Abstract
Il4 and Il13 encode the canonical T helper 2 (TH2) cytokines responsible both for promoting immune responses against extracellular pathogens and, when misregulated, causing allergic and autoimmune disease. The expression potential of these genes undergoes developmentally programmed repression and enhancement during commitment of naïve CD4+ T cells to the mature T helper 1 (TH1) and TH2 fates, respectively. Thus, like the globin locus, the TH2 cytokine locus provides a highly tractable system to study a developmental fate choice leading to alternative transcriptional states of either silence or permissivity. We used quantitative chromatin immunoprecipitation and RT-PCR to correlate changes in the transcriptional states of Il4 and Il13 with markers of permissive chromatin across the Il4–Il13 locus in naïve CD4+ T cells undergoing TH1 and TH2 differentiation. We provide evidence that DNaseI hypersensitive site V in the Il4 3′ enhancer is the likely target for signals maintaining Il4 and Il13 transcriptional permissivity in naïve cells. We also demonstrate rapid acquisition of differences in H3 acetylation between TH1- and TH2-primed cells, indicating a developmentally early role for cytokine signaling in the process of TH cell fate determination. Finally, we show that transcriptional repression correlates with the disappearance of permissive H3 modifications from everywhere in the Il4–Il13 locus except hypersensitive site IV, suggesting a critical role for this element in the maintenance of transcriptional repression. Our findings are consistent with a progressive regulatory element activation/deactivation model of TH1/TH2 development.
Depending on the nature of signals encountered during initial activation, naïve CD4+ T cells can be instructed to differentiate toward a variety of effector and memory cell fates, distinguished primarily by the potential to express different patterns of cytokine genes. Those that have received the most attention are called T helper 1 (TH1) and TH2. The physically clustered TH2 cytokine genes Il4, Il13, and Il5 are transcriptionally repressed in TH1 cells and permissive in TH2 cells; the converse is true for the unlinked TH1 cytokine gene Ifng. Because different microbes and tissue environments require different effector strategies, proper regulation of this developmental fate choice during immune responses in vivo is critical for the effective control of infectious pathogens, as well as for preventing harmful autoimmune and allergic diseases (1, 2).
Elucidation of the molecular pathways regulating the developmentally programmed changes in Il4 and Il13 transcriptional state that occur with TH development will benefit from a detailed understanding of the cis-acting genetic elements that coordinate their transcriptional behavior. There are 13 hypersensitive site (HS) clusters that have been mapped within the Il4–Il13 locus (3, 4). These can be classified into three groups based on their lineage specificity and requirement for cellular activation: (i) TH2-specific/constitutive, (ii) TH2-specific/activation-dependent, and (iii) naïve/TH1/TH2-shared/constitutive. It is likely that these HSs (and perhaps others more distal; ref. 5) comprise the complete set of regulatory elements responsible for integrating upstream signals into coordinated responses that developmentally alter the transcriptional potential of Il4 and Il13. These responses are likely to involve the recruitment and deployment of both chromatin modifying and RNA polymerase II recruitment/transcription-promoting activities. Targeted deletions have recently demonstrated that several of the HSs (V/Va and S1/S2) are indeed important for Il4 and Il13 expression in TH2 cells (6, 7). However, it is still not known, for each successive development stage, which elements are active, the upstream signaling pathways to which each responds, and the downstream activities each coordinates.
The flexible amino termini of nucleosomal histones harbor multiple residues that can undergo a variety of posttranslational modifications, including acetylation, methylation, and phosphorylation. Combined with the octameric structure of the nucleosomal core, this flexible modification system harbors tremendous combinatorial diversity and information coding potential (the so-called histone code) that can endow discrete genetic intervals with specific functional properties (8). For histone H3, posttranslational modifications can be classified into those associated with chromatin that is transcriptionally repressed (lysine 9 or 27 di- and trimethylation) or transcriptionally permissive [lysine 9/14 acetylation (AcK9/14), lysine 4 dimethylation (2MeK4) and serine 10 phosphorylation (PhS10)] (reviewed in ref. 9). Along a given genetic interval, histone-modifying factors recruited to specific regulatory elements create a dynamic pattern of histone modification specificities and intensities. These, in turn, influence the transcriptional behavior of associated genes by creating or destroying binding platforms for transcriptional activators, repressors, nucleosome remodeling, and chromatin packaging machinery (10). Thus, by characterizing the dynamic chromatin landscape of the Il4–Il13 locus during TH1 and TH2 development and correlating this to the evolving transcriptional states of Il4 and Il13 one may be able to infer for each given element: (i) the developmental stages at which it is active; (ii) the nature of histone-modifying enzymes it may recruit; and (iii) the transcription-modifying activities it may regulate.
Recent evidence indicates that the ability of naïve CD4+ T cells to express Il4 is epigenetically programmed during thymic development coincident with CD4/CD8 lineage commitment (11). During subsequent peripheral maturation steps, this potential is reported to undergo partial repression through mechanisms that involve Il4–Il13 locus DNA methylation (11). Despite the accumulation of repressive DNA methylation marks, the Il4–Il13 locus in naïve CD4+ T cells remains transcriptionally permissive. However, permissive H3 modifications at the Il4–Il13 locus of naïve CD4+ T cells have not been detected (12, 13), begging the question: how is permissivity maintained? Our analysis of the dynamic pattern of permissive H3 modifications at the Il4–Il13 locus during TH1 and TH2 development implicates the TH2-specific/constitutive HSV 3′ of the Il4 gene as the likely recipient of permissivity-inducing signals. In addition, we show that locus-wide changes in permissive modifications that have occurred 48 h after the onset of activation differ greatly in magnitude and stability depending on priming conditions. TH2-priming is associated with high-level/stable and TH1-priming with low-level/transient increases in permissive H3 modifications, indicating that cytokine signaling influences chromatin structure earlier than had previously been suggested (12, 13). We also discovered a significant delay in the occurrence of TH1-priming-dependent Il4 and Il13 transcriptional repression that correlated with the disappearance of permissive H3 modifications from everywhere in the Il4–Il13 locus except HSIV, suggesting a role for this element in the maintenance of transcriptional repression. We discuss our findings in the context of a progressive regulatory element activation/deactivation model of TH1/TH2 development.
