Abstract
An E2 ubiquitin-conjugating enzyme, Rad6, working with an E3 ubiquitin ligase Bre1, catalyzes monoubiquitylation of histone H2B on a C-terminal lysine residue. The rad6 mutant of Saccharomyces cerevisiae shows a meiotic prophase arrest. Here, we analyzed meiotic defects of a rad6 null mutant of budding yeast. The rad6 mutant exhibits pleiotropic phenotypes during meiosis. RAD6 is required for efficient formation of double-strand breaks (DSBs) at meiotic recombination hotspots, which is catalyzed by Spo11. The mutation decreases overall frequencies of DSBs in a cell. The effect of the rad6 mutation is local along chromosomes; levels of DSBs at stronger hotspots are particularly reduced in the mutant. The absence of RAD6 has little effect on the formation of ectopic DSBs targeted by Spo11 fusion protein with a Gal4 DNA-binding domain. Furthermore, the disruption of the BRE1 as well as substitution of the ubiquitylation site of histone H2B also reduces some DSB formation similar to the rad6. These results suggest that Rad6-Bre1, through ubiquitylation of histone H2B, is necessary for efficient recruitment and/or stabilization of a DSB-forming machinery containing Spo11. Histone tail modifications might play a role in DSB formation during meiosis.
The RAD6 gene encodes an E2 ubiquitin-conjugating enzyme required for a variety of biological processes (1, 2). Together with a specific partner named E3, Rad6 mediates mono- and polyubiquitylation of target proteins. One of the other known targets of Rad6-dependent ubiquitylation is histone H2B (3, 4). A C terminus lysine residue of histone H2B is monoubiquitylated in a Rad6-dependent manner. Histone H2B ubiquitylation requires a specific E3 partner, Bre1 (5, 6). The ubiquitylation of histone H2B is involved in the silencing of gene expression as well as transcription (5–8). Rad6 is recruited to promoter regions of constitutively transcribed genes in a Bre1-dependent manner (6). However, the role of Rad6-dependent histone ubiquitylation in transcription remains largely unknown (6, 7, 9).
RAD6 function is necessary for meiosis (3). A rad6 null mutant shows a meiotic prophase arrest (3). This meiotic function of Rad6 seems to be mediated through histone H2B ubiquitylation (3). It is shown that a histone H2B mutant in which lysine 123, a site for Rad6 ubiquitylation, is replaced by arginine recapitulates a meiotic phenotype of the rad6, i.e., prophase arrest (3). However, the exact role of Rad6 during meiosis remains to be determined.
Here, we characterized a rad6 null mutant in detail and found that it shows pleiotropic defects during meiosis; in particular, Rad6 is necessary for efficient Spo11-dependent formation of double-strand breaks (DSBs) at meiotic recombination hotspots. Furthermore, the bre1 mutation as well as substitution of a ubiquitylation site of histone H2B also reduces the levels of some DSBs, similar to the rad6. We propose that modifications of histone tails play a role in efficient recruitment and/or stabilization of DSB-forming machinery.
Materials and Methods
Strain. All yeast strains used in this study were derivatives of SK1 and are listed in Table 1, which is published as supporting information on the PNAS web site. rad6 and rad18 null alleles were constructed by adaptamer mediated PCR (10). The bre1 null allele was introduced by PCR-mediated gene disruption using pU6H3HA (11). spo11-Y135F and rad50S (rad50-K81I) were described previously (12, 13). The htb1-K123R and htb2-K123R were created by the PCR-mediated site-directed mutagenesis and marked with KlURA3 and KanMX6, respectively. The mutation sites were confirmed by DNA sequencing. Primer sequences used for strain construction are described in Table 2, which is published as supporting information on the PNAS web site.
Southern Blotting. Preparation of genomic DNAs and Southern analysis were carried out as described (14, 15). For DSB and crossover recombinants at the HIS4-LEU2 locus, DNAs were digested by using PstI and XhoI, respectively. For DSBs at the YCR048w, GAT1, CYS3, and GAL2 loci, BglII, HindIII, BglII, and XbaI were used, respectively. Probes were prepared by using PCR-amplified fragments specific to each locus as a template. Primer sequences are described in Table 2.
Clamped homogeneous electric field (CHEF) gel electrophoresis was performed by using CHEF DRIII (Bio-Rad, Hercules, CA), as described (16).
Fluorescence-Activated Cell Sorter (FACS) Analysis, 4′,6-Diamidino-2-Phenylindole (DAPI) Staining, and Immunostaining of Rad51. FACS analysis and DAPI staining were carried out as described (13). Chromosome spreads were prepared and immunostained as described (17).
