Abstract
Metastasis to secondary sites such as the lung, liver and bone is a traumatic event with a mortality rate of approximately 90% 1. Of these sites, the lung is the most difficult to assess using intravital optical imaging due to its enclosed position within the body, delicate nature and vital role in sustaining proper physiology. While clinical modalities (positron emission tomography (PET), magnetic resonance imaging (MRI) and computed tomography (CT)) are capable of providing noninvasive images of this tissue, they lack the resolution necessary to visualize the earliest seeding events, with a single pixel consisting of nearly a thousand cells. Current models of metastatic lung seeding postulate that events just after a tumor cell's arrival are deterministic for survival and subsequent growth. This means that real-time intravital imaging tools with single cell resolution 2 are required in order to define the phenotypes of the seeding cells and test these models. While high resolution optical imaging of the lung has been performed using various ex vivo preparations, these experiments are typically single time-point assays and are susceptible to artifacts and possible erroneous conclusions due to the dramatically altered environment (temperature, profusion, cytokines, etc.) resulting from removal from the chest cavity and circulatory system 3. Recent work has shown that time-lapse intravital optical imaging of the intact lung is possible using a vacuum stabilized imaging window 2,4,5 however, typical imaging times have been limited to approximately 6 hr. Here we describe a protocol for performing long-term intravital time-lapse imaging of the lung utilizing such a window over a period of 12 hr. The time-lapse image sequences obtained using this method enable visualization and quantitation of cell-cell interactions, membrane dynamics and vascular perfusion in the lung. We further describe an image processing technique that gives an unprecedentedly clear view of the lung microvasculature.
Keywords: Cancer Research, Issue 116, Intravital imaging, vacuum window, multiphoton microscopy, lung, time-lapse, cancer biology
Introduction
High resolution intravital optical imaging has proven to be crucial to understanding many biological processes, allowing single-cell and sub-cellular parameters to be measured and quantified. In cancer research, intravital imaging of tumor and stromal cells has led to the discovery of many microenvironmental interactions 6-11 that are only present in the intact animal.
Discoveries about microenvironments associated with intravasation and dissemination of tumor cells in breast cancer using single cell resolution optical imaging in vivo has even led to novel markers for prognosis and response to treatment in breast cancer patients 12-16. The best imaging technologies available for viewing deep within intact internal vital organs are the clinical modalities (MRI, PET, CT) which offer excellent views of the entire organ and can reveal pathologies even before they produce clinical symptoms. They are unable, however, to reveal the earliest stages of metastasis and the cellular mechanisms driving tumor progression due to their lack of single cell resolution. By the time lung metastases are visible in these modalities, they are well established and proliferating. Given the estimate that 90% of disseminated tumor cells that arrive to the lung either do not survive 17 or initially remain dormant 18, and the observation that they arrive much earlier than previously expected 19, imaging the earliest steps of arrival and survival becomes crucial to understanding the process of metastatic seeding and recurrence of tumor growth at distant sites.
Performing these observations in the lung has proven extremely difficult however; the vast majority of imaging studies have utilized ex vivo or explant preparations 20-23, which only give a view into the lung at single time points. While these preparations do provide useful information, they do not give a complete understanding of the interactions, cause and effect relationships, and dynamics that occur between the various components of the microenvironment. The lack of a proper circulatory system (and concomitant imbalance of homeostasis) and the disconnection from the rest of the body's immune system makes it desirous to validate the conclusions that these preparations generate in intact tissue in vivo.
Many groups have performed intravital imaging of the intact lung 2,4,5,24-33 with Wearn and German being the first to surgically expose the pleural layer 24 and Terry the first to utilize an implantable imaging window 25.
High resolution imaging in the lung is greatly hindered by the lung's constant motion and several techniques have been developed to overcome this limitation. Wagner and Filley 27 studied the natural motion of the canine lung and designed their surgical protocol to locate their implanted window over a relatively stationary region while Wagner utilized vacuum in his window surgical preparation to immobilize the tissue 28. Since that time, a variety of techniques have been utilized to image the lung including: bronchus clamping, sequential apnea and gated imaging, oversampled acquisition, gluing of the lung lobe and vacuum 34. Each of these has its advantages and disadvantages and no one technique has emerged as superior to another 34. For example, bronchus clamping and sequential apnea alter the normal exchange of gases in the lung and may cause atelectasis. Gated imaging and oversampled acquisition do not suffer from these disadvantages but require high-speed or specialized imaging equipment not widely accessible. Finally both gluing of the lung and the vacuum technique avoid both of the above mentioned drawbacks, but may exhibit shear force induced injury if care is not taken. In recent years, the vacuum window has been miniaturized and adapted for use in mice using confocal and multiphoton microscopy 4,5,33 and excellent high-resolution imaging has been attained 2. Table 1 summarizes this rich history and highlights those papers which describe novel advancements in the use of intravital lung imaging windows.
This protocol describes the use of extended time-lapse multiphoton intravital microscopy to image metastasis in the live, intact lung with the highest subcellular resolution possible. Images are acquired for up to 12 hr using a multiphoton microscope equipped with a high numerical aperture objective lens and multiple photomultiplier tube (PMT) detectors. Transgenic mouse models are utilized to fluorescently label native macrophages along with fluorescent high molecular weight dextran and fluorescent protein transfected tumor cells (to label the vasculature and tumor cells respectively). While this choice of fluorescently labeled cells enables visualization of tumor cell-endothelial cell-macrophage interactions and dynamics, this protocol will work for any strain of fluorescent or non-fluorescent mouse. After acquisition, residual drift motion (if any) is eliminated using a Fiji plugin 35,36 and custom macros time average the vascular channel to eliminate flashing caused by unlabeled circulating blood cells.
While this protocol focuses on imaging metastasis, the techniques are applicable to many other biological processes observable with high-resolution single-cell imaging in the lung.
Protocol
All procedures described in this protocol have been performed in accordance with guidelines and regulations for the use of vertebrate animals, including prior approval by the Albert Einstein College of Medicine Institutional Animal Care and Use Committee.
1. Generating Fluorescently Labeled Mouse Model and Tumor Cells
Prepare 100 ml of 0.1 % (w/v) bovine serum albumin/phosphate buffered saline (BSA/PBS) buffer by mixing 0.1 g of BSA with 100 ml of PBS.
- Prepare fluorescently labeled tumor cells by stable transfection. NOTE: Here we use E0771-LG cells, a highly metastatic derivative of E0771 mouse mammary adenocarcinoma cells 37 that were isolated from metastatic tumors developed in the lung of C57BL/6 mouse intravenously injected with parental E0771 cells 38.
- 24 hr before transfection, plate 1 x 105 EO771-LG cells on a 60 mm tissue culture dish on 2 ml of antibiotic free 10% FBS (fetal bovine serum) DMEM (Dulbecco's Modified Eagle Medium) and incubate at 37 °C and 5 % CO2.