Materials and Methods
Cell Lines. D10.G4 (14) and A.E7 (15) were grown in RP10 (RPMI medium 1640 supplemented with 2 mM l-glutamine/25 mM Hepes/0.05 mM 2-mercaptoethanol/50 units/ml penicillin/50 μg/ml streptomycin/10% FBS). Both clones were maintained by periodic stimulation with irradiated AKR/J splenocytes and antigen (D10.G4, conalbumin; and A.E7, pigeon cytochrome c). Resting clones were harvested at least 5 days after the last stimulation. Resting cells were activated by 4-h culture with phorbol 12-myristate 13-acetate (PMA) (5 ng/ml) and ionomycin (250 ng/ml). NIH3T3 cells were grown in DMEM10 (DMEM supplemented with 4 mM l-glutamine/4.5 g/liter glucose/1.5 g/liter sodium bicarbonate/50 units/ml penicillin/50 μg/ml streptomycin/10% FBS).
Naïve T Cell Purification. CD4+CD62LHi naïve T cells were generated by two sequential AutoMACS purification steps. First, CD4+ T cells were isolated from combined lymph node and spleen cell suspensions stained with FITC-anti-CD4 antibody (GK1.5, BD/Pharmingen) followed by anti-FITC MultiSort MicroBeads (Miltenyi Biotec, 130-058-701). After cleavage of the MultiSort MicroBeads, CD4+CD62LHi cells were purified from CD4+ T cells stained with anti-CD62L MicroBeads (Miltenyi Biotec, 130-049-701). In Fig. 2D, initial purification of CD4+ T cells was performed by negative selection using the CD4+ T cell Isolation kit (Miltenyi Biotec, 130-090-860). Purity of CD4+CD62LHi T cells as determined by fluorescence-activated cell sorter (FACS) analysis was ≥98% (positive selection for CD4+) and ≥95% (negative selection for CD4+).
Fig. 2.
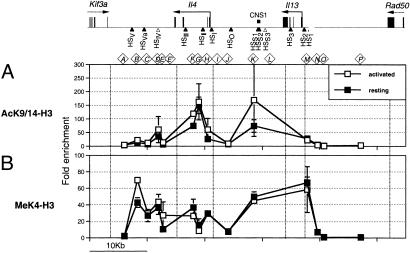
Permissive histone H3 modification at the Il4–Il13 locus during TH1 development. Shown are ChIP plots depicting fold-enrichment for AcK9/14 (circles) and 2MeK4 (squares) across the Il4–Il13 locus in CD4+CD62LHi naïve T cells (A), 48-h TH1-primed CD4+ T cells (B), and resting A.E7 cells (C). Error bars are SEM of n ≥ 2 except for the following where n = 1: naïve (AcK9/14, sites F, G, H and P; 2MeK4, sites A, E′, and J) and A.E7 (AcK9/14, sites B, C E′, F, I, and L; 2MeK4, sites E′, I, and L). 2MeK4 data are not shown for 48-h-primed cells. At the top of the figure is a physical map of the chromosome 11 Kif3a–Il4–Il13–Rad50 locus. Horizontal black arrows represent complete (solid lines) or partial (dotted lines) transcription units. Tall black rectangles represent exons. Arrowheads indicate the location of TH2-specific/constitutive (filled symbols) and TH2-specific/activation-dependent (open symbols) DNase I HSs. The small black rectangle represents a region of high sequence conservation between mouse and human (CNS1). Diamond-encased letters (A–P) represent the names and locations of PCR primer pairs. Dotted vertical lines in each plot bracket the locations of transcription units.
TH1 and TH2 Cultures. Primary TH cultures were generated by using 4- to 6-week-old BALB/c mice (The Jackson Laboratories) housed under specific pathogen-free conditions at the University of Washington, Seattle. CD4+ T cells, AutoMACS purified from combined lymph node and spleen cell suspensions stained with anti-CD4 MicroBeads (Miltenyi Biotec, 130-049-201), were ≥98% CD4 positive as determined by fluorescence-activated cell sorter analysis. Splenic antigen-presenting cells (APCs) were prepared by complement-mediated lysis using anti-Thy1 (J1j, American Type Culture Collection) and a combination of rabbit and guinea pig complement and given 3,000 rads before use. TH cell cultures were seeded at a ratio of 1:5 (T cell/APC) with anti-CD28 (25 μg/ml, 37N) and anti-T cell receptor β (2.5 μg/ml, H57–597). In addition, TH1 cultures contained recombinant mouse IL12 (1 ng/ml, R&D Systems) and anti-IL4 (10 μg/ml, 11B11) whereas TH2 cultures contained recombinant mouse IL4 (30 ng/ml, Leinco Technologies), anti-IL12 (4.5 μg/ml, C17.8), and anti-IFNγ (10 μg/ml, R46A2). Cultures, harvested at 48 h, were stimulated as described above for the cloned cell lines.
