SUMMARY
Multiple sclerosis (MS) is an inflammatory disease characterized by myelin loss and neuronal dysfunction. Despite the aggregation observed in some families, pathogenic mutations have remained elusive. In this study we describe the identification of NR1H3 p.Arg415Gln in seven MS patients from two multi-incident families presenting severe and progressive disease, with an average age at onset of 34 years. Additionally, association analysis of common variants in NR1H3 identified rs2279238 conferring a 1.35-fold increased risk of developing progressive MS. The p.Arg415Gln position is highly conserved in orthologs and paralogs, and disrupts NR1H3 heterodimerization and transcriptional activation of target genes. Protein expression analysis revealed that mutant NR1H3 (LXRA) alters gene expression profiles, suggesting a disruption in transcriptional regulation as one of the mechanisms underlying MS pathogenesis. Our study indicates that pharmacological activation of LXRA or its targets may lead to effective treatments for the highly debilitating and currently untreatable progressive phase of MS.
INTRODUCTION
Multiple sclerosis (MS) is a chronic inflammatory demyelinating and neurodegenerative disease of the central nervous system (CNS). Immune-mediated destruction of myelin sheaths and oligodendrocytes is considered the primary pathology in MS (Meffre et al., 2015). However, axonal pathology and neuronal loss is responsible for the onset of the progressive phase of the disease and neurological dysfunction (Dutta and Trapp, 2007). MS affects over two million people worldwide, and is more than twice as prevalent in females as males. At disease onset, the majority of patients present relapsing-remitting MS (RRMS) which is characterized by sudden onset of clinical symptoms followed by partial or complete recovery/remission. In contrast, patients suffering from primary progressive MS (PPMS) have an accumulation of irreversible neurological symptoms from clinical onset (Weinshenker et al., 1989).
MS was originally considered an autoimmune disease triggered by exposure to environmental agents; however, family and twin studies have clearly demonstrated the existence of a genetic component implicated in the disease (Fagnani et al., 2015; Sadovnick, 1993). Some genetic risk factors, primarily related to the immune system, have already been identified through association studies (Beecham et al., 2013). However, associated variants have a minor effect on overall disease risk and cannot account for the clustering of biological relatives with MS in families.
The identification of genes and mutations responsible for Mendelian forms of disease provide mechanistic insight into disease ontology, spur the generation of physiologically relevant cellular and animal models, and the development of novel therapeutics to better treat and halt disease progression. To identify pathogenic mutations for MS, we applied exome sequencing analysis to a multi-incident family (MS1) consisting of 11 individuals over three generations, with DNA available for nine family members, including five diagnosed with MS (Figure 1). In addition, we analyzed the functional consequences of the identified variant and its implication in the mechanism of MS pathogenesis.
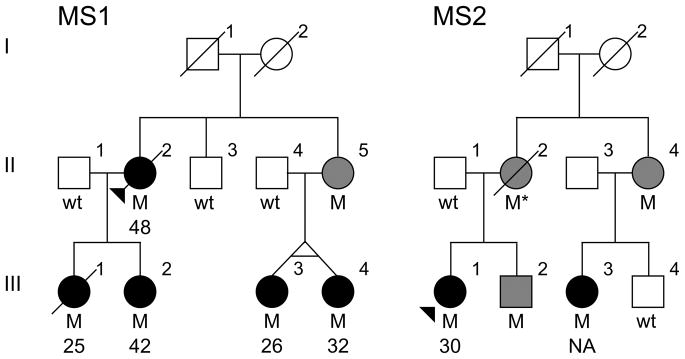
Figure 1.
Simplified pedigree for families presenting the NR1H3 p.Arg415Gln mutation. Males are represented by squares and females by circles, the proband is arrowed and a diagonal line indicates deceased subjects. Patients diagnosed with MS have black filled symbols and mutation carriers of unknown phenotype have grey filled symbols. Both families are of Caucasian descent. Heterozygote mutation carriers (M) with corresponding age at onset of disease and wild-type (wt) genotypes are indicated. An asterisk indicates an inferred mutation carrier. NA; not available.
RESULTS
Identification of NR1H3 p.Arg415Gln in MS patients
Exome sequencing analysis on MS1 III-1 and III-3 (Figure 1) identified 48,333 and 47,681 variants respectively. Of those, 37 missense substitutions with a minor allele frequency (MAF) below 1% in public and proprietary databases of variants were found in both patients (Table S1). Sanger sequencing of amplicons containing these variants in 185 controls and all nine MS1 family members for whom DNA was available resulted in the exclusion of 33 variants. These variants did not segregate with disease (n = 27) or were identified at a frequency over 1% in healthy controls (n = 6). The remaining four variants were genotyped in a case-control series consisting of 2,053 MS patients and 799 unrelated healthy controls; three variants (rs34326043, rs146468598 and rs138130331) were identified at similar frequency in patients and controls and were excluded from further analysis (Table S1). Only one variant, rs61731956 (NM_005693.3; c.1244G>A, p.Arg415Gln) in nuclear receptor subfamily 1, group H, member 3 (NR1H3) was not identified in controls but was found in one additional proband from an unrelated family (MS2; Figure 1). Segregation analysis in MS2 identified the NR1H3 p.Arg415Gln mutation in one additional family member diagnosed with MS, one unaffected sibling and two obligate carriers of unknown affection status. Haplotype analysis of NR1H3 p.Arg415Gln using an Illumina MEGA array and microsatellite markers spanning 3.3Mb between D11S1385 and D11S1326 confirmed a common haplotype in both families (Table S2), indicating that they are likely to descend from a common ancestor in whom the mutation originated. To ensure that additional mutations in this shared haplotype were not overlooked, exons with fewer than 5 exome sequencing reads were analyzed by Sanger sequencing. SNP array data within this haplotype was used to exclude copy number variants as potential disease cause.
