Abstract
We demonstrate that direct analysis in real time-high resolution mass spectrometry can be used to produce mass spectral profiles of botanical material, and that these chemical fingerprints can be used for plant species identification. The mass spectral data can be acquired rapidly and in a high throughput manner without the need for sample extraction, derivatization or pH adjustment steps. The use of this technique bypasses challenges presented by more conventional techniques including lengthy chromatography analysis times and resource intensive methods. The high throughput capabilities of the direct analysis in real time-high resolution mass spectrometry protocol, coupled with multivariate statistical analysis processing of the data, provide not only class characterization of plants, but also yield species and varietal information. Here, the technique is demonstrated with two psychoactive plant products, Mitragyna speciosa (Kratom) and Datura (Jimsonweed), which were subjected to direct analysis in real time-high resolution mass spectrometry followed by statistical analysis processing of the mass spectral data. The application of these tools in tandem enabled the plant materials to be rapidly identified at the level of variety and species.
Keywords: Chemistry, Issue 116, direct analysis in real time mass spectrometry, forensic chemistry, drug identification, plant-based drugs of abuse, Kratom, Mitragyna speciosa, Datura, Jimsonweed, mass spectrometry, ambient ionization, species identification, high-throughput analysis
Introduction
For millennia, psychoactive natural products have been used in shamanic rituals, abused for their mind-altering attributes, and consumed for their medicinal properties. The ingestion of these plants and related plant-based substances can be significant in areas where they are endemic, and they have social and economic importance. Recently however, there has been a dramatic rise in the use of these "natural" drugs due to the ease of accessibility through internet commerce. The perception that these substances are safe to use, coupled with an increased crackdown on possession and use of more traditional drugs of abuse and synthetic substances, have contributed to the spike in abuse of plant-based drugs. It is generally difficult by visualization to distinguish between these products and harmless plant material, and therefore there is interest in developing methods that can be used identify these products. However, conventional analytical methods for plant species identification are time consuming and impractical to perform. Moreover, the methods development process is time and resource intensive. These factors have made the crafting of legislation to curb the use of these substances lag far behind the rate of escalation in their abuse. Thus, there are few laws that regulate the production, manufacture, sale and consumption of many of these natural psychoactives and as such, there are hundreds of plants of abuse available to users in thousands of different forms.1,2
Two such plant-based drugs of abuse are Mitragyna speciosa, commonly known as Kratom, and plants from the Datura genus, namely D. stramonium, D. ferox and D. inoxia. Kratom and Datura are unscheduled in the United States, but the Drug Enforcement Administration has listed both as drugs of concern.3,4 Kratom is characterized by the presence of the psychoactive compounds mitragynine and 7-hydroxymitragynine, as well as other non-psychoactive alkaloids including mitraphylline, paynantheine, corynoxeine, and rhynchophylline.4-8 The psychoactive properties of Datura spp. are attributed to atropine and scopolamine, but a variety of other tropane alkaloids have been identified in the plants.9-12 Both Kratom and Datura have been implicated in poisonings and fatalities, and their identification is increasingly necessary in both forensic and toxicological contexts, as the abuse of these products is on the rise.13-16
By and large, traditional methods used for the analysis of forensic drug material, such as color tests, microscopy, and Raman and infrared spectroscopy, are used in a rule-in/rule-out presumptive capacity. Hyphenated techniques such as GC-MS and liquid chromatography-mass spectrometry (LC-MS), are confirmatory analysis methods based on comparison of the profiles of detected analytes to Scientific Working Group on Analysis of Seized Drugs (SWGDRUG) library standards.17 The sample treatment steps that are performed prior to analysis including pulverization, extraction, derivatization and evaporation, can add hours to the run time and adulterate the sample,9,11,18,19 making analysis of botanical drugs less than straightforward compared to that for other traditional drugs of abuse like cocaine or heroin. Moreover, individual chromatographic programs need to be developed for every product of interest, which makes the implementation of standard operating protocols for each species or variety of plant-based drug of abuse highly impractical for routine casework.
Direct analysis in real time-high resolution mass spectrometry is an ambient ionization mass spectrometric technique that circumvents some of the challenges associated with conventional analytical methods. Gases, liquids, solids, powders, TLC plates and plant material can all be analyzed directly using direct analysis in real time-high resolution mass spectrometry, and both polar and non-polar compounds can be easily detected in complex matrices.20-22 Furthermore, studies have shown that psychoactive compounds can be rapidly identified in plant material by direct analysis in real time-high resolution mass spectrometry, and species-specific information can be gleaned from statistical processing of the mass spectral data.22-26
Here, we demonstrate that direct analysis in real time-high resolution mass spectrometry can be used to quickly assess various plant materials (i.e., plants, powders, extracts and seeds) for their psychoactive components, and that the species and varieties of the plant-derived products can be determined in a rapid and high-throughput manner. The analysis of forensically relevant botanical material without the need for sample preparation steps or long chromatographic analysis run times, in addition to plant species identification is reported.
Protocol
1. Preparation of Plant Materials
- Kratom Fresh Leaf Material
- Use a 6 mm diameter hole punch to create uniform chads of Kratom leaf material from M. speciosa plant. Repeat 5 times.
- Kratom Powder Extraction
- Mix 5 ml of ethanol and 5 ml of distilled water to create 1:1 solvent mixture for extraction.
