Abstract
In an approach to clone and characterize centromeric DNA sequences of Candida albicans by chromatin immunoprecipitation, we have used antibodies directed against an evolutionarily conserved histone H3-like protein, CaCse4p (CENP-A homolog). Sequence analysis of clones obtained by this procedure reveals that only eight relatively small regions (≈3 kb each) of the Can. albicans genome are selectively enriched. These CaCse4-bound sequences are located within 4- to 18-kb regions lacking ORFs and occur once in each of the eight chromosomes of Can. albicans. Binding of another evolutionarily conserved kinetochore protein, CaMif2p (CENP-C homolog), colocalizes with CaCse4p. Deletion of the CaCse4p-binding region of chromosome 7 results in a high rate of loss of the altered chromosome, confirming that CaCse4p, a centromeric histone in the CENP-A family, indeed identifies the functional centromeric DNA of Can. albicans. The CaCse4p-rich regions not only lack conserved DNA motifs of point (<400 bp) centromeres and repeated elements of regional (>40 kb) centromeres, but also each chromosome of Can. albicans contains a different and unique CaCse4p-rich centromeric DNA sequence, a centromeric property previously unobserved in other organisms.
Centromeres are cis-acting chromosomal domains that direct the formation of kinetochores that subsequently attach to spindle microtubules, enabling faithful chromosome segregation during mitosis and meiosis. A complete understanding of how the centromere functions during chromosome segregation requires identification of the centromere DNA structural components. The structure–function relationship of centromeres has been extensively studied in a variety of organisms, including Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster, Caenorhabditis elegans, and humans. These studies indicate that this important chromosomal domain, found in all eukaryotes, is functionally conserved, but the centromeric DNA sequences are widely diverged among different organisms and sometimes even between chromosomes of the same organisms. Functional centromere DNA sequences in the budding yeast Saccharomyces cerevisiae and five other budding yeasts are known to consist of short AT-rich regions (≈125–400 bp) termed point centromeres (1, 2). These point centromeres contain highly conserved DNA sequences important for specific protein binding. In contrast, regional centromeres of other eukaryotes, including the fission yeast Schizosaccharomyces pombe, contain much larger (40–4,000 kb) repetitive, AT-rich, and heterochromatic DNA regions (3). Moreover, no conserved DNA sequence motifs common to all centromeres exist in higher eukaryotes. Instead, several lines of evidence suggest that a heritable epigenetic “marking” by a trans-acting non-DNA sequence determinant distinguishes centromere chromatin from bulk chromatin (4, 5).
The exact mechanism or the factors involved in epigenetic regulation of centromere determination are not yet identified, but members of two key, evolutionarily conserved protein families, CENP-A and CENP-C, are present throughout the cell cycle only at functional centromeres (6, 7). These proteins, found in species as diverse as yeasts and humans, are essential and are required for chromosome segregation in all eukaryotes studied to date. Although depletion of either CENP-A or CENP-C causes an identical phenotype, the loss of kinetochore structural integrity and function, studies in C. elegans indicate that CENP-A acts upstream of CENP-C (8–12). CENP-A, which is a variant of histone H3, appears to substitute for histone H3 in some or all centromeric nucleosome octamers and, thus, may epigenetically mark centromere DNA as the primary site for kinetochore formation (7, 13, 14).
The human pathogenic yeast Candida albicans grows in a unicellular budding form and in filamentous forms (pseudohyphae and true hyphae) and, thus, its mechanism of chromosome segregation is of special interest. Despite nearly complete DNA sequence information of the Can. albicans genome (15), almost nothing is known about this organism's centromeres. Therefore, to develop a molecular tool to identify centromeres of this medically important organism, we previously cloned the Can. albicans CENP-A homolog, CaCSE4. CaCse4p, like other CENP-A proteins, was shown to be essential for cell viability and required for proper chromosome segregation (16). In the present study, we report the isolation and characterization of the centromeric DNA from all eight Can. albicans chromosomes. Our results reveal a very high degree of DNA sequence heterogeneity among the Can. albicans centromeres.
Materials and Methods
Strains, Media, and Transformation Procedures. Can. albicans strains and their construction are listed in Table 1 and Supporting Text, which are published as supporting information on the PNAS web site. Can. albicans strains were grown in yeast extract/peptone/dextrose (YPD), yeast extract/peptone/succinate (YPSucc), or supplemented synthetic/dextrose (SD) minimal media at 30°C as described (16). Can. albicans cells were transformed by standard techniques (17, 18).
Chromosome 5 Loss Assay. Equal numbers of RM1000AH or RM1000AH/ΔCEN7 cells were added to 10 tubes containing YPD plus 50 μg/ml uridine or SD lacking uridine and grown ≈10 generations at 30°C. Approximately (3–6) × 105 cells were plated on synthetic minimal medium plus sorbose (2%) and incubated at 37°C for 4 days. The rate of chromosome 5 loss equals the number of colonies growing on sorbose medium divided by the total number of cells plated.
