Abstract
SapB is a morphogenetic peptide that is important for aerial mycelium formation by the filamentous bacterium Streptomyces coelicolor. Production of SapB commences during aerial mycelium formation and depends on most of the genes known to be required for the morphogenesis of aerial hyphae. Furthermore, the application of purified SapB to mutants blocked in morphogenesis restores their capacity to form aerial hyphae. Here, we present evidence that SapB is a lantibiotic-like peptide that is derived by posttranslational modification from the product of a gene (ramS) in the four-gene ram operon, which is under the control of the regulatory gene ramR. We show that the product of another gene in the operon (ramC) contains a region that is similar to enzymes involved in the biosynthesis of lantibiotics, suggesting that it might be involved in the posttranslational processing of RamS. We conclude that SapB is derived from RamS through proteolytic cleavage and the introduction of four dehydroalanine residues and two lanthionine bridges. We provide an example of a morphogenetic role for an antibiotic-like molecule.
Aspecial feature of the developmental cycle of the filamentous soil bacterium Streptomyces coelicolor is the formation, at the start of differentiation, of an aerial mycelium. This structure consists of hyphae that grow up from the substrate mycelium into the air (1, 2). SapB is a small, secreted morphogenetic peptide intimately involved in this process when growth occurs on complex medium (3). It functions as a biological surfactant, releasing the surface tension at the air-water interface, thereby allowing the hyphae to escape the aqueous milieu of the colony surface and grow upright (4). When they are formed, the multigenomic aerial hyphae, which impart a characteristic white fuzzy appearance to the developing colonies, differentiate into long chains of uninucleoid exospores.
Genetic dissection of the developmental program in S. coelicolor reveals that at least two classes of genes are required for the formation of an aerial mycelium, the bld (as reviewed in ref. 5) and the ram genes (6-9). All bld mutants are blocked in the ability to form aerial hyphae, and all (with the exception of bldF) regain the ability to form aerial structures upon the extracellular addition of SapB (3). However, these aerial filaments fail to metamorphose into chains of spores, implying that SapB functions only in a structural capacity (4). Another class of secreted hydrophobic proteins important for the erection of aerial hyphae is the chaplins (10, 11).
The S. coelicolor ram genes were discovered by their ability to induce rapid aerial mycelium formation when introduced on a low-copy plasmid into the closely related species Streptomyces lividans (6). Subsequently, several other investigators independently cloned these genes based on their ability to overcome the dependence of S. lividans morphogenesis on copper (12), and by their ability to rescue aerial hyphae formation by several S. coelicolor bld mutants (9). The ram gene cluster consists of four genes, ramCSAB, which appear to constitute an operon (7-9), and the convergently transcribed gene ramR (Fig. 1A). RamR is a response regulator that has been shown to bind to the ramC promoter to activate transcription of ramCSAB (7, 8). A ramR mutant is blocked in SapB production and is delayed in morphogenesis (9). Conversely, overexpression of ramR results in the overproduction of SapB and the biosynthesis of SapB by WT strains under conditions when peptide production is normally repressed (9). The N terminus of ramC shows similarity to Ser Thr protein kinases (13), whereas ramAB encode components of an ATP-binding cassette transporter (6). The ramS gene product is predicted to be a 42-aa peptide of unknown function. A homologous amf gene cluster exists in Streptomyces griseus (14), and null mutants for both ramS (9) and its homolog, amfS (15), display a Bld phenotype. Interestingly, an octapeptide corresponding to the C-terminal end of the predicted AmfS peptide extracellularly rescues aerial hyphae formation in the amfS null strain, although only weakly and at much higher concentrations than those at which SapB is active (15).
Fig. 1.
The ram gene cluster and sequence analysis of ramC.(A) The arrangement of the five genes in the ram cluster of S. coelicolor.(B) Amino acid alignment of the C terminus of RamC of S. coelicolor with the C termini of the lantibiotic modification enzymes CinM and MrsM, from Streptomyces cinnamoneus and Bacillus spp., respectively. Dashed lines indicate conserved sequences commonly found in Lan cyclases.