- In a 1.5 ml microcentrifuge tube, suspend a small amount (~5 mg) of Kratom Bali powder in 1 ml of the 1:1 EtOH:H2O solvent mixture. Repeat 5 times.
- Sonicate the Kratom Bali powder extract samples in an ultrasonic bath for 30 min at ambient temperature.
- Centrifuge the Kratom Bali powder extract samples for 2 min at 750 x g at ambient temperature.
- Decant the solvent from residual powder for subsequent analysis.
- Datura seed preparation
- Slice a D. stramonium seed in half across the transverse plane using a razor blade. Repeat using 5 different seeds.
- Repeat for D. inoxia and D. ferox seeds.
2. Direct Analysis in Real Time Ion Source Parameters
- Gas Heater Temperature
- Set the gas heater temperature of the ion source to 350 °C.
- Ion Mode
- Conduct the analysis in positive ion mode with grid voltage of 250 V.
- Helium Gas Flow Rate
- Set the helium gas flow rate to 2.0 L/sec.
3. Time-of-flight Mass Spectrometer Parameters
- Orifice Voltages
- Set the orifice 1 voltage to 20 V and the orifice 2 voltage to 5 V.
- Ring Lens and Peak Voltage
- Adjust the ring lens voltage to 5 V and change the peaks voltage to 600 V.
- Mass Spectral Acquisition
- Set the mass spectral acquisition rate to 1 spectrum per second over a mass range of m/z 60-800.
- Mass Spectrometer Resolving Power
- Set the resolving power of the mass spectrometer to 6000 FWHM.
4. Analysis of Plant Materials
- Analysis of Kratom Leaf
- Press "Start Run" in mass spectrometer control software. Suspend the chad of plant material between the ion source and mass spectrometer inlet (approximately 2 cm from the inlet) with tweezers until a spectrum is obtained. Repeat 5 times with separate chads of plant material.
- Calibrate the spectrum with polyethylene glycol 600 (PEG).
- Dip the closed end of a melting point capillary tube into the PEG standard. Suspend the coated capillary between the ion source and the mass spectrometer inlet.
- After analyzing the PEG standard, select the "Stop" button to end the analytical run.
- Press "Start Run" in mass spectrometer control software. Suspend a small amount of dried leaf material between the ion source and mass spectrometer inlet with tweezers until a spectrum is obtained. Repeat 5 times, analyzing new plant material each time.
- Calibrate the spectrum with PEG.
- Dip the closed end of a capillary into the PEG standard. Suspend the coated capillary between the ion source and the mass spectrometer inlet.
- After analyzing the PEG standard, select the "Stop" button to end the analytical run.
- Analysis of Kratom Powder
- Press "Start Run" in mass spectrometer control software. Dip the closed end of a melting point capillary into the Kratom powder.
- Suspend the coated capillary between the ion source and mass spectrometer inlet until a spectrum is obtained. Repeat the analysis 5 times with a new capillary each time.
- Calibrate the spectrum with PEG.
- Dip the closed end of a capillary into the PEG standard. Suspend the coated capillary between the ion source and the mass spectrometer inlet.
- After analyzing the PEG standard, select the "Stop" button to end the analytical run.
- Analysis of Kratom Extract
- Immerse the closed end of a capillary tube into the extract.
- Suspend the capillary tube in the 12-sample holder on the linear rail attached to the mass spectrometer. Repeat 5 times with a different extract each time.
- Press "Start Run" in mass spectrometer control software. Using the control panel, select the ">" button to advance the linear rail through the ion stream at rate of 1 mm/sec to collect spectra.
- Calibrate the spectrum with PEG.
- Dip the closed end of a capillary into the PEG standard. Suspend the coated capillary between the ion source and the mass spectrometer inlet.
- After analyzing the PEG standard, select the "Stop" button to end the analytical run.
- Analysis of Datura seeds
- Press "Start Run" in mass spectrometer control software. Suspend the Datura seed half between the ion source and mass spectrometer inlet with tweezers until a spectrum is collected. Ensure that the cut side is oriented to face the ion source. Repeat 5 times, analyzing a new seed half each time.
- Calibrate the spectrum with PEG.
- Dip the closed end of a capillary into the PEG standard. Suspend the coated capillary between the ion source and the mass spectrometer inlet.
- After analyzing the PEG standard, select the "Stop" button to end the analytical run.
- Repeat steps 4.4.1-4.4.3 for each Datura species.
5. Data Processing
- Translating Data Files
- In the data processing software, select File, "Translate DART Files" with "automatic calibration" turned on, to create calibrated data files.
- Left click and drag a box around the first peak in the chromatogram and select "Average" to create an averaged spectrum.
- Right click and drag a box around an area where no sample was collected and select "Average entire box as background" to subtract background from the averaged spectrum.
- Save the mass spectrum as a .txt file.
- Repeat steps 5.1.1-5.1.4 for each peak in the chromatogram to create an averaged spectrum for each replicate in the file.
- Repeat steps 5.1.2-5.1.5 for each file collected.
6. Statistical Analysis
- Principal Component Analysis
- In the Classify section of the spectral analysis software (See List of Materials), under the "Set up" tab, create classes for data processing by selecting "Add Class".
- Import the text files of data by selecting "Add File(s)".