- At the time of transfection, incubate 2 µg of the fluorescent protein vector with 10 µl of transfection reagent for 30 min before adding 190 µl of reduced serum media.
- Wash EO771-LG cells with Dulbecco's phosphate buffered saline (D-PBS) once and add the transfection mix gently.
- Incubate at 37 °C, 5% CO2 for 6 hr.
- Wash the transfected cells and culture in 2 ml of complete DMEM (10% FBS, 1mM pyruvate, 100 U/ml penicillin, 100 µg/ml streptomycin) for 2 days.
- Wash cells with 2 ml of sterile PBS, add 500 µl of 0.25 % trypsin-EDTA and incubate for 2 min at 37 °C.
- Collect cell suspension and mix with at least 1 volume of complete DMEM and spin down at 280 x g.
- Resuspend the cells in 1 ml of complete DMEM and expand the cell culture into a 10 cm tissue culture dish.
- Start selection of transfected cells by adding 700 µg/ml of G418 selective antibiotic to 8 ml of complete DMEM and culture for a week, changing medium every three days.
- Enrich the fluorescent population from transfected cells that have been under selection in G418 by fluorescence-activated cell sorting (FACS) 39,40.
- Wash cells with 5 ml of PBS and add 1.5 ml of 0.25% trypsin.
- Incubate for 2 min at 37 °C and mix with at least one volume of complete DMEM.
- Collect cell suspension and spin down at 280 x g and re-suspend cells in 1 mL of sterile 0.1 % (w/v) BSA/PBS buffer.
- Filter the cell suspension through a 40 µm mesh and adjust the volume to 1 ml with 0.1 % BSA/PBS buffer for sorting.
- FACS-sort the 10% brightest population based on fluorescence spectrum using the FACS sorter 41.
- Culture the sorted cells for another week under selection (700 µg/ml G418 in complete DMEM).
- Reselect fluorescently labeled cells by flow cytometry by repeating steps 1.3.1 to 1.3.6 once again.
- After a second round of selection, trypsinize, filter and re-suspend cells in 0.1% BSA/PBS as described (steps 1.3.1-1.3.4). Adjust concentration to 2 x 106 cells/ml for single cell sorting into 96 well plates using the FACS sorter.
- Prepare 3-5 96 well plates with 100 µl of full DMEM for collection. To improve survival after sorting, add 100 µl of filtered culture medium from dishes where the EO771-LG cells were grown 42.
- After sorting the cells into the 96-well plates, return cells to culture for 2 days (37 °C, 5% CO2).
- Identify wells with viable single clones by examination under an inverted microscope at 5x magnification.
- Trypsinize and expand viable clones and freeze cells for backup.
- Wash wells with growing clones with 100 µl of sterile PBS. Add 50 µl of 0.25% w/v trypsin and incubate 2 min at 37 °C. Add 50 µl of complete DMEM and transfer to sterile V-bottom or round bottom 96-well plate to spin down at 280 x g for 5 min.
- Resuspend cells in 100 µl of 700 µg/ml G418 complete DMEM and plate each clone in 12-well plates. Add 400 µl of complete DMEM with G418 and culture until confluent.
- Wash wells with 500 µl of PBS, add 100 µl of 0.25% trypsin and incubate 2 min at 37 °C. Add 100 µl of complete DMEM and spin down for 5 min at 280 x g. Resuspend cells in 100 ul of complete DMEM with G418 and plate cells in 6 well plates until confluent.
- Once confluent, trypsinize cells, spin down and resuspend in 10% DMSO in FBS to freeze cell stocks.
- Keep 1/10 of the cell suspensions in culture by plating them in 100 mm tissue culture dishes for evaluation of their metastatic potential.
- Test metastatic potential of selected clones.
- Trypsinize and re-suspend cells in saline to a concentration of 2.5 x 106 cells/ml and inject 200 µl into C57BL/6 mice intravenously through the tail vein.
- After 2 weeks, collect lung tissue as previously described 23 and quantify tumor burden by surface count or stereological method 43. Select clones with high metastatic potential for intravital imaging.
Raise MacBlue (a myeloid specific promoter driving cyan fluorescent protein (CFP) expression (Csf1r-GAL4VP16/UAS-ECFP16)) reporter mice 44. NOTE: Typically, mice between 8 and 12 weeks are used, but mice as early as 7 and as late as 20 weeks have been tested to work as well.
2. Multiphoton Microscope Set up and Imaging Preparation
NOTE: While this protocol can be performed on any multiphoton microscope, the system used to acquire the data shown in this protocol has been described previously in detail 45.
Turn on all microscope and laser components including two-photon lasers and the detectors at least one hour ahead of the desired imaging time.
Just before surgery, measure the power of the light input to the microscope by placing the head of the optical power meter in the beam path just before the microscope and adjust the laser end cavity mirror knobs until the maximum light intensity at 880 nm is read on the optical power meter.
3. Vacuum System Set up
Glue the coverslip into the imaging window (Supplemental Figure 1) with cyanoacrylate. Allow at least 4 hr for the glue to completely dry. NOTE: This step may also be done ahead of time.
Cut the 100 µl pipette tip at the first line from the small opening and connect the uncut end to the thin vacuum hose.
Connect the vacuum system according to Figure 1 and Supplemental Figure 2.
With the vacuum on and the open end blocked, adjust the vacuum regulator for <3 inHg. NOTE: Final adjustment of the vacuum level will be performed by observation of the vasculature in vivo.
Apply a thin film of petroleum grease to the underside of the imaging plate to prevent the objective lens immersion medium from being wicked away by the imaging plate.
Place the imaging plate on the microscope stage. NOTE: The custom imaging plate (Supplemental Figure 3) is a sheet of 1/8" thick aluminum machined both to fit in the stage insert space and with a through-hole in the center for holding the imaging window.
Insert the vacuum window into the imaging plate with the coverslip down.
Sterilize all surfaces and instruments including imaging stage plate, and imaging window with 70% ethanol.
Connect the cut end of the pipette tip to the vacuum window and tape the hose down to the imaging plate.
Bring the objective close to the imaging window and place a large drop of water between the objective and the coverslip.
Ensure that there are no leaks from the coverslip by blocking the central opening of the vacuum window and verifying that the water drop between the objective and coverslip does not get aspirated. NOTE: An old style computer mouse ball placed over the window is useful for blocking the central opening.
4. Surgery
- Prepare the sterile surgical area.
- Place all instruments within easy reach. Sterilize all surfaces and instruments including surgical area, and surgical tools with 70% ethanol.
Tie a 3 inch length of 2-0 suture to the catheter, ¼ inch above the bushing with a double knot.
Prepare tail vein catheter following the published protocol by Harney et al. 46.
Using an infrared (IR) heat lamp, warm the animal in its cage for ~5 min to increase the blood flow in the tail vein and to aid in catheter insertion. It is recommended to keep the animal warmed to physiological temperatures throughout the surgical procedure using either a heat lamp or a warming pad.
