Abstract
Free-solution capillary electrophoresis (CE) separates analytes, generally charged compounds in solution through the application of an electric field. Compared to other analytical separation techniques, such as chromatography, CE is cheap, robust and effectively requires no sample preparation (for a number of complex natural matrices or polymeric samples). CE is fast and can be used to follow the evolution of mixtures in real time (e.g., chemical reaction kinetics), as the signals observed for the separated compounds are directly proportional to their quantity in solution.
Here, the efficiency of CE is demonstrated for monitoring the covalent grafting of peptides onto chitosan films for subsequent biomedical applications. Chitosan's antimicrobial and biocompatible properties make it an attractive material for biomedical applications such as cell growth substrates. Covalently grafting the peptide RGDS (arginine - glycine - aspartic acid - serine) onto the surface of chitosan films aims at improving cell attachment. Historically, chromatography and amino acid analysis have been used to provide a direct measurement of the amount of grafted peptide. However, the fast separation and absence of sample preparation provided by CE enables equally accurate yet real-time monitoring of the peptide grafting process. CE is able to separate and quantify the different components of the reaction mixture: the (non-grafted) peptide and the chemical coupling agents. In this way the use of CE results in improved films for downstream applications.
The chitosan films were characterized through solid-state NMR (nuclear magnetic resonance) spectroscopy. This technique is more time-consuming and cannot be applied in real time, but yields a direct measurement of the peptide and thus validates the CE technique.
Keywords: Chemistry, Issue 116, capillary electrophoresis, reaction monitoring, real time, grafting, peptide, chitosan, films, epithelial cells, solid-state nuclear magnetic resonance (NMR) spectroscopy
Introduction
Free solution capillary electrophoresis (CE) is a technique that separates compounds in solutions based on their charge-to-friction ratio1,2. Charge-to-size ratio is often mentioned in the literature, but this simplification does not apply to polyelectrolytes, including polypeptides in this work, and was also shown not to be appropriate for small organic molecules3. CE differs from other separation techniques in that it does not have a stationary phase, only a background electrolyte (usually a buffer). This allows the technique to be robust in its ability to analyze a large range of samples with complex matrices4 such as plant fibers5, fermentation brews6 grafting onto synthetic polymers7, food samples8, and hardly soluble peptides9 without tedious sample preparation and purification. This is especially significant for complex polyelectrolytes which have dissolution issues (such as chitosan10 and gellan gum11) and therefore exist as aggregated or precipitated in solution and have been successfully analyzed without sample filtration. Further, the analysis of sugars in breakfast cereals involved injecting samples with particles of breakfast cereal samples precipitated in water8. This also extends to the analysis of branched polyelectrolytes or copolymers12,13. Extensive work has also been completed in the development of CE techniques specifically for the analysis of proteins for proteomics14, chiral separation of natural or synthetic peptides15 and microchip separations of proteins and peptides16. Since the separation and analysis take place in a capillary, only small volumes of sample and solvents are used which enables CE to have a lower running cost than other separation techniques including chromatography5,6,17. Since the separation by CE is fast, it allows the monitoring of reaction kinetics. This was demonstrated in the case of the grafting of peptides onto chitosan films for improved cell adhesion18.
Chitosan is a polysaccharide derived from the N-deacetylation of chitin. Chitosan films can be used for various biomedical applications such as bioadhesives19 and cell growth substrates18,20, due to chitosan's biocompatibility21. Cell attachment to specific extracellular matrix proteins, such as fibronectin, collagens and laminin, is directly linked to the survival of the cells22. Notably, different cell types often require attachment to different extracellular matrix proteins for survival and proper function. Cell attachment to chitosan films was shown to be enhanced through the grafting of fibronectin23; however, preparation, purification and grafting of such large proteins is not economically viable. Alternately a range of small peptides have been shown to be able to mimic the properties of large extracellular matrix proteins. For example, peptides such as the fibronectin mimetics RGD (arginine - glycine - aspartic acid) and RGDS (arginine - glycine - aspartic acid - serine) have been used to facilitate and increase cell attachment24. Covalent grafting of RGDS onto chitosan films resulted in improved cell attachment for cells known to attach to fibronectin in vivo18. Substituting larger proteins likes fibronectin with smaller peptides that have the same functionality provides a significant cost reduction.