- Assign data files to the appropriate class of plant by selecting text files and "Set Class for Selected Files".
- Select feature masses for discrimination from MS from Training Set and set Threshold % to 1%.
- Set a mass tolerance (mmu) to 10 and select "Build Vectors from Data Files".
- In the "Compute" section, perform principal component analysis by checking the box for 3D PCA Graph and select "Calculate".
- Perform leave-one-out cross validation by selecting "Validate (SLOW!)."
- Generate Heat Map
- In the Frequency Plot tab of the spectral analysis software, generate a heat map of data by selecting "Heat Map."
- Select "Threshold saved data" to set abundance threshold to 1%.
- Export the heat map to a spreadsheet by selecting "Save Heat Map to Excel."
- In the spreadsheet program, save the exported heat map as a .txt file.
- Hierarchical Clustering Analysis
- Import the heat map as a .txt file into Cluster 3.0 software.
- In the hierarchical tab of Cluster 3.0, under Genes and Arrays, check boxes "Cluster" and "Calculate weights". Set cutoff at 0.1 and exponent to 1. Select single linkage clustering to perform analysis.
- View the generated .cdt data file in Java Treeview.
Representative Results
Representative soft ionization positive-ion mode direct analysis in real time-high resolution mass spectrometry data of Kratom products and Datura seeds are shown in Figures 2 and 3. Various compounds previously isolated from M. speciosa, including mitragynine (C23H30N2O4 + H+, m/z 399.2284) and 7-hydroxymitragynine (C23H30N2O5 + H+, m/z 415.2233), were detected in all four samples and the corresponding mass measurement data are presented in Table 1.4-8 Representative Datura data are shown in Figure 3 and signature biomarkers including atropine (C17H23NO3 + H+, m/z 290.1756) and scopolamine (C17H21NO4 + H+, m/z 304.1549) were detected in the three species. Mass measurement data associated with Figure 3 are presented in Table 2.9-12 Compound identities were confirmed through elemental composition determination, isotope matching, and comparisons to reports in literature.23-24
Heat map renderings of the direct analysis in real time-high resolution mass spectrometry spectra of Kratom and Datura are illustrated in Figure 4. The data presented were used in principal component analysis (PCA) to differentiate between the two classes of plant-based drugs of abuse (Figure 5). The PCA plot was constructed using ten feature masses (listed in Table 3), with blue circles representing Kratom data and red squares representing Datura data. The feature masses selected corresponded to various alkaloids present in Datura or Mitragyna spp., including the psychoactive compounds atropine, scopolamine, mitragynine and 7-hydroxymitragynine.23-24 Three principal components accounted for 75.26% of the variance and leave-one-out cross validation (LOOCV) was 100%. The PCA plot clearly shows that the Kratom data and Datura data are well resolved from one another. PCA analysis also revealed that the individual varieties of Kratom and the different species of Datura could be identified and distinguished from one another (Figure 6). LOOCV was 94.29% with three principal components covering 75.26% of the variance. The two varieties of Kratom (Bali in blue circles and Rifat in red squares) cluster together, indicating that they belong to the species M. speciosa, but are resolved from one another, demonstrating that they represent different varieties. Furthermore, the Datura species group together and separated from the M. speciosa data, but each of the individual species of Datura (D. inoxia in green triangles, D. stramonium in pink squares and D. ferox in turquoise circles) are clearly differentiated. Despite a data point for D. stramonium appearing to be an outlier, the seed is correctly classified as D. stramonium and not D. inoxia using PCA. Most importantly, the difference in seed color between D. stramonium and D. inoxia confirms they are different species and that the data point in question cannot be D. inoxia.
Hierarchical clustering (Figure 7) was performed without the a priori selection of feature masses. Instead, the entire group of spectral data sets spanning a mass range of m/z 60-800 was imported into open source genomic clustering software and a dendrogram featuring this data was produced. The results of HCA also revealed class, species and variety differentiation solely based on direct analysis in real time-high resolution mass spectrometry derived data and confirmed those of the PCA analysis. The two classes of plant-based drugs of abuse, Kratom and Datura, were separated into individual clades of the dendrogram. Moreover, the Rifat and Bali varieties of Kratom were each isolated into individual sub-clades within the Kratom class. Similarly, D. inoxia, D. ferox and D. stramonium were resolved into their own clades by species within the Datura class.
 Figure 1.Images of M. speciosa (Kratom) products and Datura spp. seeds.a: Bali Kratom dried leaf; b: Bali Kratom powder; c: Rifat Kratom live plant; d: D. stramonium seeds; e: D. inoxia seeds; f: D. ferox seeds. Please click here to view a larger version of this figure.
Figure 1.Images of M. speciosa (Kratom) products and Datura spp. seeds.a: Bali Kratom dried leaf; b: Bali Kratom powder; c: Rifat Kratom live plant; d: D. stramonium seeds; e: D. inoxia seeds; f: D. ferox seeds. Please click here to view a larger version of this figure.