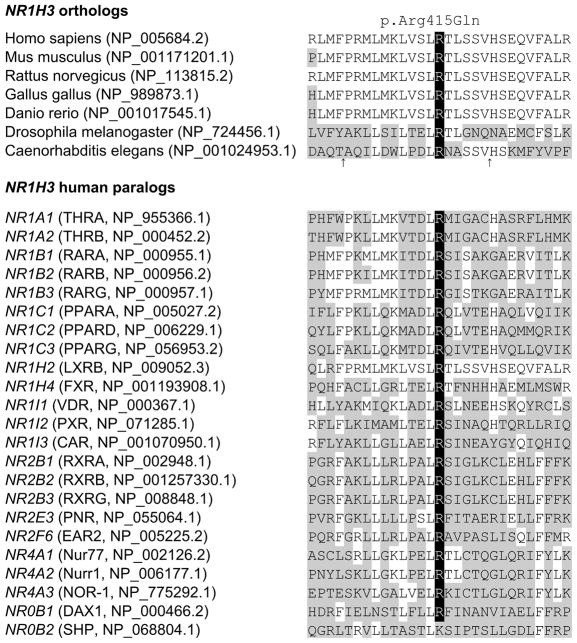
Despite the relatively small size of these two pedigrees, linkage analysis resulted in a maximum logarithm of odds (LOD) score of 2.20 (θ = 0) for NR1H3 p.Arg415Gln, thus highlighting the co-segregation of this mutation with disease, and suggesting this genomic region as the disease locus for these families. Pathogenicity is also supported by a high level of conservation in orthologs and paralogs resulting in a genomic evolutionary rate profiling (GERP) score of 5.13 (Figure 2). Additionally, a glutamine substitution is predicted deleterious on protein structure and function (SIFT = 0.00; Polyphen-2 = 1.00). Despite the linkage, conservation and in-silico predictions, the presence of seemingly unaffected obligate carriers and one male sibling at age 55 indicates that the penetrance of the mutation is incomplete, and additional genetic or environmental factors may be required for the onset of MS.
Figure 2.
NR1H3 p.Arg415Gln conservation in orthologs and human paralogs. Protein homologs were aligned via ClustalO. Amino acid position for NR1H3 p.Arg415Gln is highlighted in black. Protein homologs with amino acid positions differing from those of the human NR1H3 sequence are indicated in gray. RefSeq accession numbers is provided for orthologs, and gene, protein and RefSeq accession numbers for paralogs. An arrow indicates the exclusion of two or three amino acids from Caenorhabditis elegans at these positions.
Clinical Phenotype
Age at onset of disease for MS1 NR1H3 p.Arg415Gln carriers ranges between 25 and 48 years, with an average age of 34.6 ± 10.1 years (SD) (Table S3). Detailed clinical information was available for three family members (II-2, III-1 and III-2; Figure 1). Clinical course at the onset of disease for II-2 and III-1 was consistent with PPMS. Although III-2 was initially diagnosed with RRMS, her disease became progressive within three years of clinical onset. The disease progression and severity for NR1H3 mutation carriers is quite acute. II-2 presented an expanded disability status scale (EDSS) score of 7 after six years and was deceased at age 76, 28 years after the onset of clinical symptoms. III-1 was paraplegic 16 years after onset with an EDSS score of 8 and died at age 44. The EDSS score for III-2 was 5.5 two years after the onset of disease, and 6.5 at year four. Detailed clinical information was also available for MS2 III-1. The clinical phenotype was similar to that of MS patients from MS1 as she was diagnosed with PPMS at age 30. Her EDSS was 5.5 at age 42 (12 years after onset). Clinical evaluations for seemingly healthy mutation carriers were not available, therefore we cannot exclude the possibility that these family members developed MS at a later age or present a forme fruste not clinically ascribed to MS, such as radiologically isolated syndrome (Okuda et al., 2009).
Genetic Analysis of NR1H3
To assess whether additional pathogenic mutations in NR1H3 exist, we sequenced all coding exons in 836 probands diagnosed with PPMS or from multi-incident families. This analysis identified three previously reported silent changes (rs2279238, p.Ser99Ser; rs145536921, p.Gly137Gly; and rs141865811, p.Pro276Pro) and two missense variants (rs778927489, p.Pro199His; and rs149895806, p.Ser440Cys) in three probands. Segregation analyses of p.Pro199His and p.Ser440Cys do not support a role in disease for either variant (Figure S1).