Here, peptide grafting to chitosan was performed as previously published18. As previously demonstrated, this approach provides simple and efficient grafting by using the coupling agents EDC-HCl (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide) and NHS (N-hydroxysuccinimide) to functionalize the carboxylic acid of the RGDS to be grafted onto the chitosan film. Two advantages of this grafting method are that it does not require any modification of the chitosan or of the peptide, and it is undertaken in aqueous medium to maximize compatibility with future cell culture applications18,20. As the coupling agents and the peptide can be charged, CE is a suitable method for the analysis of the reaction kinetics. Importantly, analysis of the reaction kinetics via CE enables real-time monitoring of the grafting reaction, and thus enables both optimizing and quantifying the degree of grafting.
While it is not routinely necessary, the results of the CE analysis can be validated off-line by a direct measurement of the peptide grafting onto the chitosan films using solid-state NMR (nuclear magnetic resonance) spectroscopy25,26 to demonstrate the covalent grafting of the peptide onto the film18. However, compared with solid-state NMR spectroscopy, the real-time analysis provided by CE enables the quantification of the peptide consumption in real time and thus the ability to assess the kinetics of the reaction.
The above mentioned method is simple and allows the real-time analysis of peptide grafting onto chitosan films with indirect quantification of the extent of the grafting. The demonstrated method can be extended to the real time quantitative assessment of different chemical reactions as long as the reactants or the products to be analyzed can be charged.
Protocol
1. Preparation of Chitosan Films
Weigh out 2 g of glacial acetic acid, complete to 100 ml with ultrapure water.
Weigh out 1.7 g of chitosan powder, add 100 ml of the 2% m/m acetic acid aqueous solution. Stir for 5 days with stirring bar and magnetic stirring plate at room temperature either covered with aluminum foil or in the dark.
Centrifuge the chitosan dispersion at 1,076 x g at 23 °C for 1 hr. Collect the supernatant with a syringe and discard the precipitate.
For each film, aliquot 10 ml of the chitosan suspension into a 9 cm plastic Petri dish at room temperature. Leave the films covered to dry for at least 7 days.
Using scissors cut the dry films into 1 x 1 cm squares. Note: The experiment can be paused at this stage.
2. Preparation of Phosphate-buffered Saline (PBS)
Weigh out 8 g sodium chloride, 0.2 g potassium chloride, 1.44 g disodium hydrogen phosphate and 0.24 g potassium dihydrogen phosphate.
Dissolve these weighed out chemicals in 800 ml of ultrapure water and titrate the solution with concentrated hydrochloric acid to pH 7.4. Note: The experiment can be paused at this stage.
3. Preparation of 75 mM Sodium Borate Buffer at pH 9.2
Weigh out 3.0915 g of boric acid. Dissolve it in 75 ml of ultrapure water.
Titrate the boric acid solution to a pH of 9.2 with a sodium hydroxide solution at a concentration of 10 M or higher. Caution: Concentrated sodium hydroxide solutions are corrosive and should be handled with gloves.
Complete with ultrapure water to obtain 100 ml of solution. This yields a 500 mM sodium borate buffer at pH 9.2.
Dilute the 500 mM sodium borate buffer with ultrapure water to 75 mM sodium borate buffer. Note: The experiment can be paused at this stage.
4. Preparation of Chitosan Films for the Grafting Reaction
Rinse 10 square chitosan films (1 x 1 cm) in 5 ml of PBS for 2 hr in a Petri dish at room temperature.
During this time, prepare and validate the capillary electrophoresis instrument (step 5).
5. Preparation and Validation of the Capillary Electrophoresis Instrument
- Prepare a 43.5 cm bare fused silica capillary with an internal diameter of 50 µm (43.5 cm is the total length, the effective length to the detection window is typically 35 cm) by weakening the polymer outer coating of the capillary at the set length with a blunt utensil then snapping the capillary.
- Create a window for the capillary by using a lighter to burn the polymer coating at 8.5 cm from the inlet and after it cools wipe it clean with ethanol. Burn the coating of the capillary at each end for a few millimeters with a lighter, and after it cools wipe it clean with ethanol.