RT-PCR. RNA was isolated with STAT60, and contaminating genomic DNA was removed by using RNase-Free DNase (Ambion, 1906). Random hexamer-primed cDNA was generated by using the Superscript II RNase H– Reverse Transcriptase kit (Invitrogen, 18064-014). Real-time PCRs were performed and analyzed on a Stratagene MX4000. The PCR buffer contained 10 mM Tris (pH 8.3), 50 mM KCl, 4.5 mM MgCl2, 0.01% Tween 20, 0.3% DMSO, 0.0025% SYBR Green I solution (Molecular Probes, S-7563), 50 nM each primer (Table 1, which is published as supporting information on the PNAS web site), and Taq polymerase (Promega)/anti-Taq antibody (Clontech) conjugate per 25 μl of reaction. Cycling conditions were: 94°C for 4 min followed by 40 cycles of 94°C for 20 sec, 61°C for 1 min and 72°C for 40 sec. PCR efficiencies were optimized in pilot experiments. CT values for no reverse transcriptase (NRT) controls were at least 4-fold higher than experimental samples, corresponding to <6% background. More often, NRT background was <0.8%. The only exception was site N with 17% background. For a given cDNA, relative abundance of each target was normalized to Hprt according to the formula: 2–ΔCT, where ΔCT = CTTARGET – CTHPRT. Hprt-normalized target signals were expressed relative to expression in NIH3T3 fibroblasts according to the formula: 2–ΔΔCT, where ΔΔCT = ΔCTSAMPLE – ΔCTNIH3T3. Thus 1 arbitrary unit (AU) corresponds to the relative level of a given target in NIH3T3 fibroblasts.
Chromatin Immunoprecipitation (ChIP) Assays. Approximately 3 × 107 cells were used per ChIP (for additional details, see Supporting Text, which is published as supporting information on the PNAS web site). DNA recovered from an aliquot of sheared chromatin was used as the “input” sample. The remaining chromatin was precleared with protein A- and protein G-agarose (Upstate Biotechnologies, catalogue nos. 16-156 and 16-266) and then incubated with one of three antibodies overnight at 4°C [anti-AcK9/14-H3, catalog no. 06-599; anti-M3K4-H3, catalog no. 07-030; anti-PhS10-H3, catalog no. 05-598; Upstate Biotechnology; all used at 2 μg/ml). Input DNA and DNA recovered after IP were quantified by using picogreen fluorescence (Molecular Probes). Equivalent mass of IP and input DNA was analyzed by real-time PCR as described above for RT-PCR with the following modifications. Taq polymerase was from Qiagen (Hot Start) and cycling conditions were 94°C for 15 min followed by 40 cycles of 94°C for 20 sec, 61°C for 1 min, and 72°C for 40 sec. Data are presented as the ratio of 2–CTIP to 2–CT input.
Results
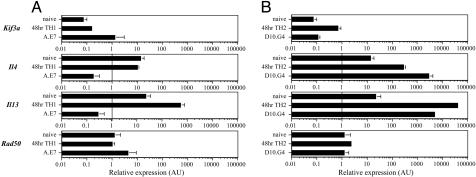
Transcriptional Response of Naïve, TH1, and TH2 Cells. We used quantitative RT-PCR to characterize the 4-h PMA/ionomycin-induced transcriptional response of Kif3a, Il4, Il13, and Rad50 in CD4+CD62LHi naïve T cells, 48-h TH1- and TH2-primed CD4+ T cells, the TH1 clone A.E7, and the TH2 clone D10.G4. A.E7 and D10.G4 are well established models of the fully polarized TH1 and TH2 fates, respectively (14, 15). As expected, naïve cells induced both cytokine genes, although to levels reduced >200-fold compared to the TH2 clone D10.G4 (Fig. 1B). Surprisingly, inducible Il4 and Il13 expression in 48-h TH1-primed cells remained at (Il4) or increased slightly above (Il13) the level detected from activated naïve cells (Fig. 1A). Transcriptional repression of Il4 and Il13 was detected only in the TH1 clone A.E7, representing a terminal stage of TH differentiation (Fig. 1A). In contrast to the developmentally delayed onset of transcriptional repression, enhancement of inducible Il4 and Il13 expression occurred rapidly with TH2-priming, approaching (Il4) or surpassing (Il13) levels detected in the TH2 clone D10.G4 by 48 h of priming (Fig. 1B). Expression of Kif3a and Rad50 were similar at all stages of development regardless of priming conditions (Fig. 1), indicating that the flanking genes are somehow insulated from the regulatory events governing the Il4–Il13 locus.
Fig. 1.
Kif3a, Il4, Il13, and Rad50 mRNA expression in developing TH1 and TH2 cells. CD4+CD62LHi naïve T cells (A and B), CD4 T cells primed for 48 h in TH1(A) or TH2(B) skewing condition, the TH1 clone A.E7 (A), and the TH2 clone D10.G4 (B) were harvested for RNA after 4 h of activation with PMA and ionomycin. Relative expression of Kif3a, Il4, Il13, and Rad50 was determined by quantitative real-time PCR. The ratio of Hprt-normalized expression in T cells and NIH3T3 cells is depicted in arbitrary units (AU). Error bars are SEM of n ≥ 2.