To evaluate whether common variants in NR1H3 influence disease risk we genotyped five tagging SNPs (tSNPs) in a MS case-control series consisting of 2,053 MS patients and 799 healthy controls. None of the variants analyzed was found to significantly associate with disease risk after correction for multiple testing (p > 0.01; Table 1). Since the clinical phenotype identified in MS1 and MS2 was consistent with progressive MS, we performed stratified association analysis by clinical course. Detailed clinical information was available for 1,687 patients, with the majority presenting a clinical phenotype consistent with RRMS (74.9%) at the onset of disease; the remainder were diagnosed with PPMS (25.1%). Analysis of RRMS patients failed to identify an association with NR1H3 tSNPs. However, a significant association between rs2279238 and PPMS was observed (p = 0.004), with carriers of the minor allele presenting a 1.35-fold increased likelihood of developing progressive MS (Table 1). Albeit not statistically significant, a trend towards association in PPMS patients was also observed for rs11039149 (p = 0.060; OR = 1.32), rs7120118 (p = 0.047, OR = 1.21) and rs7114704 (p = 0.020, OR = 1.59).
Table 1.
Genotype frequencies and statistical analysis for NR1H3 tSNPs. Significant associations after correction for multiple testing (p < 0.01) are indicated in bold. MS, multiple sclerosis; RR, relapsing-remitting; PP, primary progressive; OR, odds ratio; CI, confidence interval.
| dbSNP | Genotypes | Groups | n (%) | p-value | OR (95% CI) | ||
|---|---|---|---|---|---|---|---|
| rs11039149 | AA/AG/GG | Control | 420 (0.53) | 317 (0.40) | 51 (0.06) | ||
| MS | 1029 (0.50) | 833 (0.41) | 178 (0.09) | 0.103 | 1.12 (0.95–1.32) | ||
| RR | 657 (0.52) | 495 (0.39) | 105 (0.08) | 0.301 | 1.04 (0.87–1.25) | ||
| PP | 194 (0.46) | 197 (0.47) | 28 (0.07) | 0.060 | 1.32 (1.04–1.68) | ||
|
| |||||||
| rs12221497 | GG/GA/AA | Control | 583 (0.73) | 197 (0.25) | 16 (0.02) | ||
| MS | 1508 (0.74) | 479 (0.24) | 43 (0.02) | 0.814 | 0.95 (0.79–1.14) | ||
| RR | 909 (0.73) | 310 (0.25) | 30 (0.02) | 0.862 | 1.02 (0.84–1.25) | ||
| PP | 318 (0.76) | 94 (0.22) | 8 (0.02) | 0.659 | 0.88 (0.67–1.15) | ||
|
| |||||||
| rs2279238 | CC/CT/TT | Control | 553 (0.71) | 197 (0.25) | 26 (0.03) | ||
| MS | 1422 (0.70) | 567 (0.28) | 48 (0.02) | 0.170 | 1.07 (0.89–1.29) | ||
| RR | 898 (0.72) | 325 (0.26) | 31 (0.02) | 0.499 | 0.98 (0.81–1.20) | ||
| PP | 272 (0.65) | 141 (0.34) | 7 (0.02) | 0.004 | 1.35 (1.05–1.74) | ||
|
| |||||||
| rs7120118 | TT/TC/CC | Control | 386 (0.50) | 302 (0.39) | 83 (0.11) | ||
| MS | 995 (0.49) | 839 (0.41) | 194 (0.10) | 0.444 | 1.04 (0.88–1.23) | ||
| RR | 620 (0.50) | 496 (0.40) | 129 (0.10) | 0.931 | 1.01 (0.84–1.21) | ||
| PP | 190 (0.45) | 194 (0.46) | 35 (0.08) | 0.047 | 1.21 (0.95–1.53) | ||
|
| |||||||
| rs7114704 | CC/CT/TT | Control | 722 (0.92) | 67 (0.08) | 0 (0.00) | ||
| MS | 1828 (0.90) | 197 (0.10) | 13 (0.01) | 0.033 | 1.24 (0.93–1.65) | ||
| RR | 1138 (0.91) | 108 (0.09) | 9 (0.01) | 0.045 | 1.11 (0.81–1.52) | ||
| PP | 367 (0.87) | 54 (0.13) | 0 (0.00) | 0.020 | 1.59 (1.08–2.32) | ||
Dimerization analysis and transcriptional regulation of NR1H3 target genes
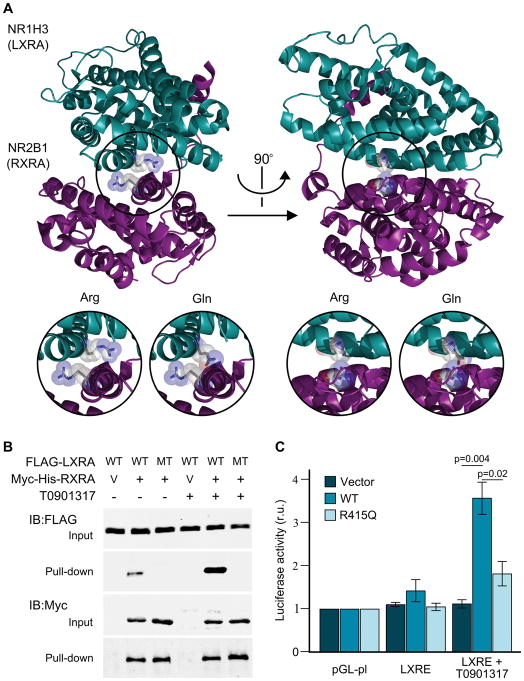
NR1H3 encodes liver X receptor alpha (LXRA) which forms obligate heterodimer complexes with retinoid X receptors (RXR) (Edwards et al., 2002). The p.Arg415Gln mutation identified in MS families is located on the surface of the heterodimerization interface on the ligand binding domain (LBD) (Figure 3A). The importance of this residue on the dimerization process with retinoid X receptor alpha (RXRA) was previously shown with an artificial p.Arg415Ala mutant, which resulted in a complete loss of function (Shulman et al., 2004). In addition, this position is homologous to p.Arg391Cys/Ser mutations in vitamin D receptor (VDR) (Figure 2), which have been shown to cause vitamin D-resistant rickets by destabilizing the dimerization with RXRA (Nguyen et al., 2006; Whitfield et al., 1996). To evaluate whether the LXRA p.Arg415Gln mutation has a similar detrimental