SapB was described in 1991 as a spore-associated peptide with a molecular mass of 2,027 Da (3). Edman degradation of SapB was blocked after the fifth cycle, and it was reported to have 18 amino acids and to be rich in hydrophobic residues (3). Until now, the genes that specify the biosynthesis of SapB have resisted identification. However, we recently discovered that the inferred product of ramC contains a region of similarity to enzymes involved in the biosynthesis of lantibiotics. Lantibiotics are ribosomally synthesized oligopeptide antibiotics produced by Gram-positive bacteria that are translated as inactive prepeptides, which undergo extensive posttranslational modification before being cleaved to yield the mature peptide (16). This observation raised the possibility that SapB might be a lantibiotic-like peptide derived from the product of ramS by RamC-mediated posttranslational modification. The formation of lantibiotics involves the dehydration of Ser and Thr residues to generate didehydroalanine (Dha) and didehydrobutyrine (Dhb), respectively. Dha and Dhb residues then react with the sulfhydryl groups of nearby Cys residues to form acid-stable thioether lanthionine (Lan) bridges and 3-methyllanthionine (MeLan) bridges, respectively. Because the number of Cys residues is usually less than that of Dha and Dhb, most lantibiotics contain some unreacted Dha and Dhb residues, which are known to block Edman degradation (17). Indeed, even after chemical modification that permits Edman degradation, only partial amino acid sequence information is generally obtained. Thus, the primary amino acid sequence and the extent of posttranslational modification of most lantibiotic peptides have been resolved by combining structural and genetic information (16). We have used such an approach to determine that SapB has a lantibiotic-like structure and is encoded by ramS.
Materials and Methods
Strains, Plasmids, and Culture Conditions. S. coelicolor J1501/pKN22 (9) was the source of SapB and was grown on R2YE agar medium (18) containing apramycin sulfate (50 μg/ml; Sigma) at 30°C.
Purification of SapB. Spores were collected, and a crude preparation of spore-associated proteins was prepared as described (19). SapB was purified from this material by preparative 2D gel electrophoresis. Preparative precast isoelectric focusing (IEF) gels (pH 3-10), were used for the first dimension. Peptide was eluted from the gels as described in ref. 19 and loaded onto precast 16% Tris-tricine gels for the second dimension. SapB was again eluted as described (19). Precast gels were purchased from Bio-Rad.
Chemical Modification of SapB for Structural Analysis. Mild acid hydrolysis was performed at 95°C in 6 M HCl for 60 min. The hydrolysate was neutralized to pH 7.0 and evaporated. The dried material was then redissolved in 20% methanol and subjected to RP-HPLC to yield the internal fragment. For HPLC analysis and fragment preparation, a Luna C8 (4.6 × 250 mm) column was used with an elution gradient of MeCN/H2O/trifluoroacetic acid (from 10:90:0.1 to 90:10:0.1) over 30 min. Material was detected at 210 nm.
Before Edman degradation, SapB was treated with ethanethiol under alkaline conditions as described in ref. 20. For reduction of Dha residues to Ala, SapB was treated with sodium borohydride at pH 8 (17). Native peptide and sodium borohydride treated material was analyzed by time-of-flight (TOF) MS.
Amino Acid Analysis. SapB (15 μg) and an internal fragment (6 μg), obtained by acid hydrolysis, were dried and hydrolyzed in gas phase 6 M HCl for 20 h at 110°C. After hydrolysis, the samples were again dried before resuspending in sample buffer. Amino acid analyses were performed on a Hewlett Packard 1090 instrument with AMINO QUANT (Agilent Technologies) chemistry software.
N Terminus Protein Sequencing. SapB was sequenced by using Applied Biosystems 492 Protein Sequencing system by using standard cycles. All reagents and solvents were purchased from Applied Biosystems.