H3 Modifications in CD4+CD62LHi Naïve T Cells. To determine whether the developmentally programmed changes in Il4 and Il13 transcriptional state are linked to chromatin-level changes, we used ChIP to assess posttranslational modification of histone H3 across the Il4–Il13 locus. We focused our analysis on the permissive chromatin modifications AcK9/14 and 2MeK4. Real-time PCR was used to quantitate the relative enrichment of specific target sequences in equivalent amounts of DNA obtained from unprecipitated and precipitated chromatin (Fig. 6, which is published as supporting information on the PNAS web site). We designed PCR primer pairs (probes A–P in Table 1) targeting 17 sites, aiming to cover the locus uniformly and to sample regions known to be functionally, structurally, or computationally implicated in cytokine regulation (Fig. 2; probes A–P). These regions include the Il4 3′ enhancer comprising the TH2-specific/constitutive HSV and TH2-specific/activation-dependent HSVa (4, 7, 16), the CNS1 enhancer (a phylogenetically conserved region encompassing the clustered TH2-specific/constitutive HSS1 and HSS2; ref. 17), and the Il4 intronic enhancer encompassing the TH2-specific/constitutive HSIII and HSII (4, 18). We also examined the TH2-specific/constitutive Il4 promoter-proximal HSI and promoter-distal HS0 (4) and Il13 promoter-proximal HS2 and promoter distal HS1 (3). Finally, we monitored HSIV and HSS3, naïve/TH1/TH2–shared/constitutive HSs immediately downstream of Il4 and Il13, respectively (3, 4, 17). Some regions were also chosen simply to provide extended coverage of the locus.
We began by isolating CD4+CD62LHi naïve T cells from spleen and lymph nodes of young BALB/c mice and processing them for ChIP with an antibody to AcK9/14. No AcK9/14 was detected at HSV and CNS1 (Fig. 2A; probes B and K), corroborating earlier studies (12, 13). Additional sites not previously examined in naïve cells (HSVa, HSIII, HSII, HS0, HS2, HS1, the Kif3a-proximal region, and the Rad50-proximal region) also lacked AcK9/14 (Fig. 2A; probes A–C, F–H, J, and M–P). HSIV 3′ of Il4 was the only region where AcK9/14 was detected (Fig. 2A; probe D). HSIV has been reported to lack AcK9/14 in naïve cells (12). Differences in the precise regions targeted for PCR analysis or in the sensitivity of the quantitative assay used here and the qualitative assay used in earlier studies may account for the discrepancy. Thus, HSIV appears to be the sole focus of preexisting AcK9/14 at the Il4–Il13 locus in naïve T cells. However, because the TH1 clone A.E7 also contains permissive H3 modifications at this site (Figs. 2C and 5, probe D), by itself, H3 modification at HSIV is insufficient to explain the transcriptional permissivity of Il4 and Il13 in naïve T cells.
Fig. 5.
Distribution of serine 10 phosphorylation on histone H3 in A.E7, D10.G4, and NIH3T3 cells. Shown are ChIP plots depicting fold-enrichment of PhS10 across the Il4–Il13 locus of resting A.E7 (circles), D10.G4 (squares), and NIH3T3 (triangles) cells. Error bars are SEM of n ≥ 3. The top of the figure is as described for Fig. 2.
A recent study reported the occurrence of AcK9/14 at the Il4 promoter in naïve CD4+ T cells and suggested that mechanistically this modification might be responsible for Il4 transcriptional permissivity (19). Although, as shown in Fig. 2A, our Il4 promoter-proximal probe J that failed to detect histone acetylation was only 2.5 kb distal to the location of the PCR primer pair used in that study, we could not exclude the possibility that we had missed a nearby peak of acetylation. To address this, we purified CD4+CD62LHi naïve T cells and analyzed these by ChIP using an AcK9/14-specific antibody and PCR primers identical to those described by Grogan et al. (19). Despite strong AcK9/14 enrichment at the control G6pd locus, we were unable to detect any signal at the Il4 promoter (Fig. 2D). Similarly, ChIP analysis of naïve cells with a 2MeK4-specific antibody also failed to reveal enrichment at this site despite a strong G6pd signal (Fig. 2D). We conclude that permissive chromatin modifications do not occur at the Il4 promoter in naïve T cells.
To search for other changes that might still explain the transcriptional permissivity of Il4 and Il13 in naïve T cells, we extended our 2MeK4 ChIP-scan to the remainder of the Il4–Il13 locus. The distribution of 2MeK4 we observed was nearly identical to that of AcK9/14, being mainly restricted to HSIV (Fig. 2A; probe D). Like AcK9/14, 2MeK4 at HSIV was also detected in TH1 cells (Fig. 2C; probe D), further indicating that, by itself, this site does not play a role in maintaining Il4 and Il13 transcriptional permissivity. By contrast, at HSV in the Il4 3′ enhancer, where no AcK9/14 had been detected, we found 2MeK4 to be enriched in naïve T cells (Fig. 2A; probe B). Permissive modifications at HSV also occurred in a TH2 but not in a TH1 clone (Figs. 2C and 3C; probe B). Furthermore, 48-h TH1-primed cells showed a significant decrease relative to naïve cells in 2MeK4 at HSV (data not shown). Thus, the developmental dynamics of 2MeK4 suggests that HSV may play a role in mediating the transcriptional permissivity of Il4 and Il13 in naïve CD4+ T cells.
Fig. 3.