effect on the dimerization with RXRA, we performed binding assays with wild-type and mutant LXRA. Pull-down studies corroborated the interaction between RXRA and wild-type LXRA; however, heterodimerization between RXRA and LXRA p.Arg415Gln was not detected (Figure 3B). Even after the addition of LXRA (T0901317) or RXRA (LG268) synthetic agonists, which strengthen the interaction between wild-type LXRA and RXRA, dimerization with mutant LXRA was not observed (Figure 3B and S2). RXRA-LXRA heterodimers bind to LXR response elements (LXRE) regulating gene expression (Lefebvre et al., 2010). Luciferase reporter assays indicate that deficient dimer formation results in the dysregulation of transcriptional activation of LXR target genes (Figure 3C). Under basal conditions, LXRE-containing reporter plasmids did not show differences in transcriptional activation between constructs (ANOVA p = 0.33, F2,9 = 1.26); however, the activation of LXRA with T0901317 led to a 3.1-fold increased expression for wild-type LXRA (ANOVA p = 0.01, F2,9 = 7.82; Fisher’s p = 0.004) whereas a non-significant increase was observed for LXRA p.Arg415Gln (Fisher’s p = 0.30), indicating that the mutation identified in LXRA results in a loss of function and impaired transcriptional regulation (Figure 3C). To assess the effect of p.Arg415Gln mutation on the expressions of endogenous LXRA target genes, wild-type and mutant LXRA were introduced into neuroblastoma SH-SY5Y cells. ATP-binding cassette, subfamily A, member 1 (ABCA1), a well-established LXRA target (Ghisletti et al., 2007), displayed ~3.3-fold increased expression in cells transfected with wild-type LXRA compared to vector or p.Arg415Gln LXRA (Figure S3). Interestingly, ABCA1 expression was comparable in cells transfected with vector, wild-type or mutant LXRA after the addition of T0901317; suggesting that endogenous LXRA or liver X receptor beta (LXRB) are sufficient to fully induce ABCA1 expression.
Figure 3.
Structural and functional analysis of NR1H3. A) Crystal structure of NR1H3 (LXRA) and NR2B1 (RXRA) heterodimer showing the highly conserved arginine residues in their corresponding ligand binding domain was generated with PyMol (PDB ID: 2ACL). Inserts provide a detailed view of the LXRA wild-type (Arg) and mutated (Gln) protein structure. B) FLAG-LXRA wild type (WT) and p.Arg415Gln mutant (R415Q) were co-expressed with RXRA-mycHis (+) or empty vector pcDNA4-myc-his-A (V) in HEK cells with or without 10μM LXRA agonist T0901317 overnight. RXRA was pulled down from the lysates with nickel-charged magnetic beads. Cell lysates and precipitates were immunoblotted with FLAG or Myc antibodies. C) Transcriptional activation of control (pGL-pl) and LXR response element-containing plasmids (LXRE) after co-transfection with empty vector, wild-type (WT) or mutant LXRA (R415Q). Firefly/Renilla luciferase activity (mean ± standard error) for the different constructs, with and without the presence of 10μM of a LXRA agonist (T0901317) normalized to pGL-pl under the same conditions are provided. Statistically significant Fisher’s Least Significant Difference (LSD) post hoc values are provided. r.u., relative units.
In addition to its role in transcriptional activation, LXRA also acts as a negative regulator of gene expression in a SUMOylation-dependent mechanism (Ghisletti et al., 2007). To assess whether LXRA p.Arg415Gln disrupts transrepression, nitric oxide synthase 2 (NOS2) was quantified in microglial BV2 cells treated with lipopolysaccharide (LPS). Under basal conditions, both wild-type and mutant LXRA transfections resulted in a similar reduction in NOS2 compared to control (Figure S3). Similarly, no differences in SUMOylation were observed between wild-type and mutant LXRA (Figure S4). The addition of T0901317 was shown to enhance transrepression of NOS2, even in cells transfected with empty vector, thus suggesting that the endogenous LXRA or LXRB are sufficient to abolish NOS2 gene expression. Taken together, this data indicates that the transrepression mechanism of LXRA is likely unaffected by the p.Arg415Gln mutation.