Matrix-Assisted Laser Desorption Ionization (MALDI)-TOF Spectrometry. SapB samples were run on a Voyager-DE STR (Applied Biosystems) MALDI-TOF MS system operated in the reflector mode. Samples were dissolved in a 50% solution of ANC/0.1% trifluoroacetic acid containing α-cyano-4-hydroxy cinammic acid (5 mg/ml) and dried on the sample plate. A nitrogen laser operating at 337 nm and a 3-ns pulse rate was used. The accelerating voltage was set at 20 kV, and a delay of 220 ns was used to accelerate ions into the flight tube of the mass spectrometer. The mass scale (m/z 500-5,000) was calibrated with a mixture of peptides, and ≈100 laser shots were used to produce each spectrum. For an accurate MS measurement of SapB molecular mass, the sample spectra were internally calibrated with the peptides Bradykinin and ACTH. The accurate mass measurement of the SapB internal fragment was carried out on the Qstar Pulsar i mass spectrometer (Applied Biosystems) with an OMALDI2 source by using external calibration. α-Cyano-4-hydroxy cinammic acid was used as the matrix. The mass resolution of the instrument was set at 9,000.
Molecular Modeling. SapB molecular modeling was carried out by using insight ii software (21). The model was constructed within the Biopolymer module and energy minimized within the Discover module, using hydrogens calculated at pH 7 and charged groups as appropriate, with a spherical water shell of radius 20 Å and the consistent valence force field. The model was validated with prostat and swisspdb viewers (www.expasy.ch/spdbv/mainpage.html) to check the Ramachandran plot.
Results and Discussion
The C Terminus of RamC Is a Possible SapB Synthetase. RamC is membrane-associated and has an N-terminal domain that resembles a Ser/Thr kinase (22) and a central dimerization domain (13). Genome database searches with the sequence of the RamC C terminus, independent of the other two domains, revealed that this domain is similar to proteins involved in the maturation of lantibiotic peptides. An alignment of the RamC C-terminal domain with the related C termini of CinM and MrsM (involved in the production of cinnamycin and mersacidin, respectively) is shown in Fig. 1B and revealed that the RamC sequence possesses seven of eight conserved motifs characteristic of these proteins. In CinM and MrsM, this domain catalyzes the cyclization step in Lan production (23), and although the amino acid residues that catalyze this reaction are not well understood, the overall similarity between the three proteins suggests that the C terminus of RamC may be a Lan cyclase.
The apparent relationship of RamC to the lantibiotic-producing enzymes suggested that the substrate of RamC might be RamS, the product of the ORF located immediately downstream of ramC in the ram gene cluster (Fig. 1A). The putative amino acid sequence of this peptide includes six Ser, four Thr, and two Cys residues (Fig. 2), suggesting that it might be a good substrate for the introduction of lantibiotic-like modifications. We note also that the two Cys residues and the five C-terminal Ser residues are conserved in the closely related AmfS peptide of S. griseus (Fig. 2), arguing that they might be important for function. Although there was little agreement between the translated RamS amino acid sequence and the apparent amino acid content of SapB (3), more recent attempts at Edman sequencing of SapB revealed the N-terminal sequence T-G-X-R-A-block, a possible match for the sequence of RamS starting at residue 22 (Fig. 2). We reasoned that SapB might be posttranslationally modified RamS that had been cleaved between positions 21 and 22 (Fig. 2). Furthermore, it seemed possible that the block in Edman degradation of SapB after the fifth cycle might be caused by dehydration of the Ser residue in position 6 to Dha (residue 27 of RamS). Finally, the molecular mass of the 21 C-terminal residues in RamS is 2,099 Da, 72 Da heavier than SapB. We postulated that the dehydration of four residues, presumably Ser and/or Thr, might account for the mass discrepancy.
Fig. 2.
Alignment of RamS of S. coelicolor with its ortholog AmfS of S. griseus. Also shown is the amino acid sequence of SapB obtained by Edman degradation after reduction with ethanethiol under alkaline conditions (20).
Structural Analysis of SapB. To determine whether SapB was a RamS-derived, Lan-containing peptide, we treated it with the reagent ethanethiol under alkaline conditions. This procedure results in the conversion of Dha and Dhb to stable S-ethylcysteinyl and β-methyl-S-cysteinyl derivatives, respectively, and Lan and MeLan bridges are also derivatized. These modifications render lantibiotic peptides susceptible to more complete Edman degradation and sequence determination (20). Indeed, such chemical modification eliminated the block in Edman degradation and permitted the determination of a partial SapB primary sequence (Fig. 2). Significantly, the 11 residues obtained aligned with the C terminus of RamS.