- Place capillary inside detection window and install it in the capillary cassette by placing it at equal lengths in the inlet and outlet and winding it around the spindles of the cassette. Then install the cassette in the capillary electrophoresis instrument.
- Set the parameters of the method for each separation. In the software menu select "method" then "edit the entire method". Set the temperature, time, voltage, and vials used for the separation (for example 25 °C, 10 min, 30 kV).
- In the pre-conditioning section, set the consecutive flushes: 10 min with 1 M sodium hydroxide (in water), 5 min with 0.1 M sodium hydroxide (in water), 5 min with ultrapure water and 5 min with 75 mM sodium borate buffer at pH 9.2 for the first method of a series of analyses.
- For the subsequent methods, set the set the consecutive flushes in the pre-conditioning section: 1 min with 1 M sodium hydroxide (in water), 5 min with 75 mM sodium borate buffer at pH 9.2.
- In the injection section, set parameters for a hydrodynamic injection with 30 mbar pressure for 10 sec for all methods. In the separation section, set the separation conditions to 30 kV at 25 °C for 9 min for all methods. NOTE: Consult user manual of specific CE instrument as procedure for operating the CE instrument may vary between manufacturers. Prepare the 1 M sodium hydroxide solution on the day.
Inject and separate a neutral internal standard (10 µl of 10% v/v dimethylsulfoxide (DMSO), in water diluted into 450 µl of 75 mM sodium borate buffer). Then inject and separate in the same way an oligoacrylate standard (dissolved in ultrapure water at 10 g∙L-1; see List of Materials) to check the validity of the capillary. Pause the sequence here until the grafting reaction is ready to start.
6. Grafting of RGDS onto Chitosan Film
Weigh out the peptide (1 mg RGDS) and the coupling agents (3 mg EDC-HCl and 2 mg NHS).
- 2 hr after the start of the chitosan film soaking in PBS, dissolve the peptide and the coupling agents in 5 ml of PBS.
- Take a 50 µl aliquot of this solution. Add 2 µl of 10% v/v DMSO in water as an internal neutral standard to the aliquot. Analyze the aliquot with CE (see step 7).
Remove the 5 ml of PBS used to rinse the chitosan films from the Petri dish. Add the 5 ml solution of peptide and coupling agents to the Petri dish containing the chitosan films.
- Cover the Petri dish with paraffin film and place it on an orbital shaker at room temperature. Take 50 µl aliquots of reaction media at set times. NOTE: The total analysis time with CE is 15 min, thus an aliquot can be taken every 15 min (or every 30 min if two reactions are monitored in parallel, etc.).
- Add 2 µl of 10% v/v DMSO in water as an internal neutral standard to each aliquot. NOTE: Aliquots should be analyzed with CE as soon as they are taken (see step 7).
After 4 hr of shaking and aliquot removal, remove the Petri dish from the shaker. Remove the reaction medium from the Petri dish. Add 5 ml of PBS to rinse the chitosan films.
Remove the PBS from the Petri dish, rinse the chitosan film with ultrapure water and allow them to dry overnight. Remove the ultrapure water and store the films at -20 °C in a plastic Petri dish.
7. Monitoring of Grafting Reaction Using CE
Inject and separate aliquots of reaction media immediately after removal from the Petri dish using the analysis conditions as in section 5.2.
Upon completion of the separations rinse the capillary with ultrapure water for 10 min. Dry it through a flush with an empty vial (air) for 10 min. NOTE: The experiment can be paused at this stage.
8. Data Treatment for CE
Check the validity of each separation, by checking that both the current during the separation and the migration time of the electroosmotic mobility marker (DMSO in this case) are similar to the ones observed for the oligoacrylate standard separation. NOTE: Up to 10-15% variation is acceptable from the expected current value of about 50 µA and migration time value of 1.3 min (electrophoretic mobility values should be used instead of migration times if a higher repeatability is required).
For each successful separation, export the raw data from the capillary electrophoresis software by selecting a specific data set, right clicking on export and selecting an appropriate signal.