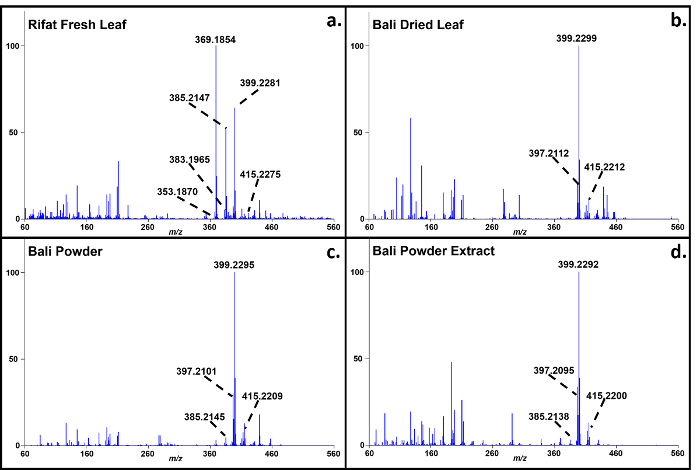 Figure 2.Direct analysis in real time-high resolution mass spectrometry positive-ion spectra of M. speciosa (Kratom) products.a: Rifat fresh leaf; b: Bali dried leaf; c: Bali powder; d: Bali powder extract. The mass measurement data associated with these spectra are shown in Table 1. Please click here to view a larger version of this figure.
Figure 2.Direct analysis in real time-high resolution mass spectrometry positive-ion spectra of M. speciosa (Kratom) products.a: Rifat fresh leaf; b: Bali dried leaf; c: Bali powder; d: Bali powder extract. The mass measurement data associated with these spectra are shown in Table 1. Please click here to view a larger version of this figure.
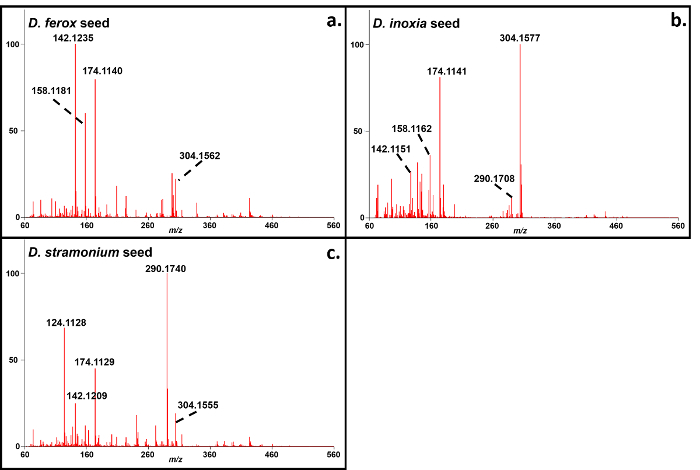 Figure 3. Direct analysis in real time-high resolution mass spectrometry positive-ion spectra of Datura spp. seeds.a: D. ferox; b: D. inoxia; c: D. stramonium. The mass measurement data associated with these spectra are shown in Table 2. Please click here to view a larger version of this figure.
Figure 3. Direct analysis in real time-high resolution mass spectrometry positive-ion spectra of Datura spp. seeds.a: D. ferox; b: D. inoxia; c: D. stramonium. The mass measurement data associated with these spectra are shown in Table 2. Please click here to view a larger version of this figure.
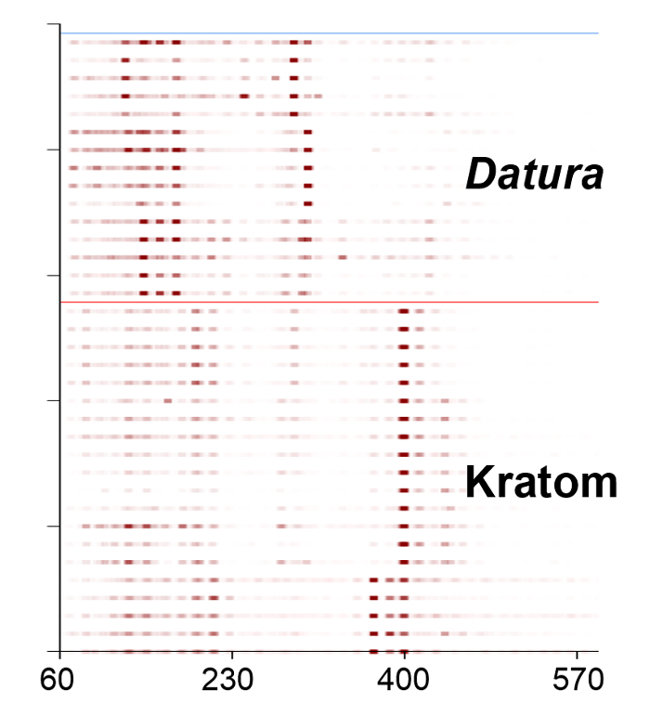 Figure 4. Heat map renderings of the direct analysis in real time-high resolution mass spectrometry spectra of Kratom and Datura plant materials. High intensity peaks are shown in dark red and lower intensity peaks are indicated in lighter shades. Please click here to view a larger version of this figure.
Figure 4. Heat map renderings of the direct analysis in real time-high resolution mass spectrometry spectra of Kratom and Datura plant materials. High intensity peaks are shown in dark red and lower intensity peaks are indicated in lighter shades. Please click here to view a larger version of this figure.