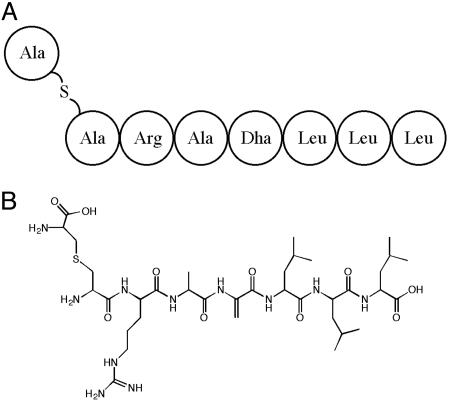
In parallel work, we carried out mild acid hydrolysis on SapB and purified an internal fragment of the peptide having a monoisotopic molecular mass of 844.4731 Da for the protonated peptide [M + H]+ (Fig. 3). Complete hydrolysis of this peptide followed by the determination of amino acid content revealed the presence of Arg (1), Ala (1), and Leu (3), with numbers in parentheses indicating the relative number of each residue. These five residues alone would yield a mass of 585.4088 Da, leaving 259.0643 Da, which we presumed corresponded to nonproteogenic residues that could not be identified. In fact, an unidentifiable molecule with the same stoichiometric ratio as that of Arg and Ala was recovered. When compared with a Lan standard, we found that LL-Lan had a retention time of 8 min, whereas that of the unknown molecule was 8.1 min. Although this result did not confirm the presence of Lan in the fragment, the theoretical addition of a Lan and a Dha residue would add 259.0627 Da. This would result in a calculated mass of 844.4715 Da (C36H66N11O10S), which is very close to the measured mass of 844.4731 Da. Consistent with this, N-terminal sequencing of this fragment by Edman degradation yielded the sequence X-R-A-Block, suggesting that the first nonambiguous residues in the fragment might correspond to Arg-Ala at positions 4 and 5 in SapB (residues 24 and 25 in RamS). If so, we reasoned that the ambiguous N-terminal residue could be Lan and the block at position 4 might be due to dehydration of the Ser in that position in RamS to Dha in SapB. Unlike Lan, which generates unidentifiable Edman products but does not block degradation, the presence of the unsaturated bond between the α and β carbons of Dha causes a block in the progression of Edman degradation (17, 24). Finally, the mass of this fragment was consistent with the predicted sequence Lan-R-A-Dha-L-L-L (Fig. 3). It was not immediately clear whether the Cys residue that gave rise to the presumed Lan was SapB residue 10 or 20 (residue 31 or 41 in RamS), however subsequent evidence was more consistent with the former.
Fig. 3.
Proposed primary (A) and covalent (B) structure of an internal SapB fragment generated by acid hydrolysis. Lan is the first and only S containing residue, whereas Dha (residue 4) is distinguishable by the double bond between the α and β carbons.
Amino acid composition analysis of full-length SapB revealed the presence of As(x) (2), Ser (1), Gly (2), Thr (3), Ala (1), Arg (1), Ile (1), and Leu (4). These data agree with the notion that SapB is a Lan-containing peptide derived from RamS. Of these amino acids, both Gly residues, the Ala, Arg, Ile, all four Leu residues, and one of the three Thr residues correspond to the Edman degradation sequence shown in Fig. 2. One of the two As(x) residues may represent the Asp at position 12 of SapB (position 33 of RamS), whereas the second As(x) could be the C-terminal Asn residue; although Edman degradation results were not reliable past the 17th cycle. No Cys residues were detected, however amino acid analysis will not detect Dha, Dhb, or Cys involved in Lan and/or MeLan bridges (24). This result was not in complete agreement with previously reported amino acid analysis (3) (presumably because of impurities in the original SapB preparation), but importantly, both analyses detected one Ser and three Thr residues. Because the C-terminal end of RamS has five Ser and three Thr residues, we inferred that none of the Thr residues is modified, but four of the Ser residues are dehydrated to Dha in the mature peptide. Edman degradation showed an unambiguous Ser at position 14 of SapB (Ser-35 of RamS), and we therefore deduced that Ser in positions 3, 6, 13, and 16 of SapB (corresponding to positions 24, 27, 34, and 37 of RamS) are dehydrated to Dha, accounting for the 72-Da discrepancy between RamS and SapB.