DISCUSSION
The application of exome sequencing analysis in a family presenting a Mendelian form of MS has led to the identification of a pathogenic mutation in NR1H3. The p.Arg415Gln substitution was identified in seven MS patients from two unrelated families sharing a common disease haplotype (Table S2). The clinical phenotype observed in these patients is consistent with an unusual form of MS with a RR onset that rapidly becomes progressive or PPMS. Although the mutation was not observed in our healthy controls, it has been described in 21 individuals from the ExAC database (MAF = 0.0002), as well as three obligate carriers and an unaffected biological family member from this study, thus suggesting that additional genetic, epigenetic or environmental factors may be necessary to develop disease. Therefore these families appear to have a highly susceptible NR1H3 genetic background for MS, but an initial damage may be necessary to trigger the onset of disease. This incomplete penetrance is not unexpected for a highly heterogeneous and multifactorial disease such as MS.
NR1H3, also known as LXRA, together with LXRB form a subfamily of nuclear receptors which control transcriptional regulation of genes involved in lipid homeostasis, inflammation and innate immunity (Joseph et al., 2003; Wang et al., 2002). The basic structure of these nuclear receptors consists of two activation domains (AF1 and AF2), a DNA binding domain, and a LBD responsible for dimerization (Edwards et al., 2002). The LXRA p.Arg415Gln mutation identified in MS families is located in a highly conserved amino acid of its LBD; which is not only conserved through speciation, but also present in the LBD of RXRs and LXRA nuclear receptor paralogs engaging in physical and functional interactions with RXRs (Figure 2) (Lefebvre et al., 2010). The crystal structure of RXRA-bound LXRA has been resolved (PDB ID: 2ACL) (Jaye et al., 2005), and the introduction of the p.Arg415Gln mutation using PyMol highlights the importance of this positively charged residue, which is located in the center of this heterodimeric complex and directly opposed to the homologous arginine on RXRs (Figure 3). Although LXRA and LXRB share significant homology (62.4% identity, 78.5% similar), analysis of LXRA knock-out mice indicates that LXRB cannot compensate for its loss (Peet et al., 1998). The activation of LXRs is dependent on the entry of naturally occurring oxysterols, reactive molecules generated from the oxidation of cholesterol, into the ligand-binding pocket of the LBD and dimerization with RXR. This activated complex binds to cis regulatory LXRE and regulates the expression of genes that control transport, catabolism, and elimination of cholesterol (Wang et al., 2002).
Cholesterol is an essential component of the CNS structure and function, and plays a key role in signal transduction, synaptogenesis and neurotransmitter release (van de Kraats et al., 2014). Cholesterol synthesis and transport in oligodendrocytes is essential for optimal myelination and remyelination, and defective homeostasis has been implicated in several neurological diseases, including MS (Chalmin et al., 2015; Leoni and Caccia, 2011).
Neuropathological examination of LXRA-LXRB double knockout mice has shown several brain abnormalities, including axonal atrophy, neuronal loss, astrogliosis, and lipid accumulation (Wang et al., 2002). These animals also present a significantly increased expression of myelin protein zero (MPZ) and peripheral myelin protein-22 (PMP22) transcripts but a decrease in myelin proteins, resulting in thinner and disorganized myelin sheaths as well as deficient remyelination (Makoukji et al., 2011; Meffre et al., 2015). The study of Schwann cell lines has shown that the downregulation of peripheral myelin genes is mediated by LXRs and oxysterols, and suggests that the LXR pathway has a critical role in myelination by repressing the basal activity of myelin genes and regulating cholesterol levels in peripheral nerves (Makoukji et al., 2011; Shackleford et al., 2013).
LXRs and their ligands are also important negative regulators of inflammatory gene expression and the innate immune response. LXRs regulate inflammation by inhibiting the expression of inflammatory mediators including interleukin-6, nitric oxide synthase and cyclooxygenase 2 (Bensinger et al., 2008; Joseph et al., 2003). Mechanistically, the anti-inflammatory effects of LXRs have been attributed to the inhibition of nuclear factor kappa-B and STAT1-mediated signaling pathways via transrepression (Glass and Saijo, 2010; Li et al., 2011). LXRA-deficient mice have been shown to be more susceptible to bacterial infection due to accelerated macrophage apoptosis and a reduced immune response (Joseph et al., 2004). In MS patients, foamy macrophages and microglia containing myelin are abundant in active lesions, and this internalized myelin has been suggested as a LXR-RXR selective agonist which activates LXRs and suppress the secretion of pro-inflammatory cytokines (Bogie et al., 2012). Although our results indicate that the p.Arg415Gln mutation identified in MS patients does not have an effect in LXRA-dependent transrepression (Figure S3); decreased auto-activation of NR1H3, one of the primary targets of LXRA (Pehkonen et al., 2012), may lead to an indirect reduction in transrepression. To further elucidate LXRA p.Arg415Gln biological outcomes and to discriminate between direct and indirect effects, analysis of patient derived cells and the characterization of knock-in cellular and animal models are necessary.