Anesthetize the animal with 5% isoflurane and verify that there is no response to a toe pinch.
Apply ophthalmic ointment to the eyes of the animal.
Apply depilatory lotion for 10-30 sec to remove hair from the left side of the animal; from the midline of the chest to ¼ of the back and from the axilla to just under the ribcage.
Clean any excess lotion and sterilize exposed skin with 70% alcohol.
Attach a sterile syringe filled with PBS to the tail vein catheter and insert and tape in place following the published protocol by Harney et al. 46. Make sure the tape is securely adhered to the needle itself and is not free in the gap between the tail and the tape.
Intubate the mouse following either the published protocol by Das et al. 47 or by DuPage et al. 48
Turn on the ventilator and set it to supply 135 breaths per min and 200 µl of isoflurane-oxygen mixture.
Connect the tracheal catheter to the ventilator.
Move the mouse to the surgical area. Take extreme care not to dislodge the catheter.
Tie the 2-0 suture around the snout of the mouse under the front teeth.
Tape the catheter to the snout.
Tape the left forelimb to the catheter to keep it out of the surgical field.
Lower the isoflurane anesthesia to a maintenance level of 2.5% and verify there is no response to a toe pinch.
Using the sharp scissors, remove 1 cm2 of skin above the left chest wall.
Lift the mammary fat pad and cauterize any exposed blood vessels with the cautery pen.
Resect the fat pad by cutting with the sharp scissors.
Remove the muscle layer down to the rib cage by cutting with the sharp scissors. Take care not to cut the axillary vein running at the base of the forelimb.
Use forceps to grab and lift the 6th rib. Using the sharp scissors held at a shallow angle (~5°) cut the rib near the edge of the opening in skin. Take extreme care not to touch the exposed lung tissue.
Widen the opening in the chest wall to expose the entire lung lobe by removing four consecutive ribs. Note: Keep the opening at least 5 mm away from the sternum to avoid the heart.
Carefully lift the mouse by grasping the tail and tracheal catheter and move the mouse to the microscope imaging stage.
With the vacuum off, fill the chamber of the vacuum window with PBS.
Invert the mouse and position the exposed lung over the vacuum imaging window.
Slowly turn on the vacuum to approximately 3-5 inches of mercury using the ball valve.
Place a restraining harness made of tissue paper folded in half twice over the chest of the mouse and tape to the stage plate as shown in Supplemental Figure 2.
Clip the thigh sensor of the pulse oximeter to the animal's upper thigh and start the software.
Place the environmental chamber on the stage and turn on the heat to maintain the mouse at a physiological temperature.
Reduce the level of isoflurane to 1-1.5 % to maintain anesthesia and maintain blood flow.
5. Intravital Imaging
Bring the 25 x 0.95 numerical aperture (NA) objective lens near to the coverslip and add a large drop of water between them.
Using epifluorescence mode, view the FITC channel and bring the lung tissue into focus.
- If not done prior to the surgery, inject tumor cells through the tail vein catheter.
- Disconnect the PBS syringe from the tail vein catheter.
- Load a sterile syringe with 100 µl of tumor cell suspension (2 x 107 cells/ml in PBS maximum). NOTE: This step can be done in advance to study cancer cell arrival to the lung at different time points.
- Connect the syringe with tumor cells onto the tail vein catheter.
- Slowly inject the tumor cells into the tail vein.
- Disconnect the syringe with the tumor cells from the tail vein catheter.
- Reconnect the PBS syringe to the tail vein catheter. NOTE: Identify the locations of all of the tumor cells before injecting the dextran as it becomes difficult to distinguish the tumor cells from the dextran signal via the ocular after injection.
- Locate tumor cells to image
- For imaging individual tumor cells, locate all tumor cells and record their locations with the multipoint panel of the software.
- Locate all fields of view to image by observing the tumor cells in the microscope ocular.
- In the software, switch to the multipoint panel by clicking on the Multi-Point button and store the location of the cell by clicking on the Add Position button.
- For mosaic imaging, locate the origin of the mosaic and set the imaging coordinates
- Locate a position in the top left corner of the structure to be captured.
- Zero the x, y and z coordinates of the stage by pushing the "Zero" button on the stage controller.
- Load up the appropriate list of mosaic coordinates by clicking on the "Load" button and selecting the list. NOTE: An example list for a 2 x 2 mosaic with a 20% overlap of a 500 µm field of view would be: Pos.1 = (0,0), Pos. 2 = (400,0), Pos. 3 = (0,400), Pos. 4 = (400,400).
Remove the syringe with PBS in the tail vein catheter and replace with the syringe containing the dextran.
Slowly inject up to 100 µl of 20 mg / ml 155 kDa rhodamine-dextran dissolved in PBS into the mouse via the tail vein catheter followed by injecting 50 µl of sterile PBS to flush the line. Do not introduce any bubbles into the line. When necessary, inject dextran at least one hour after cancer cell injection so that the total volume administered to the mouse does not exceed 4 ml/Kg/hr.
- Set up imaging parameters.
- Switch the microscope to multiphoton mode.
- Set the zoom to a factor of 2X by clicking on the Timing Signals button and updating the Zoom Factor field.
- Adjust the laser power to ~10 % (~10-15 mW at the sample) by clicking the Detectors and Laser button and then adjusting the Tsunami Power slider to 10.
Image each location to verify the presence of tumor cells and visualize the flow and integrity of the vasculature. NOTE: Vessels should appear fully perfused with flowing erythrocytes and fluorescent dextran should be contained within the vessels without leakage to the extravascular spaces. Approximately 10 - 20 tumor cells are expected to be within the clear aperture of the vacuum window.
- Adjust the starting depth of each location to be imaged.
- For each location, adjust the z position by rotating the focus knob on the stage controller to image the top slice of the tumor cell.
- Position the cell in the center of the field of view.
- Click on the Multipoint button, click on the position of the field of view to highlight it and click the Add followed by the Delete button to replace the cell's stored position in the multipoint list.
- Visually observe the relative brightness of the tumor cell in each position.
Save the new locations of each of the cells by clicking on the Save button in the Multipoint panel and specify a filename.
For imaging of individual tumor cells, pick three cells of approximately equivalent brightness and delete all other locations from the multipoint list by clicking on their location in the list and then clicking on the Delete button.
Click on the Detectors and Laser button and adjust the sliders for the PMT gain of the green and red channels 45 so that the signals are below saturation.
Adjust the slider for the blue channel 45 so that macrophages appear cyan. NOTE: Any second harmonic signal will appear only in the blue channel and can be separated from the cyan macrophages by following the channel subtraction procedure previously described 45.
Set the z-stack start depth to 0 µm and the end depth to 24 µm by moving the z stage to the location and clicking on the Start and End buttons respectively. NOTE: Cells within this depth will be visualized with the best signal to noise and resolution.
Set the z step size to 3 µm.