Convert the raw data recorded by the CE (presented as UV absorbance as a function of migration time). Convert the X-axis (migration time tm) into an electrophoretic mobility µ following Equation 1:
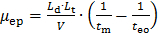 (1) where Ld is the length to the detector, Lt is the total length of the capillary, V is the voltage, and teo is the migration time of a neutral specie (the DMSO internal standard in this case)27. Convert the Y-axis of the raw data (absorbance in a.u.) to a distribution of electrophoretic mobilities W(µ) following Equation 2:28
(1) where Ld is the length to the detector, Lt is the total length of the capillary, V is the voltage, and teo is the migration time of a neutral specie (the DMSO internal standard in this case)27. Convert the Y-axis of the raw data (absorbance in a.u.) to a distribution of electrophoretic mobilities W(µ) following Equation 2:28
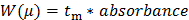 (2)
(2)
9. Additional Characterization of Peptide-grafted Films18
Insert peptide-grafted chitosan films, rolled around themselves, in a 4 mm solid-state NMR rotor. Fill the rotor with phosphate-buffered saline to swell the films, and close the rotor. Wait for a few hours.
Analyze the film with 13C NMR spectroscopy18.
Representative Results
CE is well suited to monitoring the grafting of peptides (e.g., RGDS) onto chitosan films. Suitable coupling agents include EDC-HCl and NHS which activate the peptide to be grafted onto the chitosan (Figure 1). CE is able to separate the different molecules of interest from the reaction medium. To assign the peaks on the electropherogram, pure RGDS, EDC-HCl and NHS were dissolved, injected and separated separately. After the peak assignment, the reaction medium was injected and the various reactants were identified (Figure 2). EDC-HCl reacted into a side product EDH-HCl (3-(((ethylamino)(hydroxy)methylene)amino)-N,N-dimethylpropan-1-amine). Dimethyl sulfoxide (DMSO) is used as an internal standard for the CE separations. Chitosan is present in the grafting experiment in the form of insoluble films and is thus not injected or observed in CE. Note that for all raw data, the recorded migration time axis is converted into an electrophoretic mobility axis (Equation 1) and the UV absorbance axis into a distribution of electrophoretic mobilities W(µ) (Equation 2).
As the aliquots are taken from the reaction medium they are placed into the CE instrument and injected. The extent of the reaction is monitored through the decrease of the peak associated with RGDS (Figure 3). It can also be seen that the EDC-HCl peak decreases while the EDH-HCl peak increases over time. It is important to note that there is no signal that can be assigned to a product from a side reaction of the peptide, thus it is assumed that the RGDS being removed from the reaction medium is being grafted onto the chitosan film. Overlaid electropherograms (Figure 3A) allow the quantification of peptide consumption from the start to the end of the reaction. It is to be noted that although the kinetics was initially measured for 18 hr (Figure 3B), 4 hr was deemed sufficient for the reaction to proceed to its maximal extent. To allow quantification an optimal injection volume is required to ensure the signal-to-noise ratio is high enough while preventing overloading (Figure 4A) and in the case of RGDS, injections were required to be completed in real time to prevent polycondensation (Figure 4B).
The CE-based technique described here to analyze peptide grafting to chitosan films is fast and simple; however, it does not quantify the grafting process directly. NMR spectroscopy is used to demonstrate the grafting; this measurement cannot be done in real time (it typically takes several hours) and needs to be completed post-reaction. The qualitative comparison of the chitosan films before and after grafting shows the successful grafting of the peptides through the appearance of a signal at 70 ppm in the grafted films corresponding to the amide bond between the chitosan and the peptide (Figure 5).