Results
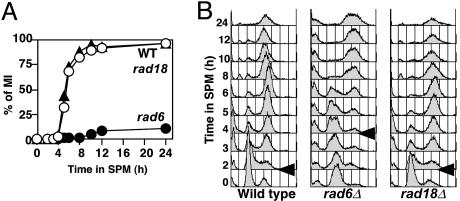
The rad6 Mutant Reduces DSB Formation During Meiosis. To determine the role of Rad6 during meiosis, we constructed a rad6 null mutant in a rapidly sporulating yeast strain, SK1, background. In addition, we characterized a null mutant of RAD18, which works together with Rad6 during postreplicative repair (18). The rad18 cells undergo meiosis I and II similar to wild type (Fig. 1). Consistent with the previous report (19), spore viability of the rad18 mutant is almost identical to that of wild type, indicating that the RAD18 is dispensable for meiosis. On the other hand, as reported previously (3), most rad6 cells show a meiotic prophase arrest (Fig. 1 A); only 10–20% of rad6 cells enter into meiosis I, and the rest of the rad6 cells never form spores. The meiotic prophase arrest of the rad6 mutant is reversible; when arrested rad6 cells are returned to mitotic growth, the mutant cells do not lose their viability (data not shown).
Fig. 1.
The rad6 but not rad18 mutant shows a meiotic prophase arrest. (A) Meiotic divisions were analyzed by 4′,6-diamidino-2-phenylindole staining. The percentage of cells entering into meiosis I is plotted versus incubation time. More than 100 nuclei were counted at each time point. Open circles, wild type (NKY1551); filled circles, rad6 (KYY1087); filled triangles, rad18 (KYY1141). (B) DNA contents in each cell were analyzed by using a fluorescence-activated cell sorter. Arrows indicate the time when premeiotic S begins in each strain.
After synchronous induction of meiosis, premeiotic S phase progression in the rad6 mutant was monitored by fluorescence-activated cell sorter analysis. A rad6 cell shows a delay in the onset of premeiotic S phase (Fig. 1B). Onset of the S phase is seen at 4-h incubation in the rad6, whereas it is seen at2hinwild type. The length of premeiotic S phase in the rad6 is similar to that in wild type. The delay in the entry into premeiotic S phase in the rad6 mutant might be related to a lack of growth of the mutant cells in medium containing acetates, e.g., YP-acetate (K.Y., unpublished results).
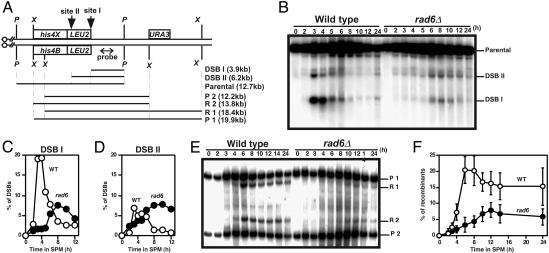
Because the rad6 mutant shows a meiotic prophase arrest as in other recombination deficient strains such as dmc1 (20), we examined the formation and repair of meiotic DSBs at various loci in the rad6 mutant (Fig. 2). In wild type, DSBs at an artificial recombination hotspot, the HIS4-LEU2 (Fig. 2 A), begin to appear around a 2-h incubation in sporulation medium, peak at 4 h, and disappear during further incubation (Fig. 2B). In a rad6 null mutant, DSBs start to appear at 5 h, a 3-h delay relative to wild type, peak at 8 h, and disappear (Fig. 2B). The delay in DSB formation in the mutant is consistent with a delay in the premeiotic S phase. In the mutant, the steady-state amount of DSBs at site I is reduced 2-fold compared to that in wild type (Fig. 2C). On the other hand, the DSB at site II is not affected by the rad6 mutation (Fig. 2D). We also analyzed DSB formation at the YCR048W and CYS3 loci, which are natural recombination hotspots. The delay in DSB formation and the reduction of DSB levels (not all DSB sites) in the mutant were also observed for YCR048W and CYS3 loci (data not shown). The levels of DSB at stronger hotspots in the YCR048W locus are particularly decreased compared with those at weaker ones (see below).
Fig. 2.
The rad6 reduces amounts of meiotic DSBs. (A) Schematic drawings of a meiotic recombination hotspot, HIS4-LEU2 locus, on chromosome III.P, PstI; X, XhoI. (B–D) DSBs at the HIS4-LEU2 locus in wild type (NKY1551) and the rad6 mutant (KYY1087) were analyzed as described in Materials and Methods. Quantification of each DSB fragment to parental fragment is shown; sites I (C) and II (D). Open circles, wild type; filled circles, rad6. (E and F) Crossover recombinants were analyzed by Southern blotting. Percentages of recombinant are a ratio of R1 and R2 recombinants to P1 and P2 parental fragments. Quantification results of three independent time courses (with error bars) are presented in F. Open circles, wild type; filled circles, rad6.
