, I. (2002) Microsc. Res. Tech. 59, 396–407. [DOI] [PubMed] [Google Scholar]
- 65.Herold, R. C., Graver, H. T. & Christner, P. (1980) Science 207, 1357–1358. [DOI] [PubMed] [Google Scholar]
- 66.Diekwisch, T. G., Berman, B. J., Anderton, X., Gurinsky, B., Ortega, A. J., Satchell, P. G., Williams, M., Arumugham, C., Luan, X., McIntosh, J. E., et al. (2002) Microsc. Res. Tech. 59, 373–395. [DOI] [PubMed] [Google Scholar]
- 67.Simmer, J. P. & Hu, J. C. (2002) Connect. Tissue Res. 43, 441–449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.