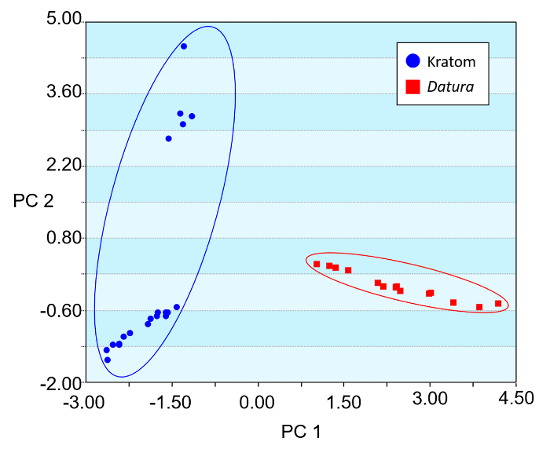 Figure 5. Principal component analysis (PCA) plot of Kratom and Datura products constructed using direct analysis in real time high resolution mass spectrometry-derived data. Three principal components (PCs) accounted for 75.26% of the variation, and the leave-one-out cross validation (LOOCV) was 100%. The feature masses used for the PCA are listed in Table 3. Please click here to view a larger version of this figure.
Figure 5. Principal component analysis (PCA) plot of Kratom and Datura products constructed using direct analysis in real time high resolution mass spectrometry-derived data. Three principal components (PCs) accounted for 75.26% of the variation, and the leave-one-out cross validation (LOOCV) was 100%. The feature masses used for the PCA are listed in Table 3. Please click here to view a larger version of this figure.
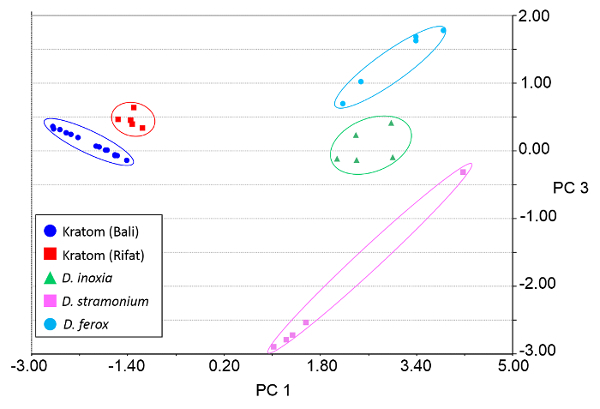 Figure 6. Principal component analysis (PCA) plot of Kratom and Datura products using direct analysis in real time-high resolution mass spectrometry data. Class assignments were based on the variety (Kratom) or species (Datura) of the plant materials. Three principal components (PCs) accounted for 75.26% of the variation and the leave-one-out cross validation (LOOCV) was 94.29%. The feature masses used for PCA are listed in Table 3. Please click here to view a larger version of this figure.
Figure 6. Principal component analysis (PCA) plot of Kratom and Datura products using direct analysis in real time-high resolution mass spectrometry data. Class assignments were based on the variety (Kratom) or species (Datura) of the plant materials. Three principal components (PCs) accounted for 75.26% of the variation and the leave-one-out cross validation (LOOCV) was 94.29%. The feature masses used for PCA are listed in Table 3. Please click here to view a larger version of this figure.
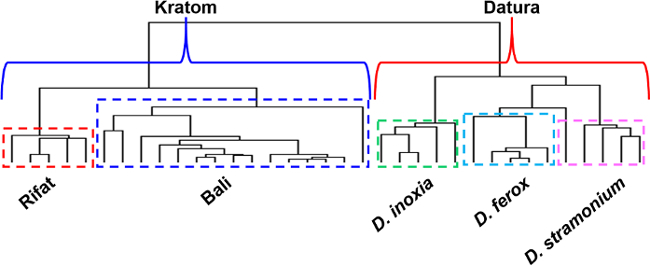 Figure 7. Hierarchical clustering results obtained using the direct analysis in real time-high resolution mass spectrometry-derived data from analysis of Kratom and Datura plant materials. The two classes of plants are clearly separated into two distinct branches in the dendrogram (shown in blue and red brackets for Kratom and Datura, respectively). The Datura seed species are also resolved from one another (shown in green, turquoise and pink dashed boxes for D. inoxia, D. ferox, and D. stramonium respectively). The Kratom plant materials are separated by variety (shown in red and blue dashed boxes for Rifat and Bali, respectively). Please click here to view a larger version of this figure.
Figure 7. Hierarchical clustering results obtained using the direct analysis in real time-high resolution mass spectrometry-derived data from analysis of Kratom and Datura plant materials. The two classes of plants are clearly separated into two distinct branches in the dendrogram (shown in blue and red brackets for Kratom and Datura, respectively). The Datura seed species are also resolved from one another (shown in green, turquoise and pink dashed boxes for D. inoxia, D. ferox, and D. stramonium respectively). The Kratom plant materials are separated by variety (shown in red and blue dashed boxes for Rifat and Bali, respectively). Please click here to view a larger version of this figure.