Consistent with reduction of DSB levels, rad6 cells decrease the amount of crossover recombinant molecules formed at the HIS4-LEU2 locus (Fig. 2E). The amount of crossover molecule in the mutant is 8% of parental fragments, one-half that in wild type (Fig. 2F).
The decrease in steady-state levels of DSBs in rad6 cells is due to either a decrease in an absolute amount of introduced DSB per se or rapid turnover of the DSBs, e.g., through rapid DSB repair. To quantify the levels of DSBs more precisely, we introduced the rad50S mutation, which blocks the resection of the DSB ends and subsequent DSB repair (12), into the rad6 mutant. We also analyzed three independent cultures for the quantification and two blots for each time course. The rad6 rad50S double mutant delays the onset of premeiotic S phase and exhibits a prophase arrest (data not shown). In rad50S cells, at the HIS4-LEU2 locus, DSBs at sites I and II accumulated to 25% and 9% over an 8-h incubation, respectively (Fig. 3 B–D). On the other hand, the rad6 rad50S mutation delayed DSB formation as in the rad6. The double mutant reduced the amount of DSB at site I to 11%, one-half that in rad50S (Fig. 3C), whereas the amount of DSB at site II in the double mutant is similar to that in the control (Fig. 3D). Thus, rad6 mutation affects DSB formation at some, but not all, loci.
Fig. 3.
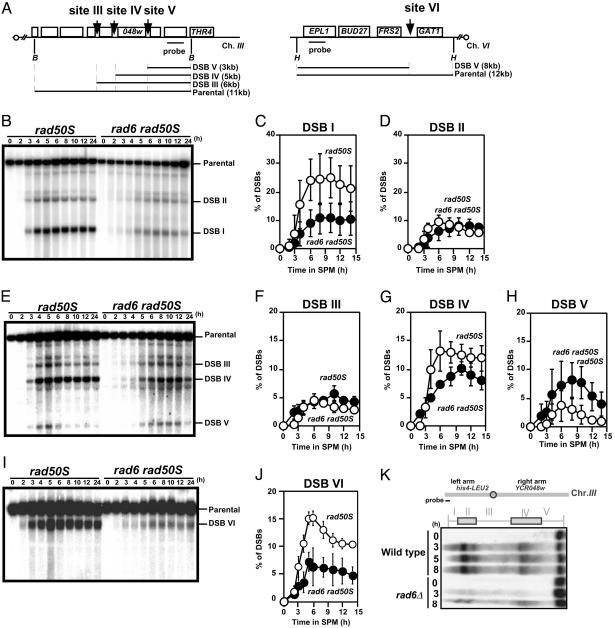
Quantification of levels of meiotic DSBs in the rad6 rad50S mutant. (A) Schematic drawings of two meiotic recombination hotspots YCR048W on chromosome III and GAT1 on chromosome VI. B, BglII; H, HindIII. DSBs at the HIS4-LEU2 (B–D), at the YCR048w (E–H) and at the GAT1 loci (I, J) in the rad50S (KYY1306) and rad6 rad50S (KYY1303) mutants were analyzed as shown above. Percentages of each DSB band to that of the parental band were quantified and an average of three independent time courses (two independent blots) with an error bar is shown. Site I (C), site II (D) of the HIS4-LEU2, site III (F), site IV (G), and site V (H) of the YCR048W, and site VI (J) at the GAT1. Open circles, rad50S; filled circles, rad6 rad50S.(K) DSBs along chromosome III were analyzed in the rad50S (KYY1306) and rad6 rad50S (KYY1303) cells. Yeast chromosomes were separated by clamped homogeneous electric field and were detected by using a probe specific to the end of the left arm. The chromosomal domains classified by Baudat and Nicolas (16) are shown (Upper) with a schematic representation of the chromosome.
The same was found to be true when the YCR048W locus was analyzed (Fig. 3 A and E). The DSB level at a strong site IV in this region is significantly reduced in rad6 rad50S cells compared with that in rad50S control cells (Fig. 3F). This difference is statistically significant (at 6, 8, 10, 12, or 14 h, P < 0.05; Student's t test). However, amounts of DSB at site III in the rad6 rad50S mutant are similar to those in the control (Fig. 3F). Interestingly, DSB at site V in the mutant is increased slightly compared with the control (Fig. 2H). We also analyzed DSB formation at the GAT1 (K.Y., unpublished results) and CYS3 loci, which are natural recombination hotspots (10). The delay in DSB formation and the reduction of DSB levels (not all DSB sites) in the mutant were also observed for GAT1 (Fig. 3 I and J) and CYS3 loci (Fig. 8, which is published as supporting information on the PNAS web site). These suggest that the function of Rad6 is required for efficient DSB formation at stronger hotspots. More importantly, the sites of DSB formation are unchanged in the rad6 mutant under this level of resolution, suggesting that selection of DSB sites is independent of the RAD6 function. Thus, Rad6 seems to determine the efficiency of DSB formation at some sites.