To verify that four of the five Ser residues were dehydrated to Dha in SapB, we reduced SapB with sodium borohydride (17). In this approach, Dha residues present in the peptide are reduced to Ala (with an associated 2-Da increase in mass for each reduction); those Dha residues that form Lan remain unmodified. If two of the four Dha residues formed Lan with the two Cys residues (see below), we predicted that this reduction of SapB should increase the monoisotopic mass of the peptide by 4 Da, from 2,027 to 2,031. Indeed, this mass increase was observed (data not shown), indicating that SapB contained two Dha residues.
Reduction of Dha to d-Ala is found in some lantibiotic peptides (16) but would not be in agreement with the accurate MALDI-TOF mass data, which determined that SapB has a monoisotopic molecular mass of 2,026.9797 ± 0.0109 Da (n = 6), which is very close to the calculated mass of 2,026.9690 Da (C84H140N25O29S2). Indeed, we have no evidence of any modification other than Ser dehydration followed by Lan cyclization. Lan cyclization does not change the peptide mass.
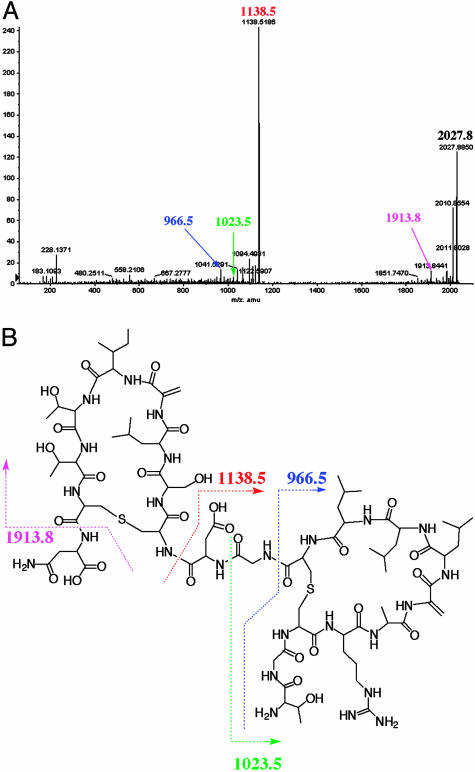
Further evidence of the structure of SapB was obtained by MALDI-TOF tandem MS (Fig. 4). The ion peak at m/z 2,027.8850 Da corresponded to SapB itself. Other ion peaks helped identify linear regions of the molecule, which are susceptible to fragmentation (in contrast to cyclic regions). For example, the ion peak at m/z 1,913.8 Da would correspond to cleavage between Cys/Lan-20 and Asn-21. Similarly, ion peaks were identified that would correspond to cleavage between Asp-12 and Dha/Lan-13 (m/z 1,138.5186 Da), Gly-11 and Asp-12 (m/z 1,023.5 Da), and Cys/Lan-10 and Gly-11 (m/z 966.5 Da). Thus, with the exception of the two N-terminal residues, ion species were detected corresponding to cleavage at each of the peptide bonds in the proposed linear regions of SapB. Notably, we did not obtain any detectable ion peaks that would correspond to fragmentation within either of the two proposed loop structures.
Fig. 4.
MALDI-TOF tandem MS analysis of SapB (A) and the proposed covalent structure of SapB (B). The cleavage site indicated by the pink arrow in the covalent structure (B) would yield a 1,913.8-Da fragmentation product, corresponding to the minor peak indicated by the pink arrow in A. Other fragmentation products are similarly labeled in both the chromatogram and covalent structure.
We attempted on several occasions to obtain additional structural information by NMR; however, this work was hampered by SapB aggregation and insolubility. Indeed, even the synthetic peptide corresponding to RamS residues 22-42, although soluble in 100% trifluoroacetic acid, was resolutely insoluble in other conditions.