Chromatin Immunoprecipitation (ChIP) and Antibodies. Detailed procedures for ChIP (19, 20), linker attachment, PCR, cloning, and slot blots (21, 22) can be found in Supporting Text. Affinity-purified polyclonal rabbit antibodies against an N-terminal peptide (amino acid positions 1–18) of CaCse4p (16) and mouse anti-c-myc (Ab-1) mAbs (Oncogene Research Products, Boston) were used for ChIP at a final concentration of 4 and 5 μg/ml, respectively.
DNA Sequence Comparisons. Plasmid DNA was sequenced by the Iowa State University DNA Sequencing Facility (Ames). Sequences were analyzed by using the GCG Wisconsin Sequence Analysis programs bestfit, repeat, stemloop, and composition. blast was used to compare the CaCse4p-rich regions against a database of the chromosome 7 CaCse4p-rich region (window size, 11–13 nucleotides; stringency, one mismatch).
Results
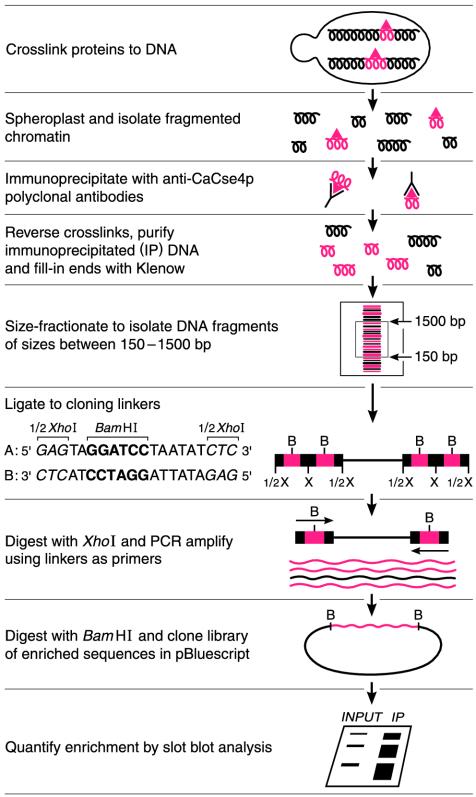
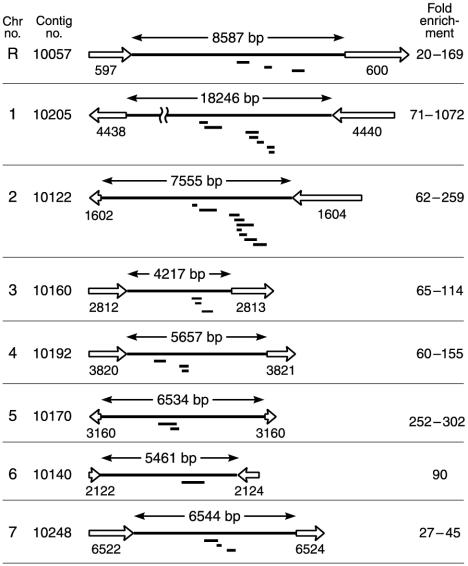
Strategy for Identification of Genome-Wide CaCse4p-Bound Regions in Can. albicans by ChIP. We have cloned enriched DNA fragments from a ChIP procedure by using an affinity-purified polyclonal antibody preparation against the first 18 aa of CaCse4p (16) as shown in Fig. 1. In brief, CaCse4p was reversibly crosslinked in vivo to its cognate DNA targets by formaldehyde. Sheared chromatin was immunoprecipitated with anti-CaCse4p antibodies and a pool of purified immunoprecipitated DNA sequences was fitted with cloning linkers, amplified by PCR, and cloned as a plasmid library in E. coli (21–23). Approximately 20% of the cloned inserts were found to be enriched significantly by the ChIP procedure and their enrichment was 20- to 1,000-fold higher than for the single-copy gene CaLEU2. DNA sequence data of the 31 enriched clones indicated that CaCse4p-bound sequences were clustered at eight relatively small regions (each 0.8–3.1 kb in length) that represent <0.2% of the Can. albicans genome (Fig. 2; see Table 2, which is published as supporting information on the PNAS web site). These eight regions localized to one in each of the eight chromosomes of Can. albicans based on the Can. albicans genome project (www-sequence.stanford.edu:8080/haploid19.html) and our experimental results. Notably, all these regions excluded ORFs and the length of the non-ORF regions ranged from 4.2 to 18.2 kb. Because most organisms carry a single functional centromere locus per chromosome and most centromeric regions are ORF-free, our data are consistent with the identification of the centromeric DNAs of Can. albicans.
Fig. 1.
Cloning and identification of CaCse4p-bound chromosomal regions. To determine enrichment, cloned DNA inserts were used individually as 32P-labeled probes against serial dilutions of total DNA (input) and the PCR-amplified, immunoprecipitated DNA pool (IP). Enrichment was calculated from the ratio of IP/input signals and was normalized to that detected with a CaLEU2 probe. Enriched DNAs were sequenced and compared against the Can. albicans database by using blast to determine the genome-wide distribution of CaCse4p. B, BamHI; X, XhoI.