We analyzed the overall distribution of DSBs along chromosome III in a rad6 rad50S cell (Fig. 3K). Along the chromosome, DSBs are clustered around the middle part of both arms (16, 21). rad6 reduced overall amounts of DSBs along the chromosome but did not change the distribution of DSBs. There are three domains rich in DSBs on the chromosome: two on the left arm and one on the right. The telomere-distal DSB domain on the left arm is more severely affected by the rad6 than the other domains.
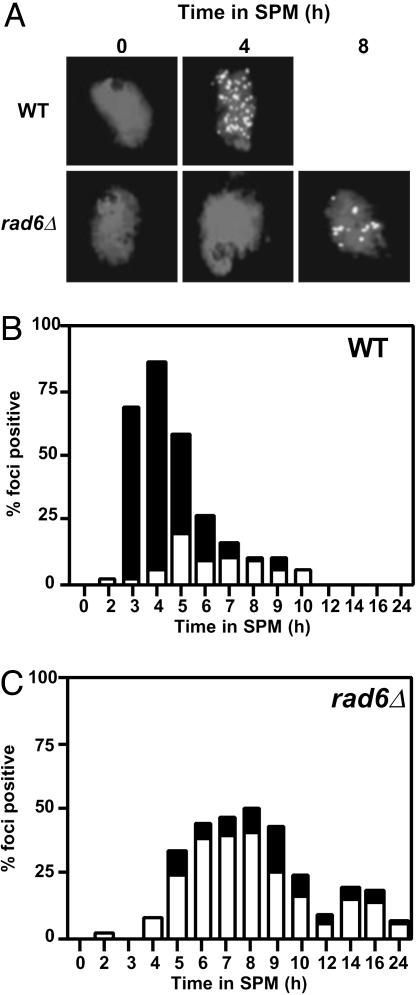
rad6 Decreases the Number of Rad51 Foci. To confirm the reduction of the total DSB level in the rad6 mutant, we examined the formation of immunostained structures containing Rad51, Rad51 foci, which mark the ongoing recombination complex (22), on chromosome spreads of rad6 cells (Fig. 4A). As expected, Rad51-focus formation in the rad6 mutant is delayed for 2 h. The average number of Rad51 foci in the rad6 is 15, one-third that in wild type, consistent with the decreased levels of DSBs in rad6 cells (Fig. 4 B and C). After a 24-h incubation in sporulation medium, most Rad51 foci in the mutant disappear, but some remain. This may be due to the asynchronous nature of rad6 meiosis.
Fig. 4.
The rad6 mutation decreases the steady-state number of Rad51 foci. (A) Chromosome spreads of wild-type (NKY1551) and rad6 (KYY1087) cells were prepared and stained with anti-Rad51 antibodies as described in Shinohara et al. (17). Representative images for each time are shown. (B and C) Numbers of Rad51 foci per a nucleoid were counted and classified into two classes; nucleoids with >20 Rad51 foci (solid bars) and those with 5–20 Rad51 foci (open bars). At least 100 nuclei were counted for numbers of the foci; wild type (B) and rad6 (C).
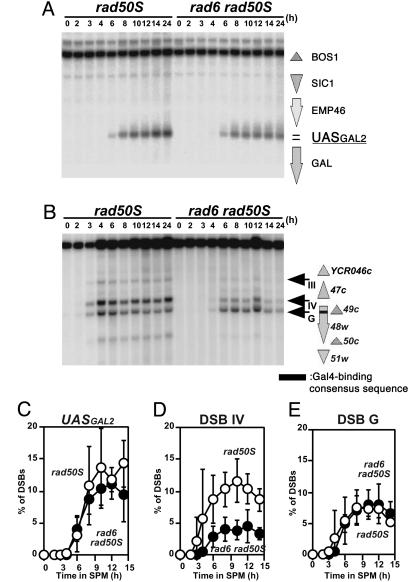
The rad6 Does Not Affect DSBs Targeted by Gal4-Spo11. Decreased levels of DSBs at some loci in the rad6 mutant suggest the impairment of recruitment (or stabilization) of a putative DSB-forming machinery. If so, we reasoned that the targeting of the machinery might suppress the defect of the rad6. Recently, Nicolas and colleagues (23) used Spo11 protein fused to a Gal4 DNA-binding domain to induce ectopic DSBs at Gal4-binding sites. They showed that the ectopic DSBs share the same properties with natural DSBs in respect of genetic requirement; all genes necessary for the formation of DSBs at a natural site are required for that of the ectopic DSBs. We examined the effect of rad6 mutation on ectopic DSB formation introduced by the Gal4-Spo11 (Fig. 5). First, we analyzed the GAL2 locus, which resides in a cold region for DSB formation in wild type. As shown previously (23), expression of Gal4-Spo11 induces a DSB near a Gal4-binding site at this locus (Fig. 5A). The rad6 mutant (in the background of rad50S) induces the DSB at a similar level to wild type, indicating that the rad6 does not affect the DSBs induced by Gal4-Spo11 (Fig. 5B). Interestingly, the ectopic DSB induced at the GAL2 locus is delayed relative to other DSBs in wild type but formed at the same rate as in the rad6 cells.