We hypothesize that at the outset of disease, damage to myelin sheaths in NR1H3 mutation carriers may produce an intensified inflammatory response due to the inability of the innate immune system to suppress the expression of pro-inflammatory mediators. In addition, these patients may present impaired remyelination of damaged axons due to the dysregulation of peripheral myelin genes resulting in chronic demyelination, neuronal loss and progressive disease. This hypothesis is supported by studies in experimental autoimmune encephalomyelitis, which have shown that mice treated with LXR agonists have reduced clinical symptoms, cellular inflammation and demyelination; whereas LXR deficiency results in exacerbated disease (Cui et al., 2011; Hindinger et al., 2006).
Pathogenic mutations in NR1H3 appear to be rare, and are unlikely to account for disease in more than one in a thousand MS patients. However, the significant association between a common NR1H3 variant in patients with PPMS (Table 1), as well as the decreased mRNA expression observed in MS patients (Liu et al., 2005), suggests that the LXRA signaling pathway is not only important for Mendelian forms of MS, but also a large portion of patients without a positive family history. Although genetic and functional evidence supports a role for NR1H3 in the pathogenesis of MS, it should be noted that exome sequencing technologies do not efficiently sequence all target regions, and cannot identify copy number variants. In addition, a change in functional readout from a mutated protein does not ensure a pathogenic outcome. Therefore replication of our findings and evaluation of NR1H3 variants in additional cohorts is warranted to confirm a role in the onset of MS.
Despite the rarity of pathogenic mutations, they provide a better understanding of the molecular mechanisms implicated in the onset of MS and its clinical course. They are also instrumental for the generation of physiologically relevant cellular and animal models in which to study the disease pathways and develop and assess therapeutic options. The central role of LXRs in the regulation of cholesterol homeostasis has already triggered a growing interest in LXR synthetic agonists for the treatment of atherosclerosis, coronary heart disease, diabetes and neurodegenerative diseases amongst others (Xu et al., 2013). Although the therapeutic use of these agonists has been hampered by serious adverse effects, agonists with a more finite biological effects are under development (Temml et al., 2014). The implementation of cell-specific drug delivery and release systems, and targeting the biological pathways disrupted specifically in NR1H3 patients, may overcome some of these adverse effects and pave the way for new MS treatments. The implication of the LXRA signaling pathway in MS, and the p.Arg415Gln mutation, has now opened novel therapeutic avenues for the currently untreatable progressive phase of the disease. Novel medications based on NR1H3 models are expected to provide symptomatic relief and halt disease progression by reducing the inflammatory response and promoting remyelination.
EXPERIMENTAL PROCEDURES
Participants
Multi-incident families and a case-control series consisting of 2,053 MS patients and 799 unrelated healthy control subjects from Canada were included in this study. All samples were collected through the longitudinal Canadian Collaborative Project on the Genetic Susceptibility to Multiple Sclerosis (CCPGSMS) (Sadovnick et al., 1998; Traboulsee et al., 2014). The ethical review board at the University of British Columbia approved the study, and all participants provided informed consent.
Genetic Analysis
We performed exome sequencing in two patients using Ion AmpliSeq exome kits and an Ion Proton Sequencer. Sanger sequencing was used to genotype amplicons containing exome variants of interest and NR1H3 coding exons. Variants segregating with disease and tSNPs were genotyped in MS patients and controls using TaqMan probes. Statistical association was determined using Fisher’s exact test, with p < 0.01 considered significant after Bonferroni correction. The disease haplotype was resolved using an Illumina MEGA array and microsatellite markers between D11S1385 and D11S1326.
Functional Analysis
HEK293 cells were co-transfected with RXRA and wild-type or mutant LXRA, and treated overnight with T0901317 when indicated. Cell lysates and precipitates were immunoblotted with FLAG or Myc antibodies. Luciferase reporter assays were used to evaluate transcriptional activation by wild-type and mutant LXRA. Cells were co-transfected with reporter plasmid containing a LXR response element (LXRE) or an empty control plasmid lacking LXRE (pGL-pl) (Cai et al., 2008), and wild-type LXRA, mutant LXRA, or empty vector. The Dual-luciferase reporter assays were performed and the luciferase activity was measured to reflect the effect on transcription activation. Protein expression analysis for ABCA1 and NOS2 were performed in SH-SY5Y cells following transfection with vector, wild-type LXRA, or p.Arg415Gln LXRA in the presence or absence of LXRA agonist T0901317.
Detailed patient demographics and comprehensive methodology are provided in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We are grateful to all individuals who generously participated in this study. We thank Kevin Atkins for data collection and extraction, and physicians who participated in the CCPGSMS. This research was undertaken thanks to funding from the Canada Research Chair and Canada Excellence Research Chair programs, Canadian Institute of Health Research, Vancouver Coastal Health Research Institute, the Milan & Maureen Ilich Foundation, and the Vancouver Foundation. W.S. is the holder of the Tier 1 Canada Research Chair in Alzheimer’s Disease. Collection of clinical information and DNA samples was funded by the MS Society of Canada Scientific Research Foundation as part of the CCPGSMS. The authors report no conflict of interest.
- In memory of José María Vilariño Vilariño.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, Supervision and Funding Acquisition, CV-G, ADS, ALT and WS; Investigation and Data Curation, ZW, JPR, CQB, ME, IMY, MdL, TG, JDL, SZ, GW and CJR; Writing - Original draft, CV-G; Writing - Review & Editing, all authors.