- Set the imaging parameters following parameters previously described 45,49.
- For imaging of individual tumor cells, click the Timing Signals button and then enter 4 V into the zoom factor field (equivalent to a zoom factor of 1.5X), enter 3 into the frame averages field and click on Time-Lapse button and enter 10 into the Time-lapse field. NOTE: These settings will acquire 1 frame every 3 sec.
- For mosaic imaging, click the Timing Signals button and enter a zoom factor of 1.5 V (equivalent to a zoom factor of 4X), enter 3 in the number of frame averages and click on the Time-Lapse button and enter 10 into the time-lapse time delay field. NOTE: These settings will acquire 1 frame every 3 sec.
Enable the multipoint, z-stack and t-lapse imaging modes by clicking on their buttons.
Press the Record button to acquire images. NOTE: Lung tissue is very delicate and susceptible to photo-damage. If after time lapse imaging blood flow stops in the imaged field, the laser is most likely too high and subsequent imaging of other fields must be done at lower power.
Every 30-45 min, slowly inject 50 µl of PBS or saline to maintain hydration of the animal.
6. Euthanasia
Increase the isoflurane to 5%.
Keep the animal under 5% isoflurane until 30 sec after it ceases to breathe and remove the animal from the stage.
Perform cervical dislocation to ensure complete euthanasia.
7. Image Analysis
- For single cell imaging experiments:
- Load images into Fiji and format them as a Hyperstack.
- For each z-slice in the Hyperstack, play the time lapse movie and look for residual x-y movement. If residual x-y movement is found, apply the plugin called StackReg 36 to the stack to eliminate the movement.
For mosaic imaging experiments:
Load images into Fiji and stitch them together by opening the Mosaic Stitching macro (Supplemental code file Mosaic Stitching) and entering information about the images such as the directory, the file base name, the number of x and y fields in the mosaic and the number of slices and time points. NOTE: Due to how Java interprets directories, folder names must have two backslashes as subfolder separators. Due to limitations in the built-in plugin Pairwise Stitching, base file names must not contain any dashes.
To obtain a clear view of the boundaries of the vasculature, average together all of the time points for the blood channel into a single image and then replicate this image as the background for each frame of the movie of the other channels. NOTE: This is done by simply running the Perform Blood Averaging macro (Supplemental code file Perform Blood Averaging).
Representative Results
To demonstrate the type of results that can be achieved with this method, we injected E0771-LG tumor cells labeled with the fluorescent protein Clover into the tail vein of MacBlue mice 44 at varying time points before surgery. After surgery, 155 kD rhodamine labeled dextran was injected IV to mark the vasculature and time-lapse imaging was performed.
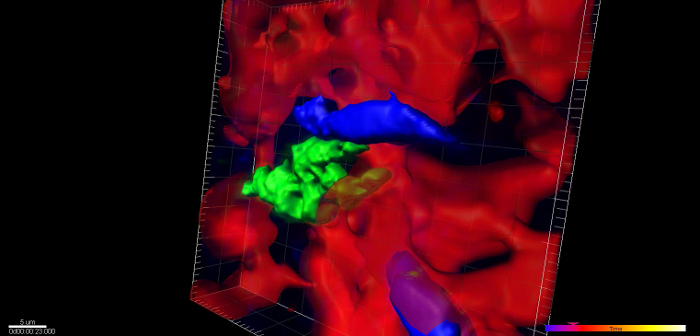
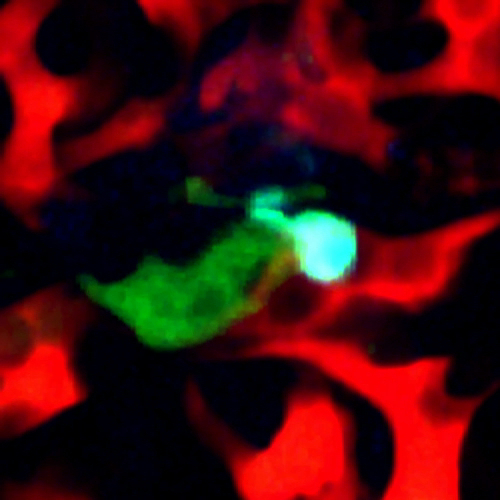
When imaging mice 24 hr post injection, single cells are visible inside of the vasculature, interacting with macrophages and monocytes. An example of this is shown in Figure 2A (Supplemental Movie 1). Here a single optical section of a solitary tumor cell (green) 12 µm deep lodged in the lung vasculature is imaged over 5 hr and 20 min as it transiently interacts with a resident macrophage (cyan). Vasculature is labeled by high molecular weight dextran. The stability of the imaging is such that sequential z-stack imaging of the field can be acquired and a 3 dimensional reconstruction can be made (Figure 2B, Supplemental Movie 2).
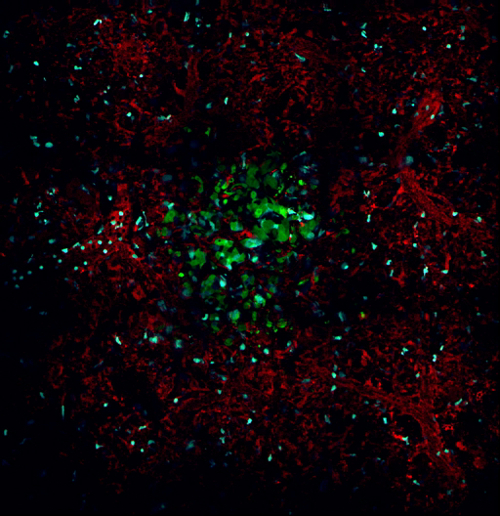
Use of the microscope settings described in the protocol for mosaic imaging allows imaging of structures larger than a single field of view. For example, Figure 3 demonstrates acquisition of a single metastatic lesion, 12 days post injection. This 5 x 5 mosaic shows an 890 µm field of view over 205 min at 15 (Figure 3A, left panel; Supplemental Movie 3) and 25 µm (Figure 3A, right panel) below the surface of the lung. Despite the large field of view, the high resolution of the underlying frames composing the mosaic enables the capture of subcellular events such as mitosis of a single cell as evidenced by chromosomal separation (Figure 3B, Supplemental Movie 4).
Intravascular injection of a high molecular weight fluorescently labelled dextran results in labeling of the vascular lumen, however unlabeled circulating erythrocytes and leukocytes occlude the dextran. In the small capillaries of the lung, the occlusion is complete resulting in a flashing of the dextran signal and a loss of definition of the vasculature boundaries (Figure 4, left; Supplemental Movie 5). The high spatial stability provided by this protocol allows time averaging of the blood channel, without blurring, thus restoring the temporary occlusions. The other signal channels can then be overlaid on the vasculature to provide a clear view of the vessel boundaries (Figure 4, right; Supplemental Movie 6).