Fig. 2.
CaCse4p-bound DNA sequences are clustered in a single, nonhomologous, gene-free region on each Can. albicans chromosome. Only one homolog is shown for each chromosome and the region is identified by its assembly 19 supercontig number. The length of each non-ORF region (long solid line) is indicated. ORFs (open arrows) are numbered with their unique assembly 19 designations. ORFs <150 aa are not shown. The location of individual, CaCse4p ChIP-enriched DNA fragments (short solid bars) are shown below each schematic illustration. DNA fragment enrichment, normalized to that for CaLEU2 DNA, is presented as a range for each chromosome. For map positions and enrichment values, see Table 2.
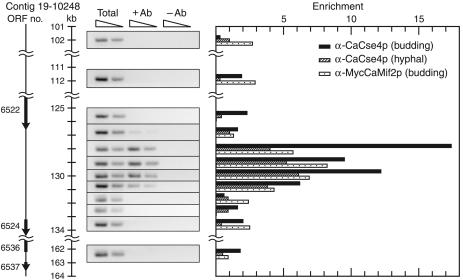
CaCse4p Binding Is Limited to a 3-kb Region on Chromosome 7 in Can. albicans Budded and Hyphal Cells. We performed a standard ChIP assay to localize more precisely the boundaries of CaCse4p binding on chromosome 7, the only Can. albicans chromosome that has been physically mapped (24). CAI4 and SC5314 strains were used for isolating chromatin from budded and hyphal cells, respectively. Chromatin was fragmented to an average size of 400 bp and immunoprecipitated with or without anti-CaCse4p antibodies. The recovered DNA was assayed with a set of 12 primer pairs (see Table 3, which is published as supporting information on the PNAS web site) to examine CaCse4p enrichment over a 62-kb region (nucleotides 101000–163000 of Contig 19-10248), which overlaps the 1.3-kb CaCse4p-binding region identified from the ChIP library (Fig. 3). This experiment revealed that CaCse4p, both in budded and hyphal Can. albicans cells, specifically binds to a 3-kb region (nucleotides 128000–131000 of Contig 19-10248) on chromosome 7. Adjacent non-ORF and ORF sequences showed background levels of enrichment comparable with those for outlying sequences located 10–30 kb away. We obtained similar results for chromosome 1 where CaCse4p binding was detected over a 2.8-kb region (data not shown) in budded cells, in agreement with our original screen. Therefore, these data suggest that the CaCse4p-binding regions of Can. albicans are ≈3 kb in length, similar in size to the 4- to 7-kb CENP-A (Cnp1p)-bound central core regions seen in fission yeast Schizosaccharomyces pombe centromeres (25).
Fig. 3.
Colocalization of two key kinetochore proteins CaCse4p and CaMif2p to a 3-kb region of Can. albicans chromosome 7. A standard ChIP assay was performed on strains CAI4, SC5314, and CAMB1 with primer pairs (Table 3) that amplify 178- to 292-bp regions spaced approximately every 1 kb between Orf19.6522.prot and Orf19.6524.prot. PCR with serial dilutions of total DNA and with or without antibody ChIP DNA fractions were performed. (Left) The ethidium bromide-stained PCR products obtained from budding CAI4 cells are shown here as negative images and are aligned with a scale bar to show the locations tested for enrichment. (Right) Enrichment of CaCse4p binding on chromosome 7 in budded CAI4 cells (black bars) and hyphal SC5314 cells (hatched bars) and enrichment of CaMif2p binding on chromosome 7 in budded CAMB1 cells (speckled bars) as determined by standard ChIP assays are graphically represented. Enrichment equals (+Ab) minus (–Ab) signals divided by the total DNA signal and is normalized to a value of 1 for CaLEU2.