Fig. 5.
Meiotic DSBs targeted by Gal4-Spo11 are not affected by the rad6. DSBs at the GAL2 (A) and the YCR048w (B) loci in the rad50S (KYY1434) and rad6 rad50S (KYY1435) mutants expressing Gal4-Spo11 were analyzed. Amounts of each DSB band to the parental band were quantified and an average of three independent time courses with an error bar is shown (C–E). The positions of DSBs relative to ORFs are shown on the right. DSBs at UASGal and site G is ectopically induced by Gal4-Spo11. UASGal (C), site IV (D), and site G (E) of the YCR048w. Open circles, rad50S; filled circles, rad6 rad50S.
The effect of the rad6 mutation is more prominent when the YCR048W locus is analyzed. In addition to normal DSBs, Gal4-Spo11 introduces a novel DSB in this region (called site G, Fig. 5C). As shown above, the introduction of the rad6 reduces the amount of DSB at site IV, a major natural DSB site in this region, 2.6-fold (Fig. 5D). However, the amount of ectopic DSB induced by Gal4-Spo11 is unaffected by the rad6 mutation (Fig. 5E). The formation of all DSBs, including the ectopic DSB in this region, is delayed in the mutant compared with wild type. These indicate that the rad6 mutation does not affect ectopic DSBs targeted by Gal4-Spo11, consistent with the idea that Rad6 is required for efficient recruitment of the DSB-formation machinery containing Spo11.
The Amount of Spo11 Is Not Reduced in a rad6 Cell. One possibility as to why the rad6 mutant is defective in DSB formation at some loci is that it is due to a reduced level of Spo11 protein in a mutant cell, i.e., there is a Spo11-dosage dependency on DSB formation. We analyzed the amount of Spo11-Flag in the rad6 mutant by Western blotting (Fig. 9, which is published as supporting information on the PNAS web site). In wild type, Spo11 protein is expressed transiently during meiotic prophase. The rad6 mutant expressed Spo11 protein at a comparable level to that in wild type, although the expression of Spo11 protein is delayed in the mutant. Thus, we concluded that a decreased level of Spo11 protein cannot explain the reduction of DSB formation at some loci in the rad6.
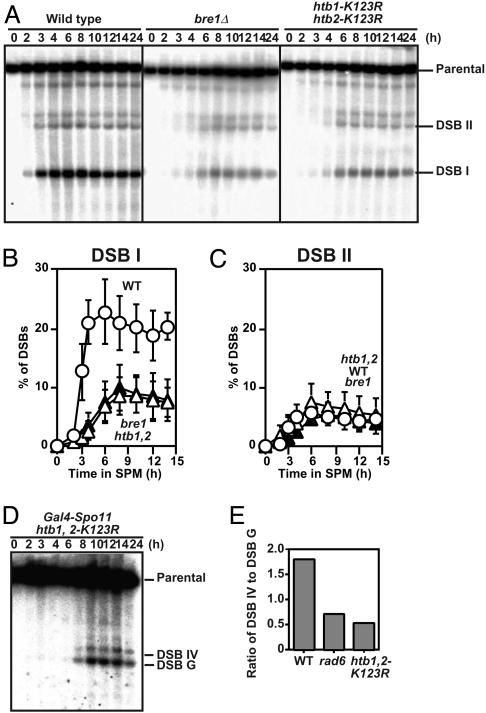
The bre1 Mutant and Histone H2B Mutant Lacking Ubiquitylation Sites Reduce DSB Formation. Rad6 forms a complex with Bre1 to promote the ubiquitylation of histone H2B (5, 6). The bre1 mutant does not form spores in S288C background, as in the rad6 (24). We constructed a bre1 null mutant in SK1 background and analyzed the DSB formation in the mutant in the presence of the rad50S mutation. As shown in Fig. 6A, the bre1 mutation reduces the formation of DSB at some loci (Fig. 6B), but not at others (Fig. 6C), as seen in the rad6 mutant. These suggest that Rad6 works with Bre1 for efficient DSB formation during meiosis.
Fig. 6.