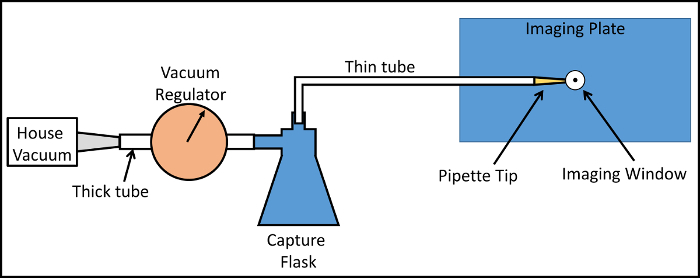 Figure 1:Layout of the Vacuum System. House vacuum is utilized and set to defined level with a vacuum regulator. A capture flask prevents contamination of the regulator and vacuum system by bodily fluids. A thin flexible tube conveys the vacuum to a pipette tip cut to fit into the vacuum port of the imaging window. The imaging window is fit into a recessed groove in the imaging plate maintaining its positional stability with respect to the microscope objective lens. Please click here to view a larger version of this figure.
Figure 1:Layout of the Vacuum System. House vacuum is utilized and set to defined level with a vacuum regulator. A capture flask prevents contamination of the regulator and vacuum system by bodily fluids. A thin flexible tube conveys the vacuum to a pipette tip cut to fit into the vacuum port of the imaging window. The imaging window is fit into a recessed groove in the imaging plate maintaining its positional stability with respect to the microscope objective lens. Please click here to view a larger version of this figure.
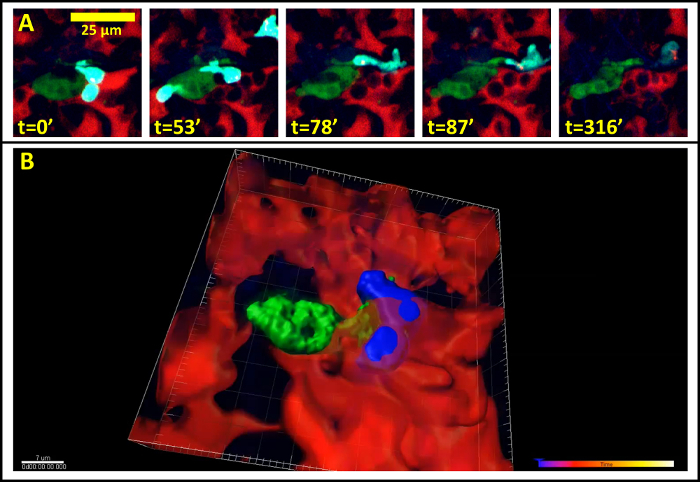 Figure 2:Single Cell Imaging in the Lung. (A) Still from a time lapse movie of a single tumor cell in the capillary bed of the lung 24 hr after tail vein injection. (Red = Blood vessels, Green = Tumor Cells, Cyan = Macrophages) (B) Stable imaging allows three dimension reconstruction of imaging data over time. Blood vessels have been time averaged for clarity. (Red = Blood vessels, Green = Tumor Cells, Blue = Macrophages). Please click here to view a larger version of this figure.
Figure 2:Single Cell Imaging in the Lung. (A) Still from a time lapse movie of a single tumor cell in the capillary bed of the lung 24 hr after tail vein injection. (Red = Blood vessels, Green = Tumor Cells, Cyan = Macrophages) (B) Stable imaging allows three dimension reconstruction of imaging data over time. Blood vessels have been time averaged for clarity. (Red = Blood vessels, Green = Tumor Cells, Blue = Macrophages). Please click here to view a larger version of this figure.
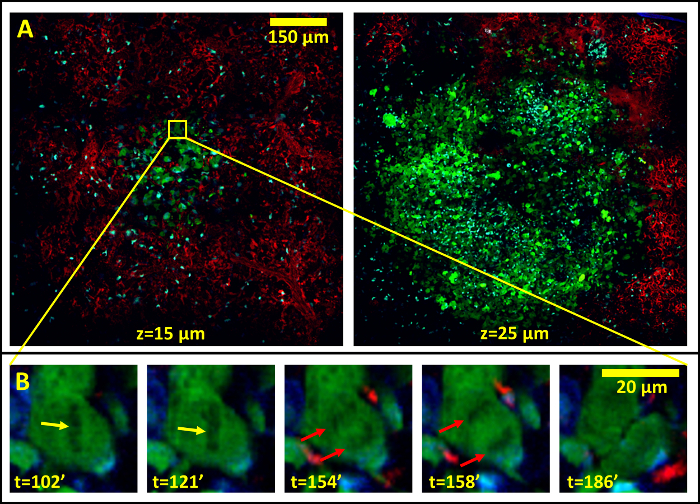 Figure 3:The Stability of the Lung Allows High-resolution, Large-field of View Imaging in the Lung by the Sequential Acquisition and Stitching Together of Multiple Low Magnification Fields. (A) 5 x 5 mosaic showing an 890 µm field of view of a metastatic lesion in the lung 12 days after tail vein injection of tumor cells taken at 15 µm below the lung surface (Left Panel). Right panel shows the same metastatic lesion at 25 µm below the lung surface. (B) The individual high resolution fields reveal subcellular processes such as chromosomal alignment (yellow arrows) and separation (red arrows) during cell division. Please click here to view a larger version of this figure.
Figure 3:The Stability of the Lung Allows High-resolution, Large-field of View Imaging in the Lung by the Sequential Acquisition and Stitching Together of Multiple Low Magnification Fields. (A) 5 x 5 mosaic showing an 890 µm field of view of a metastatic lesion in the lung 12 days after tail vein injection of tumor cells taken at 15 µm below the lung surface (Left Panel). Right panel shows the same metastatic lesion at 25 µm below the lung surface. (B) The individual high resolution fields reveal subcellular processes such as chromosomal alignment (yellow arrows) and separation (red arrows) during cell division. Please click here to view a larger version of this figure.
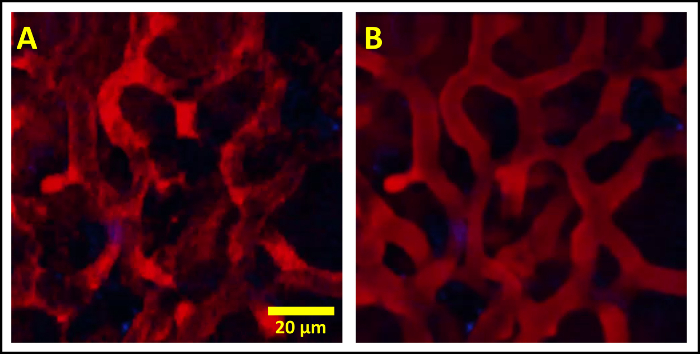 Figure 4:Intravascular Injection of Fluorescently Labeled High Molecular Weight Dextran Marks the Lumen of the Vasculature Except when Unlabeled Erythrocytes and other Circulating Cells Occlude the Fluorescence Signal. (A) Occlusion results in an incomplete labeling which moves in time resulting in a flashing effect obscuring the vessel boundaries. (B) The high spatial stability of the vacuum window allows the blood channel to be time averaged, filling in the temporary occlusions and clearly defining the vessel boundaries. The other channels are then overlaid without averaging. Please click here to view a larger version of this figure.