References
- Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, Cotsapas C, Shah TS, Spencer C, Booth D, Goris A, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45:1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensinger SJ, Bradley MN, Joseph SB, Zelcer N, Janssen EM, Hausner MA, Shih R, Parks JS, Edwards PA, Jamieson BD, Tontonoz P. LXR signaling couples sterol metabolism to proliferation in the acquired immune response. Cell. 2008;134:97–111. doi: 10.1016/j.cell.2008.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogie JF, Timmermans S, Huynh-Thu VA, Irrthum A, Smeets HJ, Gustafsson JA, Steffensen KR, Mulder M, Stinissen P, Hellings N, Hendriks JJ. Myelin-derived lipids modulate macrophage activity by liver X receptor activation. PLoS One. 2012;7:e44998. doi: 10.1371/journal.pone.0044998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Chen B, Zhou W, Zis O, Liu S, Holt RA, Honer WG, Song W. SP1 regulates a human SNAP-25 gene expression. J Neurochem. 2008;105:512–523. doi: 10.1111/j.1471-4159.2007.05167.x. [DOI] [PubMed] [Google Scholar]
- Chalmin F, Rochemont V, Lippens C, Clottu A, Sailer AW, Merkler D, Hugues S, Pot C. Oxysterols regulate encephalitogenic CD4(+) T cell trafficking during central nervous system autoimmunity. J Autoimmun. 2015;56:45–55. doi: 10.1016/j.jaut.2014.10.001. [DOI] [PubMed] [Google Scholar]
- Cui G, Qin X, Wu L, Zhang Y, Sheng X, Yu Q, Sheng H, Xi B, Zhang JZ, Zang YQ. Liver X receptor (LXR) mediates negative regulation of mouse and human Th17 differentiation. J Clin Invest. 2011;121:658–670. doi: 10.1172/JCI42974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta R, Trapp BD. Pathogenesis of axonal and neuronal damage in multiple sclerosis. Neurology. 2007;68:S22–31. doi: 10.1212/01.wnl.0000275229.13012.32. discussion S43–54. [DOI] [PubMed] [Google Scholar]
- Edwards PA, Kast HR, Anisfeld AM. BAREing it all: the adoption of LXR and FXR and their roles in lipid homeostasis. J Lipid Res. 2002;43:2–12. [PubMed] [Google Scholar]
- Fagnani C, Neale MC, Nistico L, Stazi MA, Ricigliano VA, Buscarinu MC, Salvetti M, Ristori G. Twin studies in multiple sclerosis: A meta-estimation of heritability and environmentality. Mult Scler. 2015;21:1404–1413. doi: 10.1177/1352458514564492. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Ogawa S, Pascual G, Lin ME, Willson TM, Rosenfeld MG, Glass CK. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- Hindinger C, Hinton DR, Kirwin SJ, Atkinson RD, Burnett ME, Bergmann CC, Stohlman SA. Liver X receptor activation decreases the severity of experimental autoimmune encephalomyelitis. J Neurosci Res. 2006;84:1225–1234. doi: 10.1002/jnr.21038. [DOI] [PubMed] [Google Scholar]
- Jaye MC, Krawiec JA, Campobasso N, Smallwood A, Qiu C, Lu Q, Kerrigan JJ, De Los Frailes Alvaro M, Laffitte B, Liu WS, et al. Discovery of substituted maleimides as liver X receptor agonists and determination of a ligand-bound crystal structure. J Med Chem. 2005;48:5419–5422. doi: 10.1021/jm050532w. [DOI] [PubMed] [Google Scholar]
- Joseph SB, Bradley MN, Castrillo A, Bruhn KW, Mak PA, Pei L, Hogenesch J, O’Connell RM, Cheng G, Saez E, et al. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell. 2004;119:299–309. doi: 10.1016/j.cell.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- Lefebvre P, Benomar Y, Staels B. Retinoid X receptors: common heterodimerization partners with distinct functions. Trends Endocrinol Metab. 2010;21:676–683. doi: 10.1016/j.tem.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Leoni V, Caccia C. Oxysterols as biomarkers in neurodegenerative diseases. Chem Phys Lipids. 2011;164:515–524. doi: 10.1016/j.chemphyslip.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Li N, Salter RC, Ramji DP. Molecular mechanisms underlying the inhibition of IFN-gamma-induced, STAT1-mediated gene transcription in human macrophages by simvastatin and agonists of PPARs and LXRs. J Cell Biochem. 2011;112:675–683. doi: 10.1002/jcb.22976. [DOI] [PubMed] [Google Scholar]