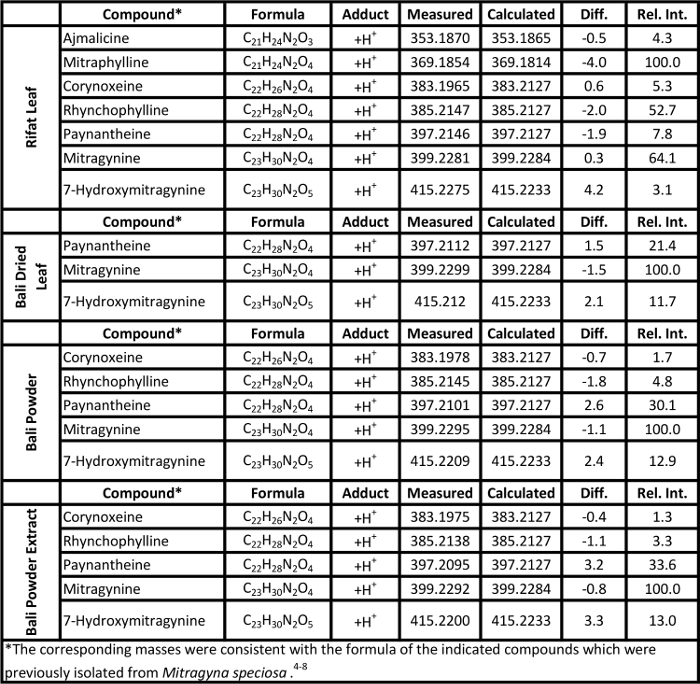 Table 1. Mass measurement data associated with the soft ionization spectra of Kratom products presented in Figure 2. Please click here to view a larger version of this table.
Table 1. Mass measurement data associated with the soft ionization spectra of Kratom products presented in Figure 2. Please click here to view a larger version of this table.
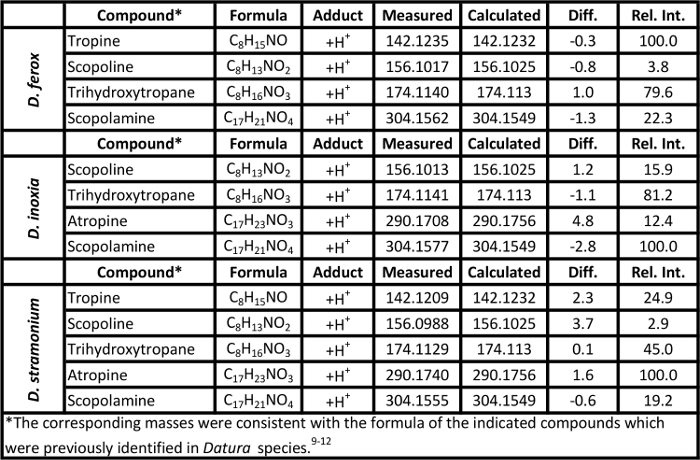 Table 2. Mass measurement data associated with the soft ionization spectra of Datura seeds presented in Figure 3. Please click here to view a larger version of this table.
Table 2. Mass measurement data associated with the soft ionization spectra of Datura seeds presented in Figure 3. Please click here to view a larger version of this table.
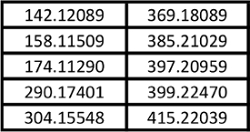 Table 3. Feature masses used for principal component analysis plot of Kratom and Datura products shown in Figures 5 and 6.
Table 3. Feature masses used for principal component analysis plot of Kratom and Datura products shown in Figures 5 and 6.
Discussion
The ability to identify plant-based drugs of abuse is of increasing necessity due to the dramatic rise in the marketing, sale and consumption of unscheduled psychoactive substances.2 Traditional methods for identification of botanical material usually involve characterization of physical features coupled with analysis of the chemical constituents by hyphenated chromatographic-mass spectrometric methods. However, both of these approaches present challenges to streamlined analysis. Physical features of the plants are often destroyed when the plants are dried, pulverized or extracted during the manufacturing process and as such, it is often difficult to distinguish one type of plant-based psychotropic product from another based on physical features alone.23 Analysis by chromatographic-mass spectrometric methods can enable identification of the psychoactive compounds in plant matrices, but the sample preparation and method development is time and resource intensive, and the creation of new protocols for each class or species of plant-based drug of abuse is impractical in many forensic chemistry labs.
Direct analysis in real time-high resolution mass spectrometry circumvents some of these challenges, as complex matrices such as leaves, powders, extracts and seeds can be analyzed with little sample preparation. Despite the complex matrices of the materials sampled here, the psychoactive constituents were easily identifiable, even at nanogram concentrations,21 due to the high sensitivity of the mass spectrometer. Seeds, leaves and powders were demonstrated to be readily analyzed by direct analysis in real time-high resolution mass spectrometry, and a variety of other types of materials can also be sampled in the same manner, including TLC plates, currency, tablets, flowers, solid phase microextraction (SPME) fibers and even insect puparial casings.21-22 Through accurate mass analysis, elemental composition determination and isotope matching, biomarkers and compounds of interest can be identified, regardless of whether the compounds are contained in a leaf, spotted on a TLC plate or coated on a capillary tube.
Direct analysis in real time-high resolution mass spectrometry methodology can be used for streamlined analysis as there are very few parameters that need to be modified from experiment to experiment. Analysis can be performed in positive-ion or negative-ion mode, and molecules of up to 3,000 amu can be detected in both cases. Ionization in positive-ion mode occurs by proton transfer from activated water clusters,21 and any compound with a proton affinity higher than that of water will be ionized. Here, positive-ion mode was used because of the high proton affinity of alkaloids, which causes them to be readily ionized by protonation. Analysis in negative-ion mode can be used for the successful detection of hydrocarbons (as O2 adducts)21, explosive materials27 and organic acids such as artesunate in malarial medications.28 Due to the ionization method of proton-transfer and the inability to produce multiply charged ions, direct analysis in real time-high resolution mass spectrometry is limited mainly to the analysis of small molecules of up to 3,000 amu.