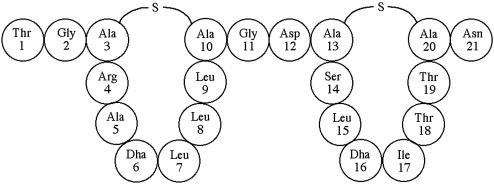
Taken together, these data suggest that mature SapB has the structure shown in Figs. 4B and 5, with two eight residue loops formed by Lan bridges between Cys and Dha residues. The placement of the second Lan is not certain; however, the absence of fragmentation between Dha-13 and Dha-16 during MALDI TOF tandem MS suggests that Cys-20 forms a Lan bridge with Dha-13 rather than Dha-16. The proposed configuration would result in a hydrophobic N-terminal loop and a less hydrophobic C-terminal loop connected by a Gly-Asp linker.
Fig. 5.
Proposed primary structure of SapB. The deduced bridging pattern yields two cyclic structures of eight residues, most of which are hydrophobic. The loops are flanked and hinged by less hydrophobic amino acids.
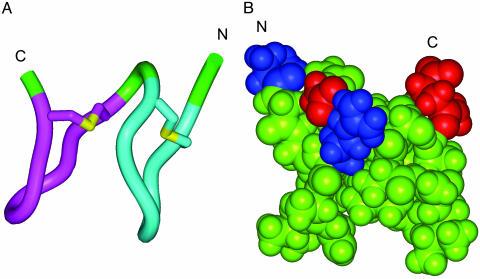
Molecular Modeling of SapB Supports an Amphiphilic Structure. To better understand the proposed structure of SapB, we carried out ab initio structural modeling by using insight ii (21), and the model is shown in Fig. 6. We found that the constraints imposed by the eight-residue rings mean that some of the hydrophobic amino acid side chains must be surface-exposed. The number of these unfavorable hydrophobic surface contacts could be reduced by stacking the two rings, using Gly-11 as a flexible linker (it should be noted, however, that the two rings could readily be more widely separated by ϕ/ψ rotations of this residue). As shown in Fig. 6, our model has a distinctly amphiphilic structure, with the charged side chains (Arg-4 and Asp-12) and the charged C and N termini aligned across one face of the molecule, and the other face dominated by surface-exposed hydrophobic side chains. SapB can reduce the surface tension of water from 72 to 27 mJ/m2 and appears to aggregate at the air-water interface (4, 25). We propose that the amphiphilic nature of SapB suggested by the model would be vital for its surfactant activity, which allows hyphae to escape from the aqueous environment of the colony surface into the air. The hydrophilic side of SapB would easily be solvated, whereas the hydrophobic side could produce a lower energy state by escaping water through projection at the air-water interface. Such alignment at the air-water boundary might also lead to self-assembly through the formation of large, stable hydrophobic interfaces between SapB monomers. A similar model also has been proposed for multimerization of the fungal surfactants, the hydrophobins, based on the 3D structure of hydrophobin HFBII (26).
Fig. 6.
Molecular model of SapB. (A) Schematic representation of the model. The C- and N-terminal macrocyclic loops are colored magenta and cyan, respectively, and the thioether (Lan bridge) sulfurs are colored yellow. (B) Spacefilling representation of the model. The view is rotated by 180° with respect to A, as indicated by the C and N termini. The basic N-terminal group and Arg-4 side chain are colored blue, and the acidic C-terminal group and Asp-12 side chain are colored red.
SapB Is an Unusual Lan-Containing Peptide. Two structural classes of lantibiotics have been described. Type A lantibiotics are elongated and flexible, whereas type B peptides are more globular. Based on the length of SapB and the potential for only two Lan bridges, SapB appears to resemble a type A lantibiotic-like peptide (16). There are several features that make SapB unlike other Lan-containing peptides. The typical features of type A lantibiotic N-terminal leader peptides are not apparent in RamS. Specifically, the RamS leader peptide is not particularly hydrophilic, and it lacks either of two characteristic cleavage sites. The RamS leader peptide resembles that of the type A lantibiotics only in that it possesses a high proportion of charged amino acids (9 of 21) and has a net negative charge (-3).