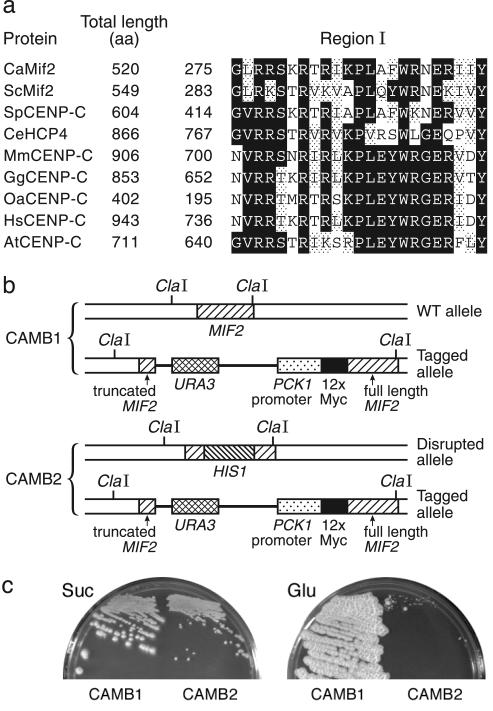
Binding of the Evolutionarily Conserved Kinetochore Protein CaMif2p (CaCENP-C) Is Preferentially Enriched at the CaCse4p-Rich Regions. CENP-C proteins are conserved kinetochore proteins important for proper chromosome segregation. With Saccharomyces cerevisiae Mif2p (CENP-C homolog) as the query sequence, a blast analysis identified two identical alleles, Orf19.5551.prot and Orf19.12997.prot, as the CENP-C homolog (CaMif2p) in Can. albicans (26% identity and 45% similarity over 471 aa). CaMIF2 codes for a 520-aa-long predicted protein containing the 23-aa C-terminal CENP-C conserved region that is important for centromere targeting (refs. 6 and 26; Fig. 4a). We used an integration strategy to express CaMif2p tagged with 12 copies of human c-myc sequence at the N terminus from a regulated promoter taken from the phosphoenolpyruvate carboxykinase (CaPCK1) gene of Can. albicans (Fig. 4b). Expression from the CaPCK1 promoter is repressed by glucose and induced by succinate (27). The inability of CAMB2 (mif2Δ/PCK1prMYCMIF2) cells to grow on glucose media shows the CaMIF2 gene is essential for viability of Can. albicans cells, whereas the ability to grow on succinate indicates myc-tagged CaMif2p functionally complements the Camif2 deletion defect (Fig. 4c). Strain CAMB2 grows slower than CAMB1 (one wild-type CaMIF2 gene) on succinate, probably because of impairment of function by the relatively large Myc domain.
Fig. 4.
CaMif2p, the CENP-C homolog in Can. albicans, is an essential protein. (a) CaMif2p contains a region that is conserved in proteins of the CENP-C family. Conserved regions of various organisms were identified by the Blockmaker server (www.blocks.fhcrc.org). Black and gray boxes indicate identical and similar amino acid residues, respectively. GenBank accession numbers: Saccharomyces cerevisiae ScMif2p, Z28089; Schizosaccharomyces pombe SpCENP-C, CAB52737; C. elegans CeHCP4, AF321299; Mus musculus MmCENP-A, U03113; Gallus gallus GgCENP-A, AB004649; Ovis aries OaCENP-A, P49453; Homo sapiens HsCENP-A, M95724; and Arabidopsis thaliana AtCENP-A, AAF71990. (b) Structural schematic illustration of diploid CaMIF2 loci in strains CAMB1 and CAMB2. CAMB1 carries two CaMIF2 genes: one under its native promoter and a myc-tagged version under CaPCK1 promoter. CAMB2 is the same as CAMB1, except the CaMIF2 gene under its native promoter is disrupted by the CaHIS1 gene. (c) Shutdown of CaMif2p expression on glucose medium (Glu) prevents CAMB2 cell growth; induction of CaMif2p expression on succinate medium (Suc) allows CAMB2 cell growth. CAMB1 cells, carrying one intact CaMIF2 allele, grow on both media. Plates were incubated at 30°C for 5 days.
To gain further evidence that CaCse4p-rich regions are indeed centromeric, we carried out ChIP experiments to see whether the Can. albicans CENP-C homolog CaMif2p colocalizes with CaCse4p. Formaldehyde-crosslinked chromatin with an average size of 400 bp was isolated from Can. albicans cells expressing myc-tagged CaMif2p in the background of one wild-type allele of CaMIF2 and was immunoprecipitated with or without mouse anti-c-myc mAbs. The recovered DNA was assayed by PCR with the same set of primers (see above) to examine whether the CaCse4p-rich regions are preferentially enriched for CaMif2p binding (Fig. 3). This experiment indicates that significant binding of CaMif2p is limited to the same 3-kb region of chromosome 7 that is enriched for CaCse4p binding. We obtained similar binding patterns of CaCse4p and CaMif2p on chromosome 1 as well (data not shown). Colocalization of two key, evolutionarily conserved, kinetochore proteins at the same regions on Can. albicans chromosomes strongly indicates that these regions are centromeric.