Histone H3 modification at the Il4–Il13 locus during TH2 development. Shown are ChIP plots depicting fold-enrichment for AcK9/14 (circles) and 2MeK4 (squares) across the Il4–Il13 locus in CD4+CD62LHi naïve T cells (A), 48-h TH1-primed CD4+ T cells (B), and resting D10.G4 cells (C). Error bars are SEM of n ≥ 2 except for the following where n = 1: naïve (AcK9/14, sites F–H and P; 2MeK4, sites A, E′, and J). In C, the AcK9/14 enrichment for probe G is 150 ± 30. 2MeK4 data are not shown for 48-h-primed cells. The top of the figure is as described for Fig. 2. The naïve plot (A) is the same as shown in Fig. 2 A.
H3 Modifications in Developing TH1 Cells. We hypothesized that the developmentally delayed onset of Il4 and Il13 transcriptional repression that occurs with TH1-priming (Fig. 1 A) would be linked to underlying features in the chromatin landscape of the Il4–Il13 locus. To test this hypothesis, purified CD4+ T cells from young BALB/c mice cultured for 48 h in TH1-priming conditions were ChIP-scanned for AcK9/14. Compared to naïve T cells, we detected small increases in AcK9/14 (all <5-fold) at the Il4 3′, intronic and CNS1 enhancers and the Il4 and Il13 promoter-proximal regions (Fig. 2 A and B; probes B, C, F–K, M, N, and P). By contrast, no change was detected at HSIV (Fig. 2 A and B; probe D). Thus, relative to naïve T cells, slightly increased Il4 and Il13 transcriptional permissivity in 48-h TH1-primed cells correlate with modest increases in AcK9/14 levels across most of the Il4–Il13 locus.
To assess the chromatin structural correlates of Il4 and Il13 transcriptional repression, we analyzed the TH1 clone A.E7. Resting cells were ChIP-scanned with antibodies that detect AcK9/14, 2MeK4, and PhS10. All three markers of permissive chromatin were almost completely absent from the Il4–Il13 locus (Figs. 2C and 5). The sole exception was HSIV, where striking enrichment for all three permissive markers was focused in a sharp peak (Figs. 2C and 5; probe D). Permissive H3 modifications at HSIV, like DNase I hypersensitivity (4), were lineage restricted as indicated by their absence from fibroblast lineage NIH3T3 cells (see Fig. 5 and data not shown for AcK9/14 and 2MeK4). Thus, the repressed transcriptional state of Il4 and Il13 correlates with the disappearance of permissive H3 modifications from everywhere in the Il4–Il13 locus, except at HSIV, where levels increased.
H3 Modifications in Developing TH2 Cells. The inverse correlation between transcriptional repression and permissive H3 modifications seen across most of the Il4–Il13 locus with TH1 development suggested that the enhanced Il4 and Il13 transcriptional response seen with TH2-priming would be positively correlated with permissive H3 modifications. To test this, purified CD4+ T cells from young BALB/c mice cultured in TH2-priming conditions for 48-hours were ChIP-scanned for AcK9/14. Relative to naïve T cells, we detected increased AcK9/14 levels at the Il4 3′, intronic and CNS1 enhancers and the upstream regions of both Il4 and Il13 (Fig. 3B; probes B, C, F–K, M, and N). Although the overall pattern of this increase was similar to that detected in 48-h TH1-primed cells (Fig. 2B), we noted two striking differences. Overall, the magnitude of the AcK9/14 increase averaged 3.5-fold higher with TH2 vs. TH1 priming, correlating with the differentially enhanced level of the induced Il4 and Il13 transcriptional response (Fig. 1). Comparable AcK9/14 signals were observed for the control gene G6pd, demonstrating the specificity of this effect (data not shown). Second, at site L near the naïve/TH1/TH2-shared/constitutive HSS3, enrichment was ≈15-fold higher in TH2- vs. TH1-primed cells, suggesting that this site might be a developmentally early target of priming condition-specific signals.
Next, to ask whether the enhanced Il4 and decreased Il13 transcriptional potential (relative to 48-h TH2-primed cells) observed in the TH2 clone D10.G4 (Fig. 1B) correlated with changes in AcK9/14 levels, we ChIP-scanned resting D10.G4 cells for AcK9/14. Interestingly, we observed some locations in the Il4–Il13 locus where AcK9/14 levels increased (Fig. 3 B and C; probes K and M) and others where they decreased (Fig. 3 B and C; probes B, C, and N). The most striking increases were at CNS1 (probe K, 4.3-fold) and the Il13 promoter proximal region (probe M, 4.7-fold), whereas decreases >2-fold occurred at HSVa (probe C, 2.5-fold) and the Il13 promoter distal region (probe N, 3.3-fold). AcK9/14 levels at other locations remained fairly constant. To compare the pattern of AcK9/14 with other permissive H3 modifications, we performed ChIP analysis of resting D10.G4 cells with antibodies to 2MeK4 and PhS10. The locus-wide distributions of 2MeK4 and PhS10 were both grossly similar to the distribution of AcK9/14 (Figs. 3C and 5). However, there were two notable differences. First, the 2MeK4 distribution was skewed toward the outer margins of the locus, showing the greatest levels of enrichment at HSV and the Il13 promoter proximal region (Fig. 3C; probes B and M). PhS10, on the other hand, appeared to diminish on the Rad50-side while remaining elevated on the Kif3a side of the locus. These subtle differences in locus-wide distribution suggest that the different permissive H3 modifications, although overlapping perhaps in some functions, may also diverge in others. In addition, the assortment of changes in AcK9/14 levels between 48-h TH2-primed cells and the TH2 clone D10.G4 suggest that the regulatory elements in the Il4–Il13 locus interact in complex ways to achieve specific degrees of Il4 and Il13 transcriptional permissivity.