Figure 4:Intravascular Injection of Fluorescently Labeled High Molecular Weight Dextran Marks the Lumen of the Vasculature Except when Unlabeled Erythrocytes and other Circulating Cells Occlude the Fluorescence Signal. (A) Occlusion results in an incomplete labeling which moves in time resulting in a flashing effect obscuring the vessel boundaries. (B) The high spatial stability of the vacuum window allows the blood channel to be time averaged, filling in the temporary occlusions and clearly defining the vessel boundaries. The other channels are then overlaid without averaging. Please click here to view a larger version of this figure.
| Year | First Author / Last Author | Animal | Surgery | Type of Imaging | Notes |
| 1926 | Wearn & German | Cats | Chest wall cut down to pleural layer. Second opening made through diaphragm down to pleura for illumination. | Bright-field microscopy. | First “window” through pleural wall. |
| 1939 | Terry | Cats | Ribs are separated, 1 in. window implanted, air removed from thorax with vacuum. | Binocular and skin microscope using polarized illumination. | First implanted optical window. Used vacuum to draw tissue to window. |
| 1963 | de Alva & Rainer | Rabbits & Dogs | One or two ribs are resected and 1 in. window inserted and sutured to chest wall. Skin was closed over window and animal allowed to heal. Before imaging, skin was dissected to expose window. | Used high speed stroboscopic cinematography through a low mag (11X) objective. | First survival window. |
| 1965 | Wagner & Filley | Dogs | Right forelimb removed, one rib cut and window inserted and sutured. | Reflection microscopy with 22X objective. | First relatively motion free window without vacuum. |
| 1969 | Wagner | Dogs | One Rib is resected, 3 in. window implanted. Threaded flange screws on window to form seal to chest wall. | Brightfield microscopy with high-magnification 100X oil immersion objective. | First window to use vacuum to stabilize tissue movement. |
| 1992 | Groh & Goetz | Rabbits | One rib resected, 1.2 in. window implanted. Threaded flange screws on window to form seal to chest wall. Vacuum applied. | Epifluorescent microscopy with 25X objective. | First use of a vacuum window with epifluorescence. |
| 1994 | Fingar & Wieman | Rats | 2 ribs are resected, 1 in. window implanted and sutured to chest wall and skin. | Epifluorescent microscopy with 40X objective. | First survival window in rats. |
| 2000 | Funakoshi & Mitsui | Mice | Entire chest wall removed. Vacuum suction ring attached to right lung. | Confocal microscopy with a 20X objective. | First use of vacuum window in mice. |
| 2005 | Lamm & Glenny | Rats | Entire chest wall removed, 1 in. window attached to lung. | Reflection and epifluorescent microscopy with a 20X objective. | First use of vacuum window in rats. |
| 2008 | Tabuchi & Keubler | Mice | 3 ribs are resected, plastic film applied to opening to seal hole in chest wall. Air removed via intrapleural catheter. | Epifluorescent microscopy with 20X objective. | First to use plastic film to seal opening in chest wall. |
Table 1: Historical Survey of the Development of Intravital Lung Imaging Windows. Many novel intravital lung imaging windows have been developed over the years with the most recent being miniaturized for use in mice, employing vacuum for tissue stabilization and attaining high enough resolution to be capable of revealing subcellular detail.
Supplemental Figure 1: Design Drawing of the Vacuum Imaging Window Please click here to download this figure.
Supplemental Figure 2: Photographs of Vacuum Setup Please click here to download this figure.
Supplemental Figure 3: Design Drawing of Stage Plate Insert Please click here to download this figure.
 Supplemental Movie 1: Movie of stills in Figure 2A. (Right click to download).
Supplemental Movie 1: Movie of stills in Figure 2A. (Right click to download).
Supplemental code file: Mosaic Stitching. Please click here to download this file.
Supplemental code file: Perform Blood Averaging. Please click here to download this file.
Discussion
High resolution in vivo optical imaging combined with fluorescently labelled functional tags such as proteins and antibodies has dramatically increased our understanding of the metastatic cascade. It has enabled direct visualization and quantification of single-cell and sub-cellular parameters in tumor cells, host cells and their microenvironment. This imaging within the primary tumor has led, for example, to the discovery of discrete microenvironments that are supportive of either growth invasion or dissemination 6,7. In the case of invasion, in vivo imaging has revealed the preferential role of co-migrational streaming of macrophages and tumor cells in intravasation 7,50.
In secondary sites like the lung, understanding the dynamics of tumor cell behavior at the earliest stages of metastasis, including the pre-micrometastasis seeding stage when single and small groups of tumor cells arrive and interact with the blood vessel endothelium, will only be accomplished using high resolution optical imaging. Standard clinical imaging modalities do not have the resolution needed to visualize either the fine structure of the capillary bed or the morphology and interaction of cells at single cell resolution. The imaging techniques presented in this protocol accomplish this challenging task.
Intravital imaging windows such as those listed in Table 1 offer a significant advantage over ex vivo lung preparations by maintaining proper lung physiology including perfusion, connection to the immune system and offering more than just a static view of cellular dynamics. The vacuum stabilized windows in particular offer a level of tissue stability that allows the acquisition of highly registered images, enabling three dimensional reconstructions (Figure 2B) and the capability for large field of view mosaic imaging (Figure 3A). Together these provide a range of views of the lung, from a histologic type low magnification view which gives the tissue morphology, to a sub cellular view that can even reveal chromosomal separation and discriminate dormant and dividing tumor cells (Figure 3B). Multiple acquisition channels allow several cell types and their interactions to be visualized simultaneously (Figure 2A).
While the protocol does take some technical skill, practice and attention to some critical steps and points will improve the success rate of the procedure and imaging times of up to 12 hr can be expected. It is critical to make sure the lung tissue is well centered over the window (step 4.25). This ensures that the vacuum is applied evenly and completely across the lung tissue (step 4.27). Failure to center the tissue will result in motion of the tissue. The restraining harness (Supplemental Figure 2) is used to reduce the motion induced by constriction of the intercostal muscles during the natural breaths the animal takes. It should be snug over the mouse, but not compress the chest. Too much compression puts pressure on all of the lobes of the lungs as well as the heart and results in a reduced viability of the mouse. If dextran is not observed flowing in the lung vasculature after injection (step 5.8), the lung tissue is either damaged from improper handling during surgery or the vacuum level is too high. Lowering the vacuum by .5-1 inch Hg can be attempted to see if flow is restored. If the flow is not restored, or if there is flow, but dextran is observed extravascularly, the lung tissue has been damaged in that region and it will be necessary to image a different field of view. Maintenance of proper blood flow in the imaging region is important to ensuring that proper physiology is being measured. Since oxygen is supplied to the tissue from inside of the alveoli and not through the vasculature, ischemic hypoxia is unlikely. Still, unperfused vasculature can potentially lead to altered oxygen/CO2 levels and will also prevent circulating leukocytes from reaching the tissue of interest. Visualization of flowing erythrocytes can be used as an indicator of the proper lung function. Lung tissue is very delicate and also susceptible to photo-damage. If after time lapse imaging blood flow stops in the imaged field, the laser is most likely too high and subsequent imaging of other fields must be done at lower power. We have found ~10-15 mW at the sample to produce sufficiently bright images without photodamage when using either GFP or Clover (which has four times the mean brightness) transfected cells. Minimum brightness levels for the fluorescent proteins are highly dependent upon microscope parameters and the expression levels in the cells and must be tested empirically.