- Liu X, Steffensen KR, Sanna A, Arru G, Fois ML, Rosati G, Sotgiu S, Link H, Gustafsson JA, Huang YM. Anti-inflammatory nuclear receptor superfamily in multiple sclerosis patients from Sardinia and Sweden. Neurobiol Dis. 2005;20:961–968. doi: 10.1016/j.nbd.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Makoukji J, Shackleford G, Meffre D, Grenier J, Liere P, Lobaccaro JM, Schumacher M, Massaad C. Interplay between LXR and Wnt/beta-catenin signaling in the negative regulation of peripheral myelin genes by oxysterols. J Neurosci. 2011;31:9620–9629. doi: 10.1523/JNEUROSCI.0761-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffre D, Shackleford G, Hichor M, Gorgievski V, Tzavara ET, Trousson A, Ghoumari AM, Deboux C, Nait Oumesmar B, Liere P, et al. Liver X receptors alpha and beta promote myelination and remyelination in the cerebellum. Proc Natl Acad Sci U S A. 2015;112:7587–7592. doi: 10.1073/pnas.1424951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M, d’Alesio A, Pascussi JM, Kumar R, Griffin MD, Dong X, Guillozo H, Rizk-Rabin M, Sinding C, Bougneres P, et al. Vitamin D-resistant rickets and type 1 diabetes in a child with compound heterozygous mutations of the vitamin D receptor (L263R and R391S): dissociated responses of the CYP-24 and rel-B promoters to 1,25-dihydroxyvitamin D3. J Bone Miner Res. 2006;21:886–894. doi: 10.1359/jbmr.060307. [DOI] [PubMed] [Google Scholar]
- Okuda DT, Mowry EM, Beheshtian A, Waubant E, Baranzini SE, Goodin DS, Hauser SL, Pelletier D. Incidental MRI anomalies suggestive of multiple sclerosis: the radiologically isolated syndrome. Neurology. 2009;72:800–805. doi: 10.1212/01.wnl.0000335764.14513.1a. [DOI] [PubMed] [Google Scholar]
- Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, Mangelsdorf DJ. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- Pehkonen P, Welter-Stahl L, Diwo J, Ryynanen J, Wienecke-Baldacchino A, Heikkinen S, Treuter E, Steffensen KR, Carlberg C. Genome-wide landscape of liver X receptor chromatin binding and gene regulation in human macrophages. BMC Genomics. 2012;13:50. doi: 10.1186/1471-2164-13-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadovnick AD. Familial recurrence risks and inheritance of multiple sclerosis. Curr Opin Neurol Neurosurg. 1993;6:189–194. [PubMed] [Google Scholar]
- Sadovnick AD, Risch NJ, Ebers GC. Canadian collaborative project on genetic susceptibility to MS, phase 2: rationale and method. Canadian Collaborative Study Group. Can J Neurol Sci. 1998;25:216–221. doi: 10.1017/s0317167100034041. [DOI] [PubMed] [Google Scholar]
- Shackleford G, Makoukji J, Grenier J, Liere P, Meffre D, Massaad C. Differential regulation of Wnt/beta-catenin signaling by Liver X Receptors in Schwann cells and oligodendrocytes. Biochem Pharmacol. 2013;86:106–114. doi: 10.1016/j.bcp.2013.02.036. [DOI] [PubMed] [Google Scholar]
- Shulman AI, Larson C, Mangelsdorf DJ, Ranganathan R. Structural determinants of allosteric ligand activation in RXR heterodimers. Cell. 2004;116:417–429. doi: 10.1016/s0092-8674(04)00119-9. [DOI] [PubMed] [Google Scholar]
- Temml V, Voss CV, Dirsch VM, Schuster D. Discovery of new liver X receptor agonists by pharmacophore modeling and shape-based virtual screening. J Chem Inf Model. 2014;54:367–371. doi: 10.1021/ci400682b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traboulsee AL, Bernales CQ, Ross JP, Lee JD, Sadovnick AD, Vilarino-Guell C. Genetic variants in IL2RA and IL7R affect multiple sclerosis disease risk and progression. Neurogenetics. 2014;15:165–169. doi: 10.1007/s10048-014-0403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Kraats C, Killestein J, Popescu V, Rijkers E, Vrenken H, Lutjohann D, Barkhof F, Polman CH, Teunissen CE. Oxysterols and cholesterol precursors correlate to magnetic resonance imaging measures of neurodegeneration in multiple sclerosis. Mult Scler. 2014;20:412–417. doi: 10.1177/1352458513499421. [DOI] [PubMed] [Google Scholar]
- Wang L, Schuster GU, Hultenby K, Zhang Q, Andersson S, Gustafsson JA. Liver X receptors in the central nervous system: from lipid homeostasis to neuronal degeneration. Proc Natl Acad Sci U S A. 2002;99:13878–13883. doi: 10.1073/pnas.172510899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshenker BG, Bass B, Rice GP, Noseworthy J, Carriere W, Baskerville J, Ebers GC. The natural history of multiple sclerosis: a geographically based study. I. Clinical course and disability. Brain. 1989;112( Pt 1):133–146. doi: 10.1093/brain/112.1.133. [DOI] [PubMed] [Google Scholar]
- Whitfield GK, Selznick SH, Haussler CA, Hsieh JC, Galligan MA, Jurutka PW, Thompson PD, Lee SM, Zerwekh JE, Haussler MR. Vitamin D receptors from patients with resistance to 1,25-dihydroxyvitamin D3: point mutations confer reduced transactivation in response to ligand and impaired interaction with the retinoid X receptor heterodimeric partner. Mol Endocrinol. 1996;10:1617–1631. doi: 10.1210/mend.10.12.8961271. [DOI] [PubMed] [Google Scholar]
- Xu P, Li D, Tang X, Bao X, Huang J, Tang Y, Yang Y, Xu H, Fan X. LXR agonists: new potential therapeutic drug for neurodegenerative diseases. Mol Neurobiol. 2013;48:715–728. doi: 10.1007/s12035-013-8461-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.