The bre1 and histone H2B mutants lacking a ubiquitylation site also show reduced levels of DSBs. DSBs at the HIS4-LEU2 locus in the bre1 (KYY1395) and htb1, 2-K123R (KYY1403) mutants with the rad50S were analyzed as described above (A). Quantification of DSBs at site I (B) and site II (C) were carried out as described above. An average of three independent time courses is shown with an error bar. Open circles, rad50S; filled triangles, bre1 rad50S; open triangles, htb1, 2-K123R rad50S. (D and E) The effect of histone ubiquitylation mutations on DSBs at sites IV and G in the YCR048w is ectopically induced by Gal4-Spo11, analyzed by using MSY5000 (D). The ratio of DSBs at site IV to those at site G of the YCR048w after an 8-h incubation was quantified in each mutant (E).
Histone H2B mutant cells with the substitution on the C-terminal lysine 123, which is ubiquitylated by Rad6-Bre1 pathway, are known to arrest before meiosis I (3). However, the previous study used a mutant cell lacking both histone H2B genes, HTB1 and HTB2, in which the substituted HTB1 gene was supplied from a plasmid. Because the dosage of histone H2B genes affects meiosis (25), we constructed a mutant strain in which both wild-type histone H2B genes were replaced with an allele with a change of lysine 123 to arginine, htb1-K123R, htb2-K123R double mutant (referred as htb1, 2-K123R hereafter). We also introduced a rad50S allele into the htb1, 2-K123R. As expected, the htb1, 2-K123R mutant recapitulates the DSB phenotypes seen in rad6 and bre1 mutants, indicating the lysine 123 of histone H2B is necessary for efficient DSB formation (Fig. 6). These suggest that Rad6-Bre1 function in DSB formation through histone H2B ubiquitylation.
We also analyzed the effect of the histone H2B ubiquitylation mutations on Gal4-Spo11-induced DSB formation. Like rad6, the ubiquitylation mutations do not affect an efficiency of the Gal4-Spo11-induced DSB but do affect that of Spo11-induced DSB (Fig. 6 D and E), consistent with the idea that histone H2B ubiquitylation is a determinant of the frequency of meiotic DSB formation.
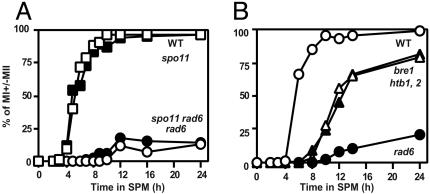
Mitotis Division I Arrest of rad6 Is SPO11-Independent. The rad6 cells show an arrest in meiosis division I (Fig. 1 A). Because the rad6 mutant can repair meiotic DSBs (Fig. 2), meiotic recombination defects conferred by the rad6 cannot explain this arrest. Therefore, we tested whether rad6-induced meiotic prophase arrest depends on the initiation of meiotic recombination. We constructed a rad6 mutant with a spo11 mutation (spo11-Y135F; ref. 13) and analyzed cell cycle progression of the double mutant. It is known that the elimination of early recombination function suppresses the prophase arrest induced by aberrant recombination. However, the spo11 mutation did not suppress the cell cycle arrest triggered by the rad6. The rad6 spo11 double mutant does arrest before meiotic division I, as in the rad6 single mutant (Fig. 7A). The same result is obtained for a rad6 mutant with the spo11 null mutation (unpublished results). These suggest that a recombination-independent function of Rad6 is required for the progression into meiosis I.
Fig. 7.
Meiosis progression in various mutants. Cells that passed meiosis I division were counted after staining with 4′,6-diamidino-2-phenylindole. (A) Open rectangles, wild type (NKY1551); filled rectangles, spo11-Y135F (NKY1170); open circles, rad6 (KYY1087); filled circles, spo11-Y135F rad6 (KYY1156). (B) Open circles, wild type (NKY1551); filled circles, rad6 (KYY1087); open triangles, htb1, 2-K123R (KYY1278); filled triangles, bre1 (KYY1442).
We also analyzed meiotic cell cycle progression in the bre1 and htb1, 2-K123R mutants. As shown in Fig. 7B, both mutant cells enter into meiosis I with delayed kinetics. After a 24-h incubation in sporulation medium, 70–75% of cells had ceased division. In addition, the mutants produce spores with a high viability (90%). These are inconsistent with the previous report for the htb1, htb2 deletion mutant with the htb1-K123R mutation on a plasmid (3). Although the strain background of the current study is different, it is likely that the dosage of histone H2B affects the progression into meiosis I, as reported (25).