To test whether the pattern of H3 modification detected in TH2 cells reflects a stable change in the transcriptional state of the locus or a dynamic response to Il4 and Il13 transcriptional activity, we compared AcK9/14 and 2MeK4 ChIP scans of resting and 4-h-activated D10.G4 cells. For both permissive H3 modifications, the average magnitude of activation-induced increases across the Il4–Il13 locus did not exceed 1.4-fold (Fig. 4), whereas, by comparison, Il4 and Il13 transcription increased >1,000-fold (Fig. 1B and data not shown). These results indicate that TH2-dependent permissive H3 modifications at the Il4–Il13 locus reflect the potential for Il4 and Il13 transcription rather than ongoing transcriptional activity itself.
Fig. 4.
Effect of activation on H3 modification at the Il4–Il13 locus of a TH2 clone. Shown are ChIP plots depicting fold-enrichment for AcK9/14 (Upper) and 2MeK4 (Lower) across the Il4–Il13 locus of D10.G4 resting (filled squares) or after 4-h culture with PMA/ionomycin (open squares). Error bars are SEM of n = 2 for activated and n ≥ 3 for resting. Resting data are from Fig. 3C. The top of the figure is as described for Fig. 2.
Discussion
In naïve T cells, only HSV and HSIV contained permissive H3 modifications. Our analysis revealed HSV to be the only region of the Il4–Il13 locus where permissive H3 modification correlated perfectly with the transcriptional states of Il4 and Il13. Thus, in naïve T cells, HSV is likely to play the key role in mediating the transcriptional permissivity of Il4 and Il13. Consistent with this, HSV is the only region of the Il4 locus in naïve CD4+ T cells to exhibit DNA hypomethylation (20). TH2-primed CD4+ T cells homozygous for a deletion encompassing HSV (HSV/Va–/–) exhibit impaired Il4 and Il13 expression (7), suggesting that the role of HSV in mediating Il4 and Il13 transcriptional permissivity may extend beyond the naïve cell stage.
How might activities recruited to HSV act? The unusual occurrence of 2MeK4 at HSV in naïve T cells without accompanying AcK9/14 (Fig. 2A; probe B) may be significant. This 2MeK4+/AcK9/14– signature has been described previously in the context of rearranging antigen-receptor gene segments and was suggested to delineate boundaries separating permissive from repressive chromatin (21). Given its status as the centromeric-most distal HS of the Il4–Il13 locus, it is possible that HSV participates in establishing a similar boundary between alternate chromatin states of Kif3a and Il4. Based on the unperturbed TH2-specific pattern of DNase I hypersensitivity at the Il4 locus of TH2-primed HSV/Va–/– CD4+ T cells, it has been suggested that HSV/Va acts without influencing chromatin structure. However, given that only HSs proximal to HSII were examined (7), it will be necessary to extend analysis of HSV/Va–/– cells to more distal regions including the CNS1 element (a deletion of which also causes impaired Il4 and Il13 expression; ref. 6) before a chromatin structural role for HSV can be ruled out. Although nuclear factor of activated T cells (NFAT) and GATA-3 have been detected by ChIP to bind the neighboring HSVa in activated TH2-primed cells (12, 16), it is not known what factors associate in vivo with HSV in naïve T cells. The occurrence of 2MeK4 and DNA hypomethylation at HSV precede the occurrence of DNase I hypersensitivity which only appears after TH2-priming (4), suggesting that regulatory elements may be activated without the appearance of overt DNase I hypersensitivity. It will be informative to test whether the postulated role of HSV in the maintenance of Il4–Il13 locus transcriptional permissivity is established developmentally or is maintained actively through continuous transduction of tonic signals.
Based on finding similar histone acetylation at the Il4–Il13 locus in 48-h TH1- and TH2-primed cells, it has been suggested that TH-development proceeds in two successive developmental stages (12, 13). The first (before 48 h) is triggered by T cell receptor (TCR) engagement and induces permissive chromatin modifications at the Il4–Il13 locus and transcription of Il4 and Il13. The second (after 48 h) requires cytokine signaling and either stabilizes/enhances (TH2-priming) or reverses (TH1-priming) the changes initiated in stage 1. Our data are inconsistent with this model as we find AcK9/14 levels across the Il4–Il13 locus (Figs. 2B and 3B) as well as Il4 and Il13 expression levels (Fig. 1 A and B) are significantly lower in 48-h-primed cells exposed to TH1- as opposed to TH2-conditions. Thus, by 48 h, cytokine signaling has already had a significant impact on both chromatin structure and gene expression. The divergent results may be due to differences in cell priming conditions or in the exact timing of cell recovery. Alternatively, the qualitative ChIP assay used in previous studies may have minimized differences in levels of chromatin modification in TH1- and TH2-primed cells. In support of the notion that cytokine receptor signals act from the very earliest stages of T cell differentiation, a recent study employing human IL4 receptor transgenic mice showed that effective TH2 differentiation depends on IL4 signals being delivered during the first 48 h of priming (22). Resolution of this issue is significant, as it bears on whether TCR and cytokine receptor signaling pathways act simultaneously or successively in the specification of chromatin states.
Our analysis also shows that TH1-priming-dependent transcriptional repression of Il4 and Il13 is developmentally delayed, requiring >48 h for its establishment. During this period, Il4 and Il13 transcriptional permissivity remain at or above the levels displayed by naïve CD4+ T cells. Transcriptional repression of Il4 and Il13 was only detected in the TH1 clone A.E7 (Fig. 1 A) and correlated with the loss of permissive H3 modifications from the majority of the Il4–Il13 locus except at HSIV. To our knowledge, a developmental delay in transcriptional repression of Il4 and Il13 has not been described, previous studies either focusing on IL4 protein expression or not directly comparing early Il4 mRNA expression between naïve and TH1-primed cells.