In this protocol, tumor cells are labeled with a bright cytoplasmic fluorescent protein that offers a clear view of the cell body and intracellular spaces that exclude the protein (i.e. nucleus). Macrophages are labeled by utilizing a transgenic mouse model syngeneic to the tumor cells. Labeling both cell types enables visualization of their direct interaction in real time. The extended duration of imaging allows quantification of the frequency, duration and extent with which cancer cells interact with macrophages in a physiologically relevant context.
When performed correctly, this procedure enables motion free, multiple-channel, high-resolution, single-cell imaging in the intact lung for periods of up to 12 hr. The use of a high numerical aperture objective lens such as the 25X 0.95NA and the multiphoton's capabilities for electronic zoom allows the highest resolution optical imaging in the lung seen to date.
Intravascularly injected fluorescent, high molecular weight dextran performs the dual role of labeling the vascular space and verifying its integrity. 155 kDa dextran is used to prevent diffusion through the interendothelial spaces. Any sign of a lack of vascular flow or of vascular leakage into the extravascular space indicates damage to the tissue due to improper handling or excessive vacuum.
Finally, unique image processing techniques can be employed that take advantage of the high spatial stability of this protocol. Since the unlabeled erythrocytes and other leukocytes exclude the fluorescent dextran when they pass through the capillaries, this signal can be averaged over time to remove the flashing that they create. This offers a well-defined view of the vasculature not possible otherwise.
Limitations of this technique include both the invasive nature of the surgery, which potentially complicates its use for study of diseases which weaken the animal (e.g. late stage metastatic cancer, acute sickle cell anemia), and the fact that the surgery is terminal which limits it use to a single, albeit long (up to 12 hr), imaging session. Further, given the long duration of the imaging session and the low hepatic glycogen reserve of mice 51, glucose supplementation may be given to avoid potential sources of bias in experiments.
This protocol could potentially be modified with injectable fluorescently labeled antibodies either in place of or in addition to the dextran in order to label other structures or cell types in real time. This will expand the capabilities for analyzing and dissecting the tumor microenvironment by directly visualizing tumor cell-host cell interactions and dynamics in real time.
Disclosures
The authors have nothing to disclose.
Acknowledgments
This research was supported by NIH-CA100324, Einstein National Cancer Institute's cancer center support grant P30CA013330, R01CA172451 to JWP and the Integrated Imaging Program. This technology was developed in the Gruss-Lipper Biophotonics Center and the Integrated Imaging Program at the Albert Einstein College of Medicine. We acknowledge the support of these Centers in this work. The authors thank Mike Rottenkolber, Ricardo Ibagon and Anthony Leggiadro of the Einstein machine shop for their skilled and timely craftsmanship, the laboratory of Matthew Krummel for generously sharing their window design drawings, Kevin Elicieri and Jeremy Bredfeldt for their expertise in microscopy and their amplifier recommendations and Allison Harney and Bojana Gligorijevic for informative discussions.
References
- Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer. 2006;6(6):449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- Entenberg D, et al. Subcellular resolution optical imaging in the lung reveals early metastatic proliferation and motility. Intravital. 2015;4(3):1–11. doi: 10.1080/21659087.2015.1086613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahl VE. A method of studying the living lung in the closed thorax, and some preliminary observations. Angiology. 1963;14:149–159. doi: 10.1177/000331976301400401. [DOI] [PubMed] [Google Scholar]
- Looney MR, et al. Stabilized imaging of immune surveillance in the mouse lung. Nat Methods. 2011;8(1):91–96. doi: 10.1038/nmeth.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presson RG, Jr, et al. Two-photon imaging within the murine thorax without respiratory and cardiac motion artifact. Am J Pathol. 2011;179(1):75–82. doi: 10.1016/j.ajpath.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gligorijevic B, Bergman A, Condeelis J. Multiparametric classification links tumor microenvironments with tumor cell phenotype. PLoS Biol. 2014;12(11):e1001995. doi: 10.1371/journal.pbio.1001995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harney AS, et al. Real-Time Imaging Reveals Local, Transient Vascular Permeability, and Tumor Cell Intravasation Stimulated by TIE2hi Macrophage-Derived VEGFA. Cancer Discov. 2015;5(9):932–943. doi: 10.1158/2159-8290.CD-15-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozluoglu M, et al. Matrix geometry determines optimal cancer cell migration strategy and modulates response to interventions. Nat Cell Biol. 2013;15(7):751–762. doi: 10.1038/ncb2775. [DOI] [PubMed] [Google Scholar]
- Suetsugu A, et al. Imaging the recruitment of cancer-associated fibroblasts by liver-metastatic colon cancer. J Cell Biochem. 2011;112(3):949–953. doi: 10.1002/jcb.23011. [DOI] [PubMed] [Google Scholar]
- Nakasone ES, et al. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer Cell. 2012;21(4):488–503. doi: 10.1016/j.ccr.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139(7):1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BD, et al. Tumor microenvironment of metastasis in human breast carcinoma: a potential prognostic marker linked to hematogenous dissemination. Clin Cancer Res. 2009;15(7):2433–2441. doi: 10.1158/1078-0432.CCR-08-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohan TE, et al. Tumor microenvironment of metastasis and risk of distant metastasis of breast cancer. J Natl Cancer Inst. 2014;106(8) doi: 10.1093/jnci/dju136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, et al. Quantitative assessment of invasive mena isoforms (Menacalc) as an independent prognostic marker in breast cancer. Breast Cancer Res. 2012;14(5):R124. doi: 10.1186/bcr3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forse CL, et al. Menacalc, a quantitative method of metastasis assessment, as a prognostic marker for axillary node-negative breast cancer. BMC Cancer. 2015;15:483. doi: 10.1186/s12885-015-1468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli J, et al. Invasive breast carcinoma cells from patients exhibit MenaINV- and macrophage-dependent transendothelial migration. Sci Signal. 2014;7(353):ra112. doi: 10.1126/scisignal.2005329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron MD, et al. Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res. 2000;60(9):2541–2546. [PubMed] [Google Scholar]