Type A peptides exert their antimicrobial effects by forming pores in the plasma membrane of the target cell. This activity depends on an amphiphilic configuration, with their hydrophobic and charged side chains aligning on opposing sides of the peptide (16). The biological activity of SapB is also thought to rely on the formation of amphiphilic structures, and the assembly of SapB on the growing aerial hyphae is thought to at least partially account for the hydrophobicity of these structures (4, 25). Although the activity of both SapB and type A lantibiotics share the requirement for amphiphilicity, SapB differs from all lantibiotics in that it is insoluble, because it readily self-aggregates (4). Also, whereas all previously characterized lantibiotic peptides function as antibiotics, some with minimum inhibitory concentration in the nanomolar range (27), SapB did not exhibit antibiotic activity. Although standard minimum inhibitory concentration assays were not possible because of the insolubility of SapB in aqueous medium, there was no inhibition of growth when 5 μg of the peptide was applied directly to growing colonies of Escherichia coli, Salmonella typhimurium, Bacillus subtilis, Bacillus cereus, Mycobacterium smegmatis, or Staphylococcus aureus. Thus, although SapB is certainly not the first lantibiotic-like peptide of streptomycete origin (16, 28), to our knowledge it is the first such peptide that fails to demonstrate antibiotic activity.
Based on the similarity of RamC to known lantibiotic synthetases, we propose that RamC is involved in the processing of RamS into SapB, and we further propose that SapB is exported from the cell by the ATP-binding cassette transporter encoded by ramAB. Whether all of the enzymatic activities required to convert RamS into mature SapB-Ser dehydration, Lan bridge formation, and proteolytic processing—are specified by RamC alone remains to be determined.
SapB Production Appears to Precede That of Other Morphogens. The chaplins are a family of hydrophobic cell wall-associated proteins that confer hydrophobicity to aerial hyphae and spores and strains lacking multiple chaplin genes fail to form aerial hyphae (10, 11). Chaplin production is blocked in all bld mutants tested, including bldN and bldM (11). In contrast, neither bldM nor bldN are required for ramC expression (7), suggesting that perhaps the action of SapB takes place before the production and localization of the chaplins. Additional mutant analysis and localization studies are needed to fully elucidate the relationship between SapB and the chaplins.
Finally, it is remarkable that streptomycetes, the source of most medicinally important antibiotics, use a compound resembling an antimicrobial peptide to drive a central developmental event. Similarly, the gray spore pigment of S. coelicolor, specified by the complex whiE locus, is a type II polyketide structurally and biosynthetically related to a major class of streptomycete antibiotics (5). Given the ubiquitous presence of ram and whiE clusters in streptomycete species and their central involvement in differentiation, it seems likely that these genes would have been present in the common ancestor of the genus. It is therefore possible that streptomycete lantibiotic and type II polyketide antibiotic clusters may have evolved from these developmental genes.
Acknowledgments
We thank Thomas Fischer, Robert Rieger, and Weiping Xie of the Proteomics Center at Stony Brook University; Prof. David Lloyd of the Department of Chemistry at Hofstra University; and Ms. Bonnie Yanchisin at Commonwealth Biotechnologies, Inc., for excellent technical assistance. We also thank Andrew Bottrill and Marie Elliot (John Innes Centre) for helpful discussions. S.K. and J.M.W. were funded by National Science Foundation Grant MCB-0211974. M.E.H. and J.R.N. were supported by Grant MOP15108 from the Canadian Institutes for Health Research.
Abbreviations: Dha, didehydroalanine; Dhb, didehydrobutyrine; Lan, lanthionine; MeLan, 3-methyllanthionine; MALDI-TOF, matrix-assisted laser desorption ionization-time of flight.
References
- 1.Kelemen, G. H. & Buttner, M. J. (1998) Curr. Opin. Microbiol. 1, 656-662. [DOI] [PubMed] [Google Scholar]
- 2.Chater, K. F. & Horinouchi, S. (2003) Mol. Microbiol. 48, 9-15. [DOI] [PubMed] [Google Scholar]
- 3.Willey, J. M., Santamaria, R., Guijarro, J., Geistlich, M. & Losick, R. (1991) Cell 65, 641-650. [DOI] [PubMed] [Google Scholar]
- 4.Tillotson, R. D., Wösten, H. A. B., Richter, M. & Willey, J. M. (1998) Mol. Microbiol. 30, 595-602. [DOI] [PubMed] [Google Scholar]
- 5.Chater, K. F. (2000) in Prokaryote Development, eds. Brun, Y. V. & Shimkets, L. J. (Am. Soc. Microbiol., Washington, DC), pp. 33-48.