Discussion
We have shown that the rad6 mutation reduces amounts of DSBs at several loci, suggesting that Rad6 determines the efficiency of some DSB formations. Because Rad6 is involved in ubiquitilaytion of a target protein, this modification seems to play a direct role in the DSB formation, as discussed below. Although we favor this possibility, the rad6 might affect DSB formation indirectly. The rad6 mutant is partially deficient in transcriptional silencing (26) and in transcription (7, 8), thus affecting overall transcription status in a cell. A change of the transcription of genes involved in meiotic recombination might affect DSB formation in rad6 cells. Alternatively, a delay in the onset of premeiotic S phase in the mutant might affect DSB formation. It is shown that premeiotic S phase is tightly coupled with the formation of meiotic DSBs (16, 27). However, we think these indirect effects are unlikely. First, the effect of the rad6 mutation is very local and region-specific. Although one DSB is affected by the mutation, another DSB 1 kb away from the affected site is unaffected. Second, ectopic DSB formation induced by Gal4-Spo11 in the rad6 and histone ubiquitylation mutants is very similar to that in wild type. Either change in transcription of recombination genes or delayed onset of premeiotic S phase should have an effect on the ectopic DSBs as well as natural DSBs. Indeed, the ectopic DSBs targeted by Gal4-Spo11 require all genes involved in DSB formation and in the onset of premeiotic S phase (23).
Furthermore, a mutation in the BRE1 gene, encoding a novel E3 partner of Rad6 (5, 6), reduced efficiencies of some DSB formations, as seen in the rad6. We also showed that a histone H2B mutant lacking the ubiquitylation site also recapitulates the phenotypes of the rad6 and bre1 in respect to DSB formation. These strongly argue that ubiquitylation of histone H2B plays a role in meiotic DSB formation. Histone H2B ubiquitylation might promote recruitment and/or stabilization of DSB forming machinery. This is supported by the fact that the rad6 mutation does not affect the DSB formation ectopically induced by Gal4-Spo11. Based on this observation, we propose that ubiquitylated histone H2B (ubH2B) is a recognition code for a protein complex engaged in DSB formation during meiosis. Because absence of ubH2B does not eliminate DSB formation, it is likely that various other histone tail modifications, called histone code (28, 29), are recognized (and read) by the complex, as seen in transcription initiation and elongation.
Recently it has been shown that ubiquitylated histone H2B promotes histone H3 methylation at K4 and K79 (30, 31). This suggests that histone H2B ubiquitylation might influence DSB formation through methylation of histone H3. The relationship between ubiquitylation of H2B and methylation of H3 is largely unknown. It is possible that the methylation of histone H3 is another determinant for DSB formation.
The rad6 mutation more severely affects the DSB formation at sites with a higher frequency than those with a lower frequency. These indicate that DSB sites can be classified into two types, Rad6-dependent and -independent. Alternatively, the basal level of DSB formation is a Rad6-independent process, whereas an additional increase in DSB formation is mediated by a Rad6-dependent process. Because the DSB sites are unchanged in the mutants, the selection of DSBs sites during meiosis is independent of the role of Rad6-Bre1-ubiquitylated histone H2B. The global chromatin structure is likely to play a role in the site selection (32, 33).
The studies described here suggest the role of a histone modification in meiotic DSB formation. This supports the view that the DSB formation shares some properties with the initiation of transcription (34, 35).
Supplementary Material
Acknowledgments
We thank Dr. H. Oshiumi for helpful discussions. We are grateful to Drs. K. Ohta (RIKEN, Saitana, Japan) and A. Nicolas (Institute Curie, Paris) for materials used in this study. This work was supported by grants from the Ministry of Education, Science and Culture of Japan (to A.S.). K.Y. was supported by the 21st Century Center of Excellence Program from the Ministry of Education, Science and Culture of Japan.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: DSBs, double-strand breaks.
Note Added in Proof. While this paper was under review, Sollier et al. (36) reported that Set1, a histone H3 K4 methyltransferase, is required for DSB formation at the CYS3 locus, consistent with our data described here.