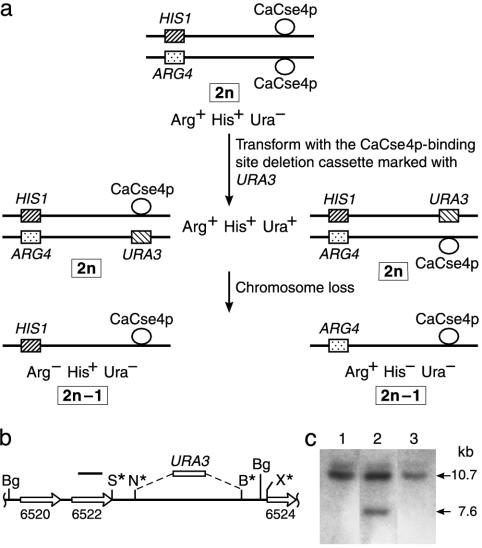
Fragment-Mediated Transplacement of the CaCse4p-Rich Region of Chromosome 7 Leads to High-Frequency Loss of Only the Altered Chromosome. If these CaCse4p-rich regions correspond to centromeric DNA, deletion of such a region from a native Can. albicans chromosome should severely impair its binding to spindle microtubules and the altered chromosome would be lost at a high frequency. For this study, Can. albicans strain RM1000AH was constructed such that the two homologs of chromosome 7 could be distinguished; one copy of the ARG4 locus on chromosome 7 was disrupted with HIS1, making the strain heterozygous for both ARG4 and HIS1 (Fig. 5a). These loci are unlinked to the locus where CaCse4p binds on chromosome 7 because they are separated by at least 460 kb (24). Deletion cassette pCEN7Δ was constructed in which 4.5 kb of the 6.5-kb non-ORF region bound by CaCse4p was replaced by URA3 (Fig. 5b). This construct carries ≈1-kb homologies to the flanking DNA sequences of the CaCse4p-binding region to allow efficient fragment-mediated transplacement. When Can. albicans strain RM1000AH was transformed with SacI/XhoI-digested plasmid pCEN7Δ carrying the deletion cassette, only 15–20 Ura+ transformants per μg of DNA were obtained. Because of the high rate of nonhomologous recombination in Can. albicans, only 2 of 17 examined tranformants arose from homologous fragment-mediated transplacement. The Ura+ loss frequency of these two transformants was 58–82% and all 1,308 Ura– segregants obtained from these two transformants were Arg–His+, as expected for the loss of one entire homolog of chromosome 7 (Fig. 5a). This rate of chromosome loss approaches that reported for deletion of one copy of CEN3 in a diploid Saccharomyces cerevisiae strain (≈90%) or for deletion of the central core of the fission yeast Schizosaccharomyces pombe centromere (88%) carried on a circular minichromosome (28, 29).
Fig. 5.
Deletion of the CaCse4p-bound DNA sequences in one homolog of chromosome 7 causes chromosome loss. (a) Construction of a strain to detect chromosome 7 loss events. (Top) The two homologs of chromosome 7 in strain RM1000AH are differentially marked on one arm with CaHIS1 and CaARG4. (Middle) Next, the CaCse4p-bound region is replaced with CaURA3 in one homolog of chromosome 7. (Bottom) Chromosome loss can be followed by phenotypic analysis for the indicated marker genes. (b) Map of the CaCse4p-bound region replaced with CaURA3. A 1.4-kb CaURA3 fragment replaces 4.5 kb of the chromosome 7 6.5-kb non-ORF region. Thus, the altered homolog carries a BglII fragment that is 3.1 kb shorter than the 10.7-kb BglII fragment found on the wild-type homolog. B, BamHI; Bg, BglII; N, NheI; S, SacI; X, XhoI (not all sites are shown). Asterisks indicate engineered restriction sites. (c) Southern blot of BglII-digested genomic DNA from parental strain RM1000AH (lane 1), an Arg+His+Ura+ strain (lane 2), and an Arg–His+Ura– 2n – 1 aneuploid segregant (lane 3). The probe was a 32P-labeled PCR product corresponding to nucleotides 125170–126192 of contig 19-10248 (black bar in b).
The chromosome 7 CaCse4p-rich locus that was replaced by URA3 was examined in these last two transformants by genomic Southern blot hybridization to confirm that Can. albicans transformants cosegregating URA3 and ARG4 carried the predicted structure as compared with parent strain RM1000AH. The native CaCse4p-rich region of chromosome 7 in RM1000AH is carried on two 10.7-kb BglII fragments (Fig. 5c, lane 1), one from each homolog. As expected for the two Arg+His+Ura+ transplacement strains (RM1000AH/ΔCEN7), the probe hybridized to a 10.7-kb fragment carrying the CaCse4p-rich native locus and also to a 7.6-kb fragment carrying the URA3 substitution (Fig. 5c, lane 2). In the Arg–His+Ura– segregants, however, the 7.6-kb band is no longer present, confirming the loss of this altered homolog (Fig. 5c, lane 3). Thus, this 4.5-kb region containing the 3-kb CaCse4p-binding region of chromosome 7 indeed acts as an essential part of an authentic centromere locus and contributes to maintenance of that homolog in Can. albicans.
Because a centromere is a cis-acting DNA sequence, deletion or inactivation of a centromere should affect maintenance of the altered chromosome only. To verify whether this high rate of nondisjunction of chromosome 7 is the consequence of a centromere-specific cis effect, we examined the loss rate of other Can. albicans chromosomes in strain RM1000AH/ΔCEN7. Wild-type Can. albicans strains cannot grow on sorbose media unless cells lose one copy of chromosome 5; thus, the rate of chromosome 5 loss can be determined by the number of sorbose-using colonies (30, 31). A fluctuation test analysis of the rate of chromosome 5 loss in strain RM1000AH/ΔCEN7 [(2.89 ± 0.29) × 10–4] and parent strain RM1000AH [(5.73 ± 0.72) × 10–4] indicates that deletion of the CaCse4p-binding region of chromosome 7 does not affect mitotic stability (or increase nondisjunction) of other Can. albicans chromosomes. Thus, this region provides chromosome-specific stabilization as is expected from a cis-acting centromere sequence.