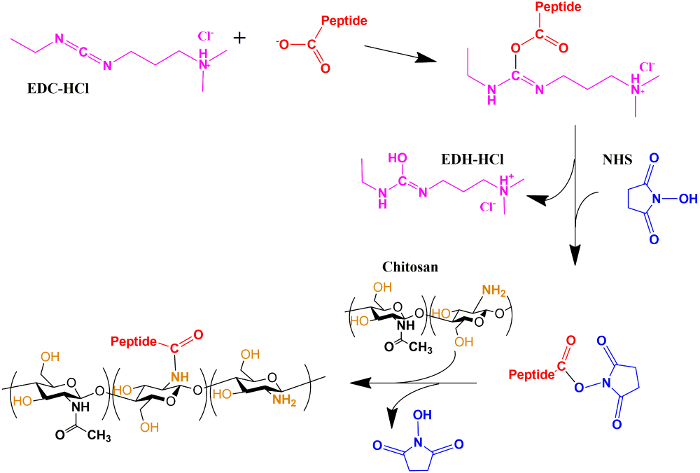 Figure 1. Scheme of the grafting reaction. Chemical reaction scheme showing the activation by EDC-HCl and NHS of the carboxylic acid functional group of RGDS followed by its grafting onto the chitosan's film surface. Please click here to view a larger version of this figure.
Figure 1. Scheme of the grafting reaction. Chemical reaction scheme showing the activation by EDC-HCl and NHS of the carboxylic acid functional group of RGDS followed by its grafting onto the chitosan's film surface. Please click here to view a larger version of this figure.
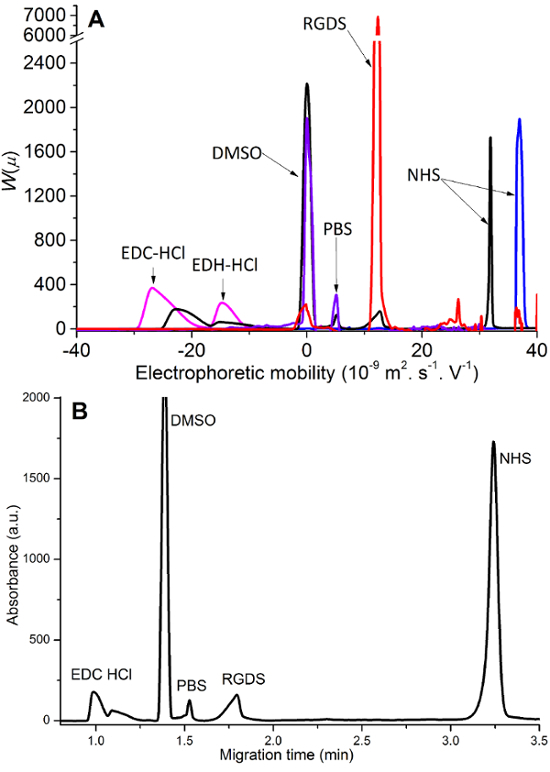 Figure 2.Peak assignment of the species present in the reaction medium. A. Separation and peak assignment for solutions of partially hydrolyzed EDC-HCl (pink), RGDS (red), NHS (blue), as well as for PBS (purple) and the reaction medium (black). B. Electropherograms of reaction media (black) presented as a function of migration time (electrophoretic mobility should be used to overcome poor repeatability in migration times). Please click here to view a larger version of this figure.
Figure 2.Peak assignment of the species present in the reaction medium. A. Separation and peak assignment for solutions of partially hydrolyzed EDC-HCl (pink), RGDS (red), NHS (blue), as well as for PBS (purple) and the reaction medium (black). B. Electropherograms of reaction media (black) presented as a function of migration time (electrophoretic mobility should be used to overcome poor repeatability in migration times). Please click here to view a larger version of this figure.