- Bragado P, Sosa MS, Keely P, Condeelis J, Aguirre-Ghiso JA. Microenvironments dictating tumor cell dormancy. Recent Results Cancer Res. 2012;195:25–39. doi: 10.1007/978-3-642-28160-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husemann Y, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13(1):58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- St Croix CM, Leelavanichkul K, Watkins SC. Intravital fluorescence microscopy in pulmonary research. Adv Drug Del Rev. 2006;58(7):834–840. doi: 10.1016/j.addr.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Al-Mehdi AB, et al. Intravascular origin of metastasis from the proliferation of endothelium-attached tumor cells: a new model for metastasis. Nat Med. 2000;6(1):100–102. doi: 10.1038/71429. [DOI] [PubMed] [Google Scholar]
- Qian B, et al. A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PLoS One. 2009;4(8):e6562. doi: 10.1371/journal.pone.0006562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian BZ, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475(7355):222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearn JT, Barr J, German W. The Behavior of the Arterioles and Capillaries of the Lung. Exp Biol Med. 1926;24(2):114–115. [Google Scholar]
- Terry RJ. A Thoracic Window for Observation of the Lung in a Living Animal. Science. 1939;90(2324):43–44. doi: 10.1126/science.90.2324.43. [DOI] [PubMed] [Google Scholar]
- De Alva WE, Rainer WG. A method of high speed in vivo pulmonary microcinematography under physiologic conditions. Angiology. 1963;14:160–164. doi: 10.1177/000331976301400402. [DOI] [PubMed] [Google Scholar]
- Wagner WW, Jr, Filley GF. Microscopic observation of the lung in vivo. Vasc Dis. 1965;2(5):229–241. [PubMed] [Google Scholar]
- Wagner WW., Jr Pulmonary microcirculatory observations in vivo under physiological conditions. J Appl Physiol. 1969;26(3):375–377. doi: 10.1152/jappl.1969.26.3.375. [DOI] [PubMed] [Google Scholar]
- Groh J, Kuhnle GE, Kuebler WM, Goetz AE. An experimental model for simultaneous quantitative analysis of pulmonary micro- and macrocirculation during unilateral hypoxia in vivo. Res Exp Med. 1992;192(6):431–441. doi: 10.1007/BF02576301. [DOI] [PubMed] [Google Scholar]
- Fingar VH, Taber SW, Wieman TJ. A new model for the study of pulmonary microcirculation: determination of pulmonary edema in rats. J Surg Res. 1994;57(3):385–393. doi: 10.1006/jsre.1994.1159. [DOI] [PubMed] [Google Scholar]
- Lamm WJ, Bernard SL, Wagner WW, Glenny RW. Intravital microscopic observations of 15-micron microspheres lodging in the pulmonary microcirculation. J Appl Physiol. 2005;98(6):2242–2248. doi: 10.1152/japplphysiol.01199.2004. [DOI] [PubMed] [Google Scholar]
- Tabuchi A, Mertens M, Kuppe H, Pries AR, Kuebler WM. Intravital microscopy of the murine pulmonary microcirculation. J Appl Physiol. 2008;104(2):338–346. doi: 10.1152/japplphysiol.00348.2007. [DOI] [PubMed] [Google Scholar]
- Funakoshi N, et al. A new model of lung metastasis for intravital studies. Microvasc Res. 2000;59(3):361–367. doi: 10.1006/mvre.2000.2238. [DOI] [PubMed] [Google Scholar]
- Fiole D, Tournier JN. Intravital microscopy of the lung: minimizing invasiveness. J Biophotonics. 2016. [DOI] [PubMed]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process. 1998;7(1):27–41. doi: 10.1109/83.650848. [DOI] [PubMed] [Google Scholar]
- Ewens A, Mihich E, Ehrke MJ. Distant metastasis from subcutaneously grown E0771 medullary breast adenocarcinoma. Anticancer Res. 2005;25(6B):3905–3915. [PubMed] [Google Scholar]
- Kitamura T, et al. CCL2-induced chemokine cascade promotes breast cancer metastasis by enhancing retention of metastasis-associated macrophages. J Exp Med. 2015;212(7):1043–1059. doi: 10.1084/jem.20141836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A, et al. Technologies for Single-Cell Isolation. Int J Mol Sci. 2015;16(8):16897–16919. doi: 10.3390/ijms160816897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Campbell HM, Dittel BN, Ray A. Purification of specific cell population by fluorescence activated cell sorting (FACS) J Vis Exp. 2010. [DOI] [PMC free article] [PubMed]
- Hauser H, Wagner R, editors. Mammalian cell biotechnology in protein production. Berlin, New York: Walter de Gruyter; 1997. [Google Scholar]
- Lim UM, Yap MG, Lim YP, Goh LT, Ng SK. Identification of autocrine growth factors secreted by CHO cells for applications in single-cell cloning media. J Proteome Res. 2013;12(7):3496–3510. doi: 10.1021/pr400352n. [DOI] [PubMed] [Google Scholar]
- Nielsen BS, et al. A precise and efficient stereological method for determining murine lung metastasis volumes. Am J Pathol. 2001;158(6):1997–2003. doi: 10.1016/S0002-9440(10)64671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov DA, et al. Expression of Gal4-dependent transgenes in cells of the mononuclear phagocyte system labeled with enhanced cyan fluorescent protein using Csf1r-Gal4VP16/UAS-ECFP double-transgenic mice. J Leukoc Biol. 2008;83(2):430–433. doi: 10.1189/jlb.0807585. [DOI] [PubMed] [Google Scholar]
- Entenberg D, et al. Setup and use of a two-laser multiphoton microscope for multichannel intravital fluorescence imaging. Nat Protoc. 2011;6(10):1500–1520. doi: 10.1038/nprot.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harney AS, Condeelis J, Entenberg D. Extended time-lapse intravital imaging of real-time multicellular dynamics in the tumor microenvironment. J Vis Exp. 2016. p. e54042. [DOI] [PMC free article] [PubMed]
- Das S, MacDonald K, Chang HY, Mitzner W. A simple method of mouse lung intubation. J Vis Exp. 2013. p. e50318. [DOI] [PMC free article] [PubMed]
- DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4(7):1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entenberg D, et al. Imaging tumor cell movement in vivo. Curr Protoc Cell Biol. 2013;Chapter 19:Unit 19.7. doi: 10.1002/0471143030.cb1907s58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsialou A, et al. Intravital multiphoton imaging reveals multicellular streaming as a crucial component of in vivo cell migration in human breast tumors. Intravital. 2013;2(2):e25294. doi: 10.4161/intv.25294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Verkman AS. Analysis of organ physiology in transgenic mice. Am J Physiol Cell Physiol. 2000;279(1):C1–C18. doi: 10.1152/ajpcell.2000.279.1.C1. [DOI] [PubMed] [Google Scholar]