Although Il4 and Il13 transcriptional repression was developmentally delayed, we still detected rapid occurrence of TH1-priming-specific effects at the chromatin structural level. As described above, 48 h after the onset of priming, levels of AcK9/14 across the Il4–Il13 locus were lower in cells exposed to TH1 as opposed to TH2 conditions, particularly at HSS3, where levels were reduced >15-fold. Whether, at a time before 48 h, AcK9/14 levels were comparable and then diminished specifically with TH1-priming or, from the outset, increased to a lesser degree with TH1-priming is not clear from our results. Regardless of the precise trajectory of AcK9/14 levels early in the course of TH1 priming, it is clear that the comparatively smaller increase in permissive H3 modifications in 48-h TH1-primed cells was insufficient to support transcriptional repression. Nonetheless, the TH1-priming-specific pattern of permissive H3 modifications observed at 48 h may be necessary for the later establishment of the transcriptionally repressed state.
HSIV is exceptional in that it is the only element at the Il4–Il13 locus where permissive H3 modifications occurred in naïve, TH1, and TH2 cells (Figs. 2, 3, and 5). In addition, levels of AcK9/14 at HSIV increased over the course of both TH1 and TH2 development (Figs. 2 and 3; probe D). Preliminary results indicate that HSIV is free of repressive H3 modifications in both TH1 and TH2 cells, but not naïve CD4+ T cells (A.B., unpublished data). Thus, if we assume that the constellation of H3 modifications at a given regulatory element reflects its activation state, HSIV is likely to be active in both TH1 and TH2 cells, but not in naïve CD4+ T cells. A property shared among TH1 and TH2 clones is irreversibility of Il4 and Il13 transcriptional states (23). Therefore, we speculate that HSIV plays a role in maintaining transcription state stability of Il4 and Il13 in both TH1 and TH2 cells. Evaluation of this hypothesis must await targeted deletion of HSIV. Interestingly, even though DNase I hypersensitivity at HSS3 is reported to be like HSIV (constitutive and shared among naïve, TH1 and TH2 cells) (3), permissive H3 modifications at HSS3 appeared to be TH2-specific (Figs. 2 and 3 B and C; probe L), suggesting a function that is not analogous to HSIV. Alternatively, it is possible that our probe L (although only 370 bp away from HSS3) may be insensitive to highly localized structures associated with HSS3.
Not all regions in the Il4–Il13 locus of TH2-primed cells showed increases in permissive H3 modifications. The exceptions were at the outermost flanks of the locus (Figs. 3 B and C and 5; probes A, O, and P) and regions 5′ and 3′ of Il4 (Figs. 3 B and C and 5; probes E′ and J). These hypomodified regions suggest structural compartmentalization of the locus into three domains: an Il4 regulatory domain (encompassing HSV through HSIV), an Il4 domain (encompassing the Il4 transcription unit through HSO), and an Il13 domain (encompassing CNS1 through HS1). A structural motif comprising a linked trio of HSs, two TH2-specific and the other shared among naïve, TH1 and TH2 cells, occurs twice within the locus: once 3′ of Il4 (HSV/HSVa and HSIV) and then again 3′ of Il13 (HSS1/HSS2 and HSS3). Given this structural parallel, we speculate that there may be an additional domain boundary between HSS3 and the Il13 transcription unit that demarcates an Il13 regulatory domain (encompassing CNS1 through HSS3) from an Il13 domain (encompassing the Il13 transcription unit through HS1). Finer resolution ChIP analysis will be necessary to address this possibility. Transcription of the genes flanking the Il4–Il13 locus (Kif3a and Rad50) is similar in naïve, TH1 and TH2 cells (Fig. 1 A and B). Thus, from the standpoint of ensuring proper regulation of adjacent genes subject to independent transcriptional control, it makes sense that the Il4–Il13 locus should be structurally segregated from the flanking Kif3a and Rad50 genes. However, the function of subdivisions within the Il4–Il13 locus is not immediately obvious. One possibility is that boundaries may be differentially activated at late stages of CD4+ T cell differentiation when, on a given chromosome, Il4 and Il13 can adopt dissimilar transcriptional states (24). Resolution of these issues will be aided by the generation and chromosome-specific analysis of Il4/Il13-expressing T cell clones in which a single gene copy of either Il4 or Il13 is transcriptionally silent.
Our data are consistent with a model of Il4 and Il13 transcriptional state specification that is driven by an ordered sequence of regulatory element activation/de-activation events. At the naïve stage, preactivation of HSV suffices to maintain low levels of Il4 and Il13 transcriptional permissivity. Synergistic activation of the 3′ Il4 and CNS1 enhancers during TH2-priming leads to high level Il4 and Il13 transcriptional permissivity. Temporally delayed deactivation of both the 3′ Il4 and CNS1 enhancers with TH1-priming leads to Il4 and Il13 transcriptional repression. Temporally delayed activation of HSIV “locks in” the transcriptional states of silence and permissivity in TH1 and TH2 cells, respectively. Definitive assessment of this model will require studies of the direct cis effects of specific regulatory element deletions on chromatin structure and transcription as well as identification of factors that mediate the functional effects of each regulatory element.