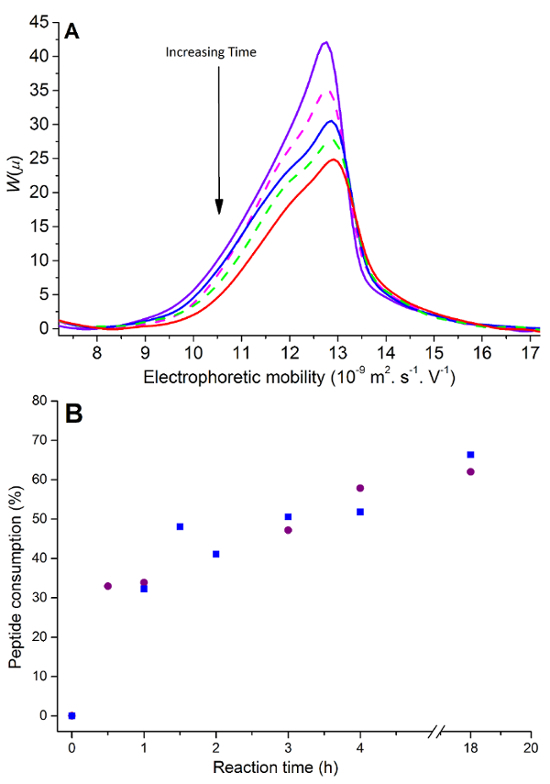 Figure 3.CE monitoring of RGDS consumption. (A) Overlaid peptide peaks at reaction time 30 min (purple solid line), 60 min (magenta dash line), 90 min (blue solid line), 120 min (green dash line), 150 min (red solid line) and (B) kinetics of grafting completed over 18 hr in replicates (square and circle). Please click here to view a larger version of this figure.
Figure 3.CE monitoring of RGDS consumption. (A) Overlaid peptide peaks at reaction time 30 min (purple solid line), 60 min (magenta dash line), 90 min (blue solid line), 120 min (green dash line), 150 min (red solid line) and (B) kinetics of grafting completed over 18 hr in replicates (square and circle). Please click here to view a larger version of this figure.
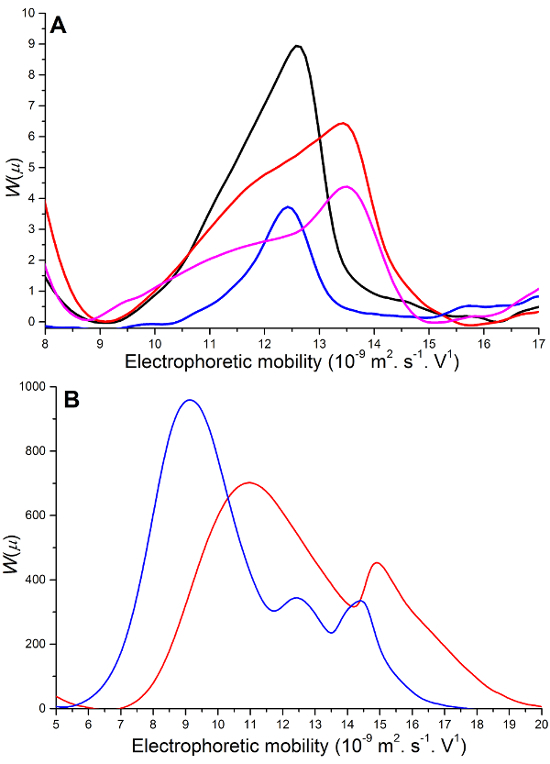 Figure 4.Overlaid peptide peaks in reaction media showing suboptimal results. (A) Varying (hydrodynamic) injection times: 5 sec (blue line), 10 sec (black line), 20 sec (red line) and 30 sec (magenta line). (B) Reaction media left in a CE vial for an extended period of time before injection: 30 min (red line) and 90 min (blue line). Please click here to view a larger version of this figure.
Figure 4.Overlaid peptide peaks in reaction media showing suboptimal results. (A) Varying (hydrodynamic) injection times: 5 sec (blue line), 10 sec (black line), 20 sec (red line) and 30 sec (magenta line). (B) Reaction media left in a CE vial for an extended period of time before injection: 30 min (red line) and 90 min (blue line). Please click here to view a larger version of this figure.
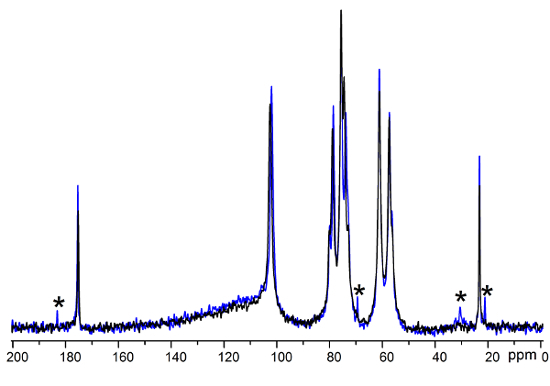 Figure 5.13C swollen-state NMR spectra of chitosan films. Comparison of the films before (black line) and after (blue line) peptide grafting. Signals present only in the spectrum recorded after grafting are indicated by asterisks. Please click here to view a larger version of this figure.