We also examined whether the CaCse4p-rich regions we identified are present only once in translocated chromosomes with altered structures reported in various Can. albicans strains. A macrorestriction map of the Can. albicans genome with the enzyme SfiI has been constructed (32). Based on the information available in the Can. albicans genome database, we have localized CaCse4p-rich regions to specific SfiI fragments in five of the eight chromosomes (IJ1, chromosome 1; 4F, chromosome 4; 5M, chromosome 5; 6C, chromosome 6; 7F, chromosome 7). Among all the translocated products of strains WO-1 and STN21 (32, 33), we find no chromosomes either lacking a CaCse4p-rich region or containing more than one such region. Thus, as would be expected if the CaCse4p-associated regions do function as active centromeres, no stable acentric or dicentric translocated products have ever been observed.
Can. albicans Centromeric DNA Sequences Are Different and Unique on Each Chromosome. Centromeric DNA in most organisms is characterized by the presence of distinctive reiterated sequences such as satellite DNAs or retrotransposon-related sequences, which apparently form a preferred substrate for kinetochore assembly. Each of the eight centromeric DNA sequences identified here appears to be unique. However, they were examined by pairwise comparisons to look for common motifs or repeat arrays. Short DNA sequence homologies (11–13 bp in length) were identified by using blast comparisions between a database of the chromosome 7 CaCse4p-rich region and any second CaCse4p-enriched region, but no sequences of even this short length were shared by any three regions. The longest chromosome-specific direct repeats that were found within the CaCse4p-rich regions are 13 bp (twice on chromosome 7: ttgatttgaattt). Tandem direct repeats also are rare; only four different instances of two repeats of 5–7 bp were found within the CaCse4p-rich region on chromosome 7 (gataatt, aattt, tttga, and atctc). Longer chromosome-specific inverted repeats were found flanking the CaCse4-rich region on chromosomes 4 and 5. The inverted repeat on chromosome 4 is 523 bp in length, unique in the Can. albicans genome, and found entirely within the non-ORF region. On chromosome 5, each arm of the inverted repeat is at least 3.8 kb in length, carries repetitive agglutinin-like sequences (ALS gene family), and spans non-ORF and ORF DNA sequences. The Can. albicans centromeric regions also lack homology to the DNA sequence motifs (centromere DNA elements I, II, and III) of budding yeast point centromeres. Like centromeric DNA of most other organisms, the AT content of the CaCse4p-bound regions is 62–67%, similar to that of human α-satellite DNA, human neocentromeres (34), and fission yeast centromeres. Our comparative analysis of these CaCse4p-rich DNA sequences and adjacent regions did not detect any short DNA sequence motifs or repeat arrays that were conserved among the eight regions. The marked sequence heterogeneity of the eight Can. albicans centromeres is remarkable because native centromeres of different chromosomes in all organisms studied to date carry some kind of species-dependent common sequence motifs or repeat sequences.
Discussion
We have identified and characterized the centromeric DNA of all eight chromosomes of Can. albicans. Each of these eight centromeres is a unique and different region lacking any common sequence motifs. A core 3-kb region of each chromosome was shown to be the binding site of two important evolutionarily conserved kinetochore proteins, CaCse4p (CENP-A homolog) and CaMif2p (CENP-C homolog) and the binding of CaCse4p is limited to this region in a cell-type-independent manner. Finally, deletion of the CaCse4p-binding region from chromosome 7 results in a high rate of loss of only the altered chromosome without affecting the stability of other chromosomes.
The CaCse4p-associated region is intermediate in size between that seen at budding yeast point centromeres and the fission yeast regional centromere. The fission yeast Schizosaccharomyces pombe centromere consists of a central core that is immediately flanked at both ends by a core-associated repeat (imr) organized in an inverted orientation (35, 36). Repeats K/dg and L/dh extend the inverted repeat and, in addition, form direct-repeat arrays at cen2 and cen3. The Schizosaccharomyces pombe centromeric central cores are 4 to 7 kb nonhomologous DNA regions that are rich in A + T (71%), are associated with CENP-A (Cnp1p), and are essential for centromere function (2, 25, 29). In Can. albicans, the CaCse4p-associated regions, which are ≈3 kb in length, 65% A + T, and important for chromosome segregation, most closely resemble the central core regions of fission yeast Schizosaccharomyces pombe centromeres, but the adjacent repeats are lacking.