Supplementary Material
Acknowledgments
We thank Yan Zhang, Jennifer Epler, Thomas Arroll, and Heather Shilling for excellent technical assistance; Mark Groudine and his laboratory for teaching us how to perform ChIP assays; Anton Krumm for thoughtful discussions; and Chris Wilson, Amy Weinmann, Joonsoo Kang, and Heather Shilling for critical reading of the manuscript. M.B. thanks L. J. Bix-Daw and D. E. Daw for their support. This study was supported by grants to M.B. from the National Institutes of Health, the Burroughs Wellcome Foundation, the Cancer Research Institute, the Cystic Fibrosis Foundation, and the Marion Smith Endowment.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ChIP, chromatin immunoprecipitation; TH, T helper; HS, hypersensitive site; PMA, phorbol 12-myristate 13-acetate.
References
- 1.Reiner, S. & Locksley, R. (1995) The Regulation of Immunity to Leishmania Major (Annual Reviews, Palo Alto). [DOI] [PubMed]
- 2.Urban, J. F., Jr., Madden, K. B., Svetic, A., Cheever, A., Trotta, P. P., Gause, W. C., Katona, I. M. & Finkelman, F. D. (1992) Immunol. Rev. 127, 205–220. [DOI] [PubMed] [Google Scholar]
- 3.Takemoto, N., Koyano-Nakagawa, N., Yokota, T., Arai, N., Miyatake, S. & Arai, K. (1998) Int. Immunol. 10, 1981–1985. [DOI] [PubMed] [Google Scholar]
- 4.Agarwal, S. & Rao, A. (1998) Immunity 9, 765–775. [DOI] [PubMed] [Google Scholar]
- 5.Lee, G. R., Fields, P. E., Griffin, T. J. & Flavell, R. A. (2003) Immunity 19, 145–153. [DOI] [PubMed] [Google Scholar]
- 6.Mohrs, M., Blankespoor, C. M., Wang, Z. E., Loots, G. G., Afzal, V., Hadeiba, H., Shinkai, K., Rubin, E. M. & Locksley, R. M. (2001) Nat. Immunol. 2, 842–847. [DOI] [PubMed] [Google Scholar]
- 7.Solymar, D. C., Agarwal, S., Bassing, C. H., Alt, F. W. & Rao, A. (2002) Immunity 17, 41–50. [DOI] [PubMed] [Google Scholar]
- 8.Strahl, B. D. & Allis, C. D. (2000) Nature 403, 41–45. [DOI] [PubMed] [Google Scholar]
- 9.Fischle, W., Wang, Y. & Allis, C. D. (2003) Curr. Opin. Cell Biol. 15, 172–183. [DOI] [PubMed] [Google Scholar]
- 10.Khorasanizadeh, S. (2004) Cell 116, 259–272. [DOI] [PubMed] [Google Scholar]
- 11.Makar, K. W., Perez-Melgosa, M., Shnyreva, M., Weaver, W. M., Fitzpatrick, D. R. & Wilson, C. B. (2003) Nat. Immunol. 4, 1183–1190. [DOI] [PubMed] [Google Scholar]
- 12.Avni, O., Lee, D., Macian, F., Szabo, S. J., Glimcher, L. H. & Rao, A. (2002) Nat. Immunol. 3, 643–651. [DOI] [PubMed] [Google Scholar]
- 13.Fields, P. E., Kim, S. T. & Flavell, R. A. (2002) J. Immunol. 169, 647–650. [DOI] [PubMed] [Google Scholar]
- 14.Kaye, J., Porcelli, S., Tite, J., Jones, B. & Janeway, C. A., Jr. (1983) J. Exp. Med. 158, 836–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecht, T. T., Longo, D. L. & Matis, L. A. (1983) J. Immunol. 131, 1049–1055. [PubMed] [Google Scholar]
- 16.Agarwal, S., Avni, O. & Rao, A. (2000) Immunity 12, 643–652. [DOI] [PubMed] [Google Scholar]
- 17.Loots, G. G., Locksley, R. M., Blankespoor, C. M., Wang, Z. E., Miller, W., Rubin, E. M. & Frazer, K. A. (2000) Science 288, 136–140. [DOI] [PubMed] [Google Scholar]
- 18.Hural, J. A., Kwan, M., Henkel, G., Hock, M. B. & Brown, M. A. (2000) J. Immunol. 165, 3239–3249. [DOI] [PubMed] [Google Scholar]
- 19.Grogan, J. L., Wang, Z. E., Stanley, S., Harmon, B., Loots, G. G., Rubin, E. M. & Locksley, R. M. (2003) J. Immunol. 171, 6672–6679. [DOI] [PubMed] [Google Scholar]
- 20.Lee, D. U., Agarwal, S. & Rao, A. (2002) Immunity 16, 649–660. [DOI] [PubMed] [Google Scholar]
- 21.Morshead, K. B., Ciccone, D. N., Taverna, S. D., Allis, C. D. & Oettinger, M. A. (2003) Proc. Natl. Acad. Sci. USA 100, 11577–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seki, N., Miyazaki, M., Suzuki, W., Hayashi, K., Arima, K., Myburgh, E., Izuhara, K., Brombacher, F. & Kubo, M. (2004) J. Immunol. 172, 6158–6166. [DOI] [PubMed] [Google Scholar]
- 23.Grogan, J. L., Mohrs, M., Harmon, B., Lacy, D. A., Sedat, J. W. & Locksley, R. M. (2001) Immunity 14, 205–215. [DOI] [PubMed] [Google Scholar]
- 24.Kelly, B. L. & Locksley, R. M. (2000) J. Immunol. 165, 2982–2986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.