Figure 5.13C swollen-state NMR spectra of chitosan films. Comparison of the films before (black line) and after (blue line) peptide grafting. Signals present only in the spectrum recorded after grafting are indicated by asterisks. Please click here to view a larger version of this figure.
Discussion
The simplicity of the protocol described here makes it ideally suited to widespread application. However, particular attention needs to be paid to of the following key steps.
Proper CE instrument preparation
It is important to separate a known standard immediately prior to the separation of unknown samples (as well as at the end of a series of separations) to check the validity of the capillary and instrument on the day. This standard can be an oligoacrylate27 or any sample known to give multiple peaks over a wide range of migration times. Monitoring the current during all separations and analyzing the migration of a neutral marker in each separation are key steps to identify issues. An unstable current or large variations (more than 10-15%) in the plateau value of the current may be due to inconsistency in the buffer (pH or concentration). If it is followed by a delayed migration time of the neutral maker it may also be due to the capillary not being sufficiently clean. Routine checks of the pH of the buffer and an extra flush of the capillary for 10 min with 1 M sodium hydroxide (freshly prepared) can be used to prevent/amend this problem. During the experiment, extra cleaning steps can be employed to ensure an optimal separation, typically including or lengthening a flush with concentrated aqueous sodium hydroxide to regenerate the fuse silica capillary surface.
Proper data treatment
At the conclusion of the experiment, appropriate treatment of the raw data is essential when comparing results. This involves the conversion of the X and Y-axis obtained from the CE instrument (step 8.3). The migration times determined in CE have a relatively poor repeatability and we recommend using electrophoretic mobilities instead, leading to a much better repeatability. The significance of correctly treating data has been shown previously with the separation and characterization of chitosan10, poly(acrylic acid)29, block copolymers13 and the detection of sugars in breakfast cereals8 through smaller standard deviations (RSD) observed between experiments for mobilities than for migration times. Further, CE has been shown to be more robust than HPLC in the separation of monosaccharides in complex matrices5. Other steps of optimization may include adjusting the injection volume (time of injection) to allow separation with good sensitivity without causing overloading which would prevent quantification. An injection time of 10 sec is deemed optimal at 30 mbar (Figure 4B): a shorter injection time leads to a reduced sensitivity while longer injection times lead to a peak shape distortion indicative of capillary overloading.
Importance of real-time monitoring
A critical strength of this CE-based method is the ability to monitor reactions in real time. This requires optimal CE conditions for detection and separation of the relevant reactant(s) and/or product(s). Furthermore, for the analysis of chemical reactions an appropriate time zero must be taken as a benchmark of the reaction; this typically consists in the separation of the measured out reactants just prior to the reaction starting. This can be done for example before one particular reactant is introduced, before the temperature is increased, or before UV irradiation is started to trigger the reaction.
In the case of the grafting of the peptide RGDS (onto chitosan or onto another substrate), the peptide is able to react with itself to produce linear oligomers or branched structures through polycondensation18. This is because RGDS contains both amine and carboxylic acid functional groups. These peptide oligomers do not have the same electrophoretic mobility as the initial peptide RGDS and therefore may cause an inaccurate quantification, through for example co-migration with other species. It is therefore important to ensure that aliquots of the reaction medium are injected and separated within a few minutes of being taken from the reaction medium (Figure 4B).
Proper preparation of the chitosan films
When specifically dealing with chitosan films there are a number of steps to adhere to. During the production of the chitosan films, the films need to be left to dry for at least 7 days (preferably more). If this is not completed, when the films are placed in PBS buffer to rinse they will dissolve rather than form a swollen film which then prevents the next steps. Additionally, it is important to neutralize the film prior to the grafting reaction to remove any remaining acetic acid which may leach out of the film and compete with the peptide for the grafting reaction18. This can be done through soaking in dilute aqueous sodium hydroxide or in PBS. The pH of the buffer used as solvent for the grafting reaction is also critical: if it is too acidic the films will partially or completely dissolve. During the grafting reaction it is important that the film is able to have maximum contact with the reaction solution. Therefore, the Petri dish containing the films and the reaction mixture are placed on a shaker. It is also imperative to prevent the evaporation of the reaction mixture to prevent uncontrolled concentration variations resulting in inaccurate quantifications; the use of paraffin film to cover the Petri dish was effective to prevent it.