References
- 1.Hicke, L. (2001) Nat. Rev. Mol. Cell. Biol. 2, 195–201. [DOI] [PubMed] [Google Scholar]
- 2.Ulrich, H. D. (2002) Eukaryot. Cell 1, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robzyk, K., Recht, J. & Osley, M. A. (2000) Science 287, 501–504. [DOI] [PubMed] [Google Scholar]
- 4.Sung, P., Prakash, S. & Prakash, L. (1988) Genes Dev. 2, 1476–1485. [DOI] [PubMed] [Google Scholar]
- 5.Hwang, W. W., Venkatasubrahmanyam, S., Ianculescu, A. G., Tong, A., Boone, C. & Madhani, H. D. (2003) Mol. Cell 11, 261–266. [DOI] [PubMed] [Google Scholar]
- 6.Wood, A., Krogan, N. J., Dover, J., Schneider, J., Heidt, J., Boateng, M. A., Dean, K., Golshani, A., Zhang, Y., Greenblatt, J. F., et al. (2003) Mol. Cell 11, 267–274. [DOI] [PubMed] [Google Scholar]
- 7.Henry, K. W., Wyce, A., Lo, W. S., Duggan, L. J., Emre, N. C., Kao, C. F., Pillus, L., Shilatifard, A., Osley, M. A. & Berger, S. L. (2003) Genes Dev. [DOI] [PMC free article] [PubMed]
- 8.Turner, S. D., Ricci, A. R., Petropoulos, H., Genereaux, J., Skerjanc, I. S. & Brandl, C. J. (2002) Mol. Cell. Biol. 22, 4011–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng, H. H., Dole, S. & Struhl, K. (2003) J. Biol. Chem. 278, 33625–33628. [DOI] [PubMed] [Google Scholar]
- 10.Reid, R. J., Lisby, M. & Rothstein, R. (2002) Methods Enzymol. 350, 258–277. [DOI] [PubMed] [Google Scholar]
- 11.Wach, A., Brachat, A., Pohlmann, R. & Philippsen, P. (1994) Yeast 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 12.Alani, E., Padmore, R. & Kleckner, N. (1990) Cell 61, 419–436. [DOI] [PubMed] [Google Scholar]
- 13.Cha, R. S., Weiner, B. M., Keeney, S., Dekker, J. & Kleckner, N. (2000) Genes Dev. 14, 493–503. [PMC free article] [PubMed] [Google Scholar]
- 14.Cao, L., Alani, E. & Kleckner, N. (1990) Cell 61, 1089–1101. [DOI] [PubMed] [Google Scholar]
- 15.Shinohara, M., Shita-Yamaguchi, E., Buerstedde, J. M., Shinagawa, H., Ogawa, H. & Shinohara, A. (1997) Genetics 147, 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baudat, F. & Nicolas, A. (1997) Proc. Natl. Acad. Sci. USA 94, 5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinohara, M., Gasior, S. L., Bishop, D. K. & Shinohara, A. (2000) Proc. Natl. Acad. Sci. USA 97, 10814–10819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailly, V., Lauder, S., Prakash, S. & Prakash, L. (1997) J. Biol. Chem. 272, 23360–23365. [DOI] [PubMed] [Google Scholar]
- 19.Dowling, E. L., Maloney, D. H. & Fogel, S. (1985) Genetics 109, 283–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bishop, D. K., Park, D., Xu, L. & Kleckner, N. (1992) Cell 69, 439–456. [DOI] [PubMed] [Google Scholar]
- 21.Gerton, J. L., DeRisi, J., Shroff, R., Lichten, M., Brown, P. O. & Petes, T. D. (2000) Proc. Natl. Acad. Sci. USA 97, 11383–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop, D. K. (1994) Cell 79, 1081–1092. [DOI] [PubMed] [Google Scholar]
- 23.Pecina, A., Smith, K. N., Mezard, C., Murakami, H., Ohta, K. & Nicolas, A. (2002) Cell 111, 173–184. [DOI] [PubMed] [Google Scholar]
- 24.Enyenihi, A. H. & Saunders, W. S. (2003) Genetics 163, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsui, K., Simon, L. & Norris, D. (1997) Genetics 145, 647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, H., Kahana, A., Gottschling, D. E., Prakash, L. & Liebman, S. W. (1997) Mol. Cell. Biol. 17, 6693–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith, K. N., Penkner, A., Ohta, K., Klein, F. & Nicolas, A. (2001) Curr. Biol. 11, 88–97. [DOI] [PubMed] [Google Scholar]
- 28.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 29.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 30.Ng, H. H., Xu, R. M., Zhang, Y. & Struhl, K. (2002) J. Biol. Chem. 277, 34655–34657. [DOI] [PubMed] [Google Scholar]
- 31.Sun, Z. W. & Allis, C. D. (2002) Nature 418, 104–108. [DOI] [PubMed] [Google Scholar]
- 32.Ohta, K., Shibata, T. & Nicolas, A. (1994) EMBO J. 13, 5754–5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu, T. C. & Lichten, M. (1994) Science 263, 515–518. [DOI] [PubMed] [Google Scholar]
- 34.Keeney, S. (2001) Curr. Top. Dev. Biol. 52, 1–53. [DOI] [PubMed] [Google Scholar]
- 35.Lichten, M. & Goldman, A. S. (1995) Annu. Rev. Genet. 29, 423–444. [DOI] [PubMed] [Google Scholar]
- 36.Sollier, J., Linm W., Soustellem C., Suhre, K., Nicolas, A., Geli, V. & De La Roche Saint-Andre, C. (2004) EMBO J. 23, 1957–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.