Although native regional centromeres usually include some repeated DNA sequence elements, these elements are not necessarily limited to centromere regions, and centromere function does not exhibit an absolute correlation with one specific DNA sequence. Studies in Schizosaccharomyces pombe show that two entirely different DNA sequences within the large centromeric domain are each sufficient for centromere function (37). Satellite repeats and transposable elements that constitute the majority of the 420-kb fully functional Drosophila minichromosome Dp1187 are also abundant elsewhere in the genome (38). Moreover, the holocentric C. elegans genome and centromere formation at unique DNA sequences (neocentromeres) on human chromosomes suggest that the primary DNA sequence cannot be the sole determinant for centromere identity in regional centromeres. A highly contrasting situation exists in point centromeres of Saccharomyces cerevisiae, where all centromeres not only contain common conserved DNA sequences but also a single point mutation can abolish centromere function, indicating a strong correlation between centromere DNA sequence and its function. Therefore, despite the absence of any associated repeat sequences, Can. albicans centromeres resemble those regional centromeres that lack an obvious correlation between the centromere DNA sequence and its function. How centromere DNA sequences with high sequence heterogeneity form active kinetochores is still an enigma. Centromere regions in these organisms must be capable of forming the necessary specialized higher-order structure even though the underlying DNA sequences are different. DNA topology, DNA modifications, specialized chromatin states, DNA–protein interactions, and other epigenetic factors may be involved in functional centromere formation in these organisms.
Despite the incentive to develop centromere-stabilized vector systems for use in Can. albicans, an artificial chromosome system has not yet been developed in this medically important organism. We have introduced into Can. albicans cells ARS plasmids carrying 6- to 180-kb DNA inserts spanning the CaCse4p-rich regions from different chromosomes, but all were found to be mitotically unstable, presumably with inactive centromeres. Several possible reasons explain why stable artificial chromosomes have not yet been obtained in Can. albicans. Although circular centromere-bearing artificial chromosomes are functional in most organisms, it is possible that Can. albicans is an exceptional organism in which the centromere is active only on linear constructs. It is also possible that relatively distant DNA sequences in addition to the CaCse4p-associated sequences we cloned into the ARS vectors are necessary for centromere function, although this seems unlikely because we assayed DNA fragments as large as 180 kb. We also tested plasmids containing known repeat sequences (RB2 and RPS; ref. 39) in association with the CaCse4p-rich regions, and these were mitotically unstable. A more interesting possibility would be that centromere function in Can. albicans may require epigenetic activation. That is, naked DNA sequences introduced by transformation may be unable to acquire the proper heterochromatic conformation. Studies in Schizosaccharomyces pombe have demonstrated that some plasmids carrying centromeric DNA sequences are subject to epigenetic activation after introduction into cells by standard transformation procedures. The centromere on these plasmids initially is in an inactive state, but switches to a stable active state after a few generations (40). Thus, centromere-bearing plasmids introduced into Can. albicans by transformation could be in a similar centromere-inactive state, whereas the same sequences would be present in a centromere-active state on the native chromosome. Identification of conditions necessary to obtain functional artificial chromosomes in Can. albicans should lead to important information on the mechanisms involved in centromere activation in vivo.
Supplementary Material
Acknowledgments
We thank Louise Clarke and Tanja Stoyan for valuable discussions and critical reading of this manuscript; Xiomara Elías Argote for designing PCR primers and help with ChIP assays; Steve Poole for assistance with sequence analysis; Yoshi Nakagawa (Nagoya University Graduate School of Medicine, Nagoya, Japan) for supplying RB2 and RPS plasmids; and Dottie McLaren for artwork. Sequence data for Can. albicans was obtained from the Stanford Genome Technology Center web site at www-sequence.stanford.edu/group/candida (the sequencing of Can. albicans was accomplished with the support of the National Institute on Dental Research and the Burroughs Wellcome Fund). This work was supported by National Institutes of Health National Cancer Institute Research Grant CA-11034.
Abbreviations: ChIP, chromatin immunoprecipitation; Can. albicans, Candida albicans.