- 6.Ma, H. & Kendall, K. (1994) J. Bacteriol. 176, 3800-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Conner, T. J., Kanellis, P. & Nodwell, J. R. (2002) Mol. Microbiol. 45, 45-57. [DOI] [PubMed] [Google Scholar]
- 8.Keijser, B. J., van Wezel, G. P., Canters, G. W. & Vijgenboom, E. (2002) J. Bacteriol. 184, 4420-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen, K. T., Willey, J. M., Nguyen, L. T., Viollier, P. H. & Thompson, C. J. (2002) Mol. Microbiol. 46, 1223-1238. [DOI] [PubMed] [Google Scholar]
- 10.Claessen, D., Rink, R. de Jong, W., Siebring, J., de Vreugd, P., Boersma, F. G. H., Dijkhuizen, L. & Wösten, H. A. B. (2003) Genes Dev. 17, 1714-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliot, M. A., Karoonuthaisiri, N., Huang, J., Bibb, M. J., Cohen, S. N., Kao, C. M. & Buttner, M. J. (2003) Genes Dev. 17, 1727-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keijser, B. J., van Wezel, G. P., Canters, G. W., Kieser, T. & Vijgenboom, E. (2000) J. Mol. Microbiol. Biotechnol. 2, 565-574. [PubMed] [Google Scholar]
- 13.Hudson, M. E. & Nodwell, J. R. (2004) J. Bacteriol. 186, 1330-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ueda, K., Miyake, K., Horinouchi, S. & Beppu, T. (1993) J. Bacteriol. 175, 2006-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ueda, K., Oinuma, K., Ikeda, G., Hosono, K., Ohnishi, Y. Horinouchi, S. & Beppu, T. (2002) J. Bacteriol. 184, 1488-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sahl, H. G. & Bierbaum, G. (1998) Ann. Rev. Microbiol. 52, 41-79. [DOI] [PubMed] [Google Scholar]
- 17.Smith, L., Novak, J. Rocca, J., McClung, S., Hillman, J. D. & Edison, A. S. (2000) Eur. J. Biochem. 267, 6810-6816. [DOI] [PubMed] [Google Scholar]
- 18.Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. (2000) in Practical Streptomycetes Genetics (John Innes Foundation, Norwich, U.K.), p. 408.
- 19.Guijarro, J., Santamaria, R., Shauer, A. & Losick, R. (1988) J. Bacteriol. 170, 1895-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer, H. E., Heber, M., Eisermann, B., Korte, H., Metzger, J. W. & Jung, G. (1994) Anal. Biochem. 223, 185-190. [DOI] [PubMed] [Google Scholar]
- 21.Accelrys (2000) insight ii (Accelrys, Cambridge, U.K.), Release 2000.1
- 22.Hudson, M. E., Zhang, D. & Nodwell, J. R. (2002) J. Bacteriol. 184, 4920-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McAuliffe, O., Ross, R. P. & Hill, C. (2001) FEMS Microbial. Rev. 25, 285-308. [DOI] [PubMed] [Google Scholar]
- 24.Mota-Meira, M., Lacroix, C., LaPointe, G. & Lavoie, M. C. (1997) FEBS Lett. 410, 275-279. [DOI] [PubMed] [Google Scholar]
- 25.Wösten, H. A. B. & Willey J. M. (2000) Microbiology 146, 767-773. [DOI] [PubMed] [Google Scholar]
- 26.Hakanpää, J., Paananen, A., Askolin, S., Nakari-Setälä, T., Parkkinen, T., Penttilä, M., Linder, M. B. & Rouginen, J. (2004) J. Biol. Chem. 279, 534-539. [DOI] [PubMed] [Google Scholar]
- 27.Wiedemann, I., Breukink, E., van Kraaij, C., Kuipers, O. P., Bierbaum, G., de Kruijff, B. & Sahl, H.G. (2001) J. Biol. Chem. 276, 1772-1779. [DOI] [PubMed] [Google Scholar]
- 28.Widdick, D. A., Dodd, H. M., Barraille, P., White, J., Stein, T. H., Chater, K. F., Gasson, M. J. & Bibb, M. J. (2003) Proc. Natl. Acad. Sci. USA 100, 4316-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]