Other than ionization mode, the temperature of the ion source is an important parameter and the appropriate temperature largely depends on the sample being analyzed. For example, it is important to use lower temperatures (~250 °C) for SPME fiber analysis to prevent destruction of the coating material on the fiber, while higher temperatures (~500 °C) should be used for amino acid analysis for desorption and subsequent ionization. Here, plant material analyses were conducted at 350 °C, as this allows for ionization of alkaloids and other compounds of interest without causing pyrolysis of compounds in the plant matrix.
Direct analysis in real time-high resolution mass spectrometry analysis not only enables identification of the psychoactive constituents of the plant material based on accurate masses, elemental composition determination and isotope matching, but it also produces unique chemical fingerprints that can be exploited for species identification using multivariate statistical analysis with highly reproducible results, even with smaller datasets. Multivariate statistical analysis has been applied to a wide variety of direct analysis in real time-high resolution mass spectrometry data, including that derived from wood, puparial casings, seeds, leaf material, and biodiesel fuelstocks, showcasing the versatility and reproducibility of the method.22-26 The high-throughput capabilities of direct analysis in real time-high resolution mass spectrometry enable the acquisition of large amounts of mass spectral data in a short time period, and the large number of replicates needed for statistical analysis are easily acquired using this method. The direct analysis in real time-high resolution mass spectrometry mass spectral dataset of Datura and Kratom was comprised of over 100 individual spectra that were collected with less than an hour of total time investment. To obtain the same number of spectra using GC-MS with an oven temperature program of 30 minutes would take approximately 50 hours, without consideration of the added time for sample preparation steps, such as extraction or derivatization.
Principal component analysis can be used to highlight variations between sets of plant substances based on the presence and intensity of selected feature masses. The statistical analysis processing provides species identification, as well as varietal information. Other methods of statistical analysis, such as hierarchical clustering analysis (HCA), can also be applied without the a priori selection of feature masses. The results of HCA of comprehensive chemical fingerprints show that unsupervised statistical analysis can be successfully applied for species identification of plant-based drugs of abuse.25
The species discrimination of forensic botanical material by direct analysis in real time-high resolution mass spectrometry was demonstrated using the identification of psychotropic compounds and other biomarkers in the soft-ionization mass spectra, and the application of multivariate statistical analysis. The application of two types of statistical analyses revealed not only that the class of a plant-based drug of abuse can be identified, but also that the variety and species of said drug can be determined based on the unique chemical fingerprints observed by direct analysis in real time-high resolution mass spectrometry. The method presented here enables the rapid, high-throughput identification of mind-altering substances in a manner that circumvents challenges encountered in conventional analytical methods, and provides forensic laboratories with a means to characterize and identify psychoactive plant material without time and resource intensive method development. This protocol can be extended to species differentiation of a variety of other plant-derived materials.22-26
Disclosures
The authors have no disclosures.
Acknowledgments
The authors gratefully acknowledge a University at Albany-SUNY Presidential Initiatives Fund for Research and Scholarship in Forensic Science and Cybersecurity grant, a National Science Foundation grant (Grant #1310350) and a National Institute of Justice grant (Grant #2015-DN-BX-K057) to RAM. We also acknowledge Justine E. Giffen for taking the photographs of the plant materials.
References
- Authority, E.F.S. EFSA Compendium of botanicals that have been reported to contain toxic, addictive, psychotropic or other substances of concern onrequest of EFSA. EFSA Journal. 2009;7:1–100. [Google Scholar]
- The challenge of new psychoactive substances: list of plant-based substances (20 Substances) UNODC; 2013. pp. 101–102. [Google Scholar]
- Kratom (Mitragyna speciosa Korth) US Drug Enforcement Administration; 2013. Available from: http://www.deadiversion.usdoj.gov/drug_chem_info/kratom.pdf. [Google Scholar]
- Jimson Weed (Datura stramonium) US Drug Enforcement Administration; 2013. Available from: http://www.deadiversion.usdoj.gov/drug_chem_info/jimson_w.pdf. [Google Scholar]
- Jansen K, Prast C. Ehtnopharmacology of Kratom and the Mitragyna alkaloids. J. Ethnopharmacol. 1988;23:115–119. doi: 10.1016/0378-8741(88)90121-3. [DOI] [PubMed] [Google Scholar]