References
- 1.Clarke, L. & Carbon, J. (1980) Nature 287, 504–509. [DOI] [PubMed] [Google Scholar]
- 2.Clarke, L. (1998) Curr. Opin. Genet. Dev. 8, 212–218. [DOI] [PubMed] [Google Scholar]
- 3.Cleveland, D. W., Mao, Y. & Sullivan, K. F. (2003) Cell 112, 407–421. [DOI] [PubMed] [Google Scholar]
- 4.Henikoff, S., Ahmad, K. & Malik, H. S. (2001) Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- 5.Smith, M. M. (2002) Curr. Opin. Cell Biol. 14, 279–285. [DOI] [PubMed] [Google Scholar]
- 6.Yu, H.-G., Hiatt, E. N. & Dawe, R. K. (2000) Trends Plant Sci. 5, 543–547. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan, B. A., Blower, M. D. & Karpen, G. H. (2001) Nat. Rev. Genet. 2, 584–596. [DOI] [PubMed] [Google Scholar]
- 8.Tomkiel, J., Cook, C. A., Saitoh, H., Bernat, R. L. & Earnshaw, W. C. (1994) J. Cell Biol. 125, 531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukagawa, T. & Brown, W. R. A. (1997) Hum. Mol. Genet. 6, 2301–2308. [DOI] [PubMed] [Google Scholar]
- 10.Kalitsis, P., Flower, K. J., Earle, E., Hill, J. & Choo, K. H. A. (1998) Proc. Natl. Acad. Sci. USA 95, 1136–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oegema, K., Desai, A., Rybina, S., Kirkham, M. & Hyman, A. A. (2001) J. Cell Biol. 153, 1209–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore, L. L. & Roth, M. B. (2001) J. Cell Biol. 153, 1199–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meluh, P. B., Yang, P., Glowczewski, L., Koshland, D. & Smith, M. M. (1998) Cell 94, 607–613. [DOI] [PubMed] [Google Scholar]
- 14.Lo, A., Craig, J., Saffery, R., Kalitsis, P., Irvine, D., Earle, E., Magliano, D. & Choo, K. (2001) EMBO J. 20, 2087–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, T., Federspiel, N. A., Chibana, H., Dungan, J., Kalman, S., Magee, B. B., Newport, G., Thorstenson, Y. R., Agabian, N., Magee, P. T., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 7329–7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanyal, K. & Carbon, J. (2002) Proc. Natl. Acad. Sci. USA 99, 12969–12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgers, P. M. & Percival, K. J. (1987) Anal. Biochem. 163, 391–397. [DOI] [PubMed] [Google Scholar]
- 18.Hull, C. M. & Johnson, A. D. (1999) Science 285, 1271–1275. [DOI] [PubMed] [Google Scholar]
- 19.Braunstein, M., Rose, A. B., Holmes, S. G., Allis, C. D. & Broach, J. R. (1993) Genes Dev. 7, 592–604. [DOI] [PubMed] [Google Scholar]
- 20.Kuo, M.-H. & Allis, C. D. (1999) Methods 19, 425–433. [DOI] [PubMed] [Google Scholar]
- 21.Kinzler, K. W. & Vogelstein, B. (1989) Nucleic Acids Res. 17, 3645–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Law, A., Hirayoshi, K., O'Brien, T. & Lis, J. T. (1998) Nucleic Acids Res. 26, 919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orlando, V. (2000) Trends Biochem. Sci. 25, 99–104. [DOI] [PubMed] [Google Scholar]
- 24.Chibana, H., Magee, B. B., Grindle, S., Ran, Y., Scherer, S. & Magee, P. T. (1998) Genetics 149, 1739–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi, K., Chen, E. S. & Yanagida, M. (2000) Science 288, 2215–2219. [DOI] [PubMed] [Google Scholar]
- 26.Suzuki, N., Nakano, M., Nozaki, N., Egashira, S., Okazaki, T. & Masumoto, H. (2004) J. Biol. Chem. 279, 5934–5946. [DOI] [PubMed] [Google Scholar]
- 27.Leuker, C. E., Sonneborn, A., Delbruck, S. & Ernst, J. F. (1997) Gene 192, 235–240. [DOI] [PubMed] [Google Scholar]
- 28.Clarke, L. & Carbon, J. (1983) Nature 305, 23–28. [DOI] [PubMed] [Google Scholar]
- 29.Baum, M., Ngan, V. K. & Clarke, L. (1994) Mol. Biol. Cell 5, 747–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rustchenko, E. P., Howard, D. H. & Sherman, F. (1994) J. Bacteriol. 176, 3231–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janbon, G., Sherman, F. & Rustchenko, E. (1999) Genetics 153, 653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu, W.-S., Magee, B. B. & Magee, P. T. (1993) J. Bacteriol. 175, 6637–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwaguchi, S.-I., Kanbe, T., Tohne, T., Magee, P. T. & Suzuki, T. (2000) Yeast 16, 411–422. [DOI] [PubMed] [Google Scholar]
- 34.Amor, D. J., Bentley, K., Ryan, J., Perry, J., Wong, L., Slater, H. & Choo, K. H. A. (2004) Proc. Natl. Acad. Sci. USA 101, 6542–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi, K., Murakami, S., Chikashige, Y., Funabiki, H., Niwa, O. & Yanagida, M. (1992) Mol. Biol. Cell 3, 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steiner, N. C., Hahnenberger, K. M. & Clarke, L. (1993) Mol. Cell. Biol. 13, 4578–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ngan, V. & Clarke, L. (1997) Mol. Cell. Biol. 17, 3305–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, X., Wahlstorm, J. & Karpen, G. H. (1997) Cell 91, 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chindamporn, A., Nakawaga, Y., Mizuguchi, I., Chibana, H., Doi, M. & Tanaka, K. (1998) Microbiology 144, 849–857. [DOI] [PubMed] [Google Scholar]
- 40.Steiner, N. C. & Clarke, L. (1994) Cell 79, 865–874. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.