- Matsumoto K. Pharmacological Studies on 7-Hydroxymitragynine, Isolated from the Thai Herbal Medicine Mitragyna speciosa: Discovery of an Orally Active Opioid Analgesic. Chiba University; 2006. [Google Scholar]
- Matsumoto K, et al. Involvement of µ-opioid receptors in antinociception and inhibition of gastrointestinal transit induced by 7-hydroxymitragynine, isolated from Thai herbal medicine Mitragyna speciosa. Eur. J. Pharmacol. 2006;549 doi: 10.1016/j.ejphar.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, et al. Antinociceptive effect of 7-hydroxymitragynine in mice: Discovery of an orally active opioid analgesic from the Thai medicinal herb Mitragyna speciosa. Life Sciences. 2004;74:2143–2155. doi: 10.1016/j.lfs.2003.09.054. [DOI] [PubMed] [Google Scholar]
- El Bazaoui A, Stambouli H, Bellimam MA, Soulaymani A. Determination of tropane alkaloids in seeds of Datura stramonium L. by GC/MS and LC/MS. Ann. Toxicol. Anal. 2009;21:183–188. [Google Scholar]
- Friedman M, Levin CE. Composition of Jimson Weed (Datura stramonium) seeds. J. Agr. Food Chem. 1989;37:998–1005. [Google Scholar]
- Philipof S, Berkhov S. GC-MS Investigation of tropane alkaloids in Datura stramonium. Z Naturforsch. 2002;57:559–561. doi: 10.1515/znc-2002-5-627. [DOI] [PubMed] [Google Scholar]
- Preissel U, Preissel H-G. Brugmansia and Datura: Angel's Trumpets and Thorn Apples. Firefly Books; 2002. [Google Scholar]
- Boumba VA, Mitselou A, Vougiouklakis T. Fatal poisoning from ingestion of Datura stramonium seeds. Vet. Hum. Toxicol. 2004;46:81–82. [PubMed] [Google Scholar]
- Kronstrand R, Roman M, Thelander G, Eriksson A. Unintentional Fatal Intoxications with Mitragynine and O-Desmethyltramadol from the Herbal Blend Krypton. J. Anal. Toxicol. 2011;35:242–247. doi: 10.1093/anatox/35.4.242. [DOI] [PubMed] [Google Scholar]
- Neerman M, Frost R, Deking J. A drug fatality involving Kratom. J. Forensic Sci. 2013;58:278–279. doi: 10.1111/1556-4029.12009. [DOI] [PubMed] [Google Scholar]
- Steenkamp PA, Harding NM, Van Heerden FR, Van Wyk B-E. Fatal Datura poisoning: Identification of atropine and scopolamine by high performance liquid chromatography / photodiode array / mass spectrometry. Forensic Sci. Int. 2004;145:31–39. doi: 10.1016/j.forsciint.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Scientific Working Group for the Analysis of Seized Drug Recommendations. Scientific Working Group for the Analysis of Seized Drugs; 2014. [Google Scholar]
- Kikura-Hanajiri R, et al. Simultaneous analysis of mitragynine, 7-hydroxymitragynine, and other alkaloids in the psychotropic plant "Kratom" (Mitragyna speciosa) by LC-ESI-MS. Forensic Toxicol. 2009;27:67–74. [Google Scholar]
- Wang M, et al. Comparison of three chromatographic techniques for the detection of mitragynine and other indole and oxindole alkaloids in Mitragyna speciosa (Kratom) plants. J. Separation Sci. 2014. [DOI] [PubMed]
- Cody R. Observation of molecular ions and analysis of nonpolar compounds with the Direct Analysis in Real Time ion source. Anal. Chem. 2009;81:1101–1107. doi: 10.1021/ac8022108. [DOI] [PubMed] [Google Scholar]
- Cody R, Laramee J, Durst HD. Versatile new ion source for the analysis of materials in open air under ambient conditions. Anal. Chem. 2005;77:2297–2302. doi: 10.1021/ac050162j. [DOI] [PubMed] [Google Scholar]
- Musah RA, et al. A High Throughput Ambient Mass Spectrometric Approach to Species Identification and Classification from Chemical Fingerprint Signatures. Sci. Rep. 2015;5 doi: 10.1038/srep11520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesiak AD, Cody RB, Dane AJ, Musah RA. Rapid detection by direct analysis in real time-mass spectrometry (DART-MS) of psychoactive plant drugs of abuse: The case of Mitragyna speciosa aka "Kratom". Forensic Sci. Int. 2014;242:210–218. doi: 10.1016/j.forsciint.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Lesiak AD, Cody RB, Dane AJ, Musah RA. Plant Seed Species Identification from Chemical Fingerprints: A High-Throughput Application of Direct Analysis in Real Time Mass Spectrometry. Anal. Chem. 2015. [DOI] [PubMed]
- Espinoza EO, et al. Distinguishing wild from cultivated agarwood (Aquilaria spp.) using direct analysis in real time and time of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2014;28:281–289. doi: 10.1002/rcm.6779. [DOI] [PubMed] [Google Scholar]
- Lancaster C, Espinoza E. Evaluating agarwood products for 2-(2-phenylethyl)chromones using direct analysis in real time time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 2012;26:2649–2656. doi: 10.1002/rcm.6388. [DOI] [PubMed] [Google Scholar]
- Sisco E, Dake J, Bridge C. "Screening for trace explosives by AccuTOF™-DART®: An in-depth validation study.". Forensic Sci. Int. 2013;232:160–168. doi: 10.1016/j.forsciint.2013.07.006. [DOI] [PubMed] [Google Scholar]
- Fernandez FM, et al. Characterizatoin of solid counterfeit drug samples by desorption electrospray ionization and direct analysis in real time coupld to time-of-flight mass spectrometry. Chem. Med. Chem. 2006;1:702–705. doi: 10.1002/cmdc.200600041. [DOI] [PubMed] [Google Scholar]