The main limitation of the CE technique is that individually it is not able to confirm the grafting process. In the context of the chemical grafting process mentioned above, the quantification of the peptide grafting is indirect. This can be overcome with the use of a complementary technique such as solid-state NMR spectroscopy as previously mentioned. Other limitations of the CE technique include that it requires the compound of interest to be charged. Therefore neutral species will migrate at the same time. In certain cases this can be overcome if the compound of interest complexes with borate. Finally if the compound of interest does not contain chromophores detection other than UV such as conductivity may need to be used. This requires the additional purchase of a conductivity detector which requires optimization.
Advantages of analyzing peptide grafting via CE vs other analytical methods
The CE method has several advantages over alternative indirect methods including high performance liquid chromatography (HPLC), amino acid analysis (AAA) and the direct method of NMR spectroscopy. Compared to AAA it is a high-throughput, robust method which allows it to analyze complex samples efficiently without tedious sample preparation. This is advantageous especially in the analysis of chemical reactions in real time. HPLC has been used previously for peptide grafting analysis30, however it was deemed only semi-quantitative. CE has a lower running cost than HPLC and does not require sample filtration prior to analysis, minimizing the risk of sample loss5. Although 13C NMR spectroscopy is able to directly measure the product of interest, it is a costly technique and is unable to measure it in real time.
The protocol descripted here provides a rapid, efficient, inexpensive and reliable method for optimizing peptide grafting to chitosan film. This new approach thus provides significant advantages for tailoring the cell attachment properties of chitosan films compared to traditionally used methods such as chromatography and AAA. This CE method can be used to monitor a number of other chemical reactions in real time, typically reactions occurring on the timeframe of several hr, for which the reactants/products of interest can be charged. In this case, it is however important to note that the CE method should be optimized prior to the analysis of a different chemical reaction to allow a successful analysis. This includes the analysis of the pure reactants and products prior to the reaction to allow them to be identified and ensure that they can be detected and separated, as well as to ensure that no contaminants may prevent quantification. The separation may be improved and the total analysis time changed by varying the capillary length, buffer composition and voltage, and potentially using a capillary with coated walls. The detection may be improved by modifying the conditions in which the sample is injected to favor the charged reactants or by injecting a larger amount of the sample into the capillary. Further, other detectors apart from UV detection can be employed including fluorescence, contactless conductivity detectors or the CE can be coupled to a mass spectrometer. The ability to monitor reactions in real time enables the grafting reaction to be performed directly in a CE vial if the substrate is in solution and is able to be analyzed by CE. This can take place directly inside the CE instrument, as we recently performed it for grafting of aminoantipyrine on poly(acrylic acid)7 Additionally, the approach is not limited to grafting reactions but can be extended to monitor various other chemical reactions. Further, the monitoring of the reactions allows optimization of the reaction and may also be used to validate the product of the reaction. As long as the compound of interest can be dissolved and charged, the CE method allows fast, cheap and robust separation, detection and quantification.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
MG, MO'C and PC thank the Molecular Medicine Research Group at WSU for Research Seed Funding, as well as Michele Mason (WSU), Richard Wuhrer (Advanced Materials Characterisation Facility, AMCF, WSU) and Hervé Cottet (Montpellier) for discussions.
References
- Muthukumar M. Theory of electrophoretic mobility of a polyelectrolyte in semidilute solutions of neutral polymers. Electrophoresis. 1996;17:1167–1172. doi: 10.1002/elps.1150170629. [DOI] [PubMed] [Google Scholar]
- Barrat JL, Joanny JF. in Advances in Chemical Physics, Vol Xciv Vol. 94 Advances in Chemical Physics. John Wiley & Sons Inc; 1996. pp. 1–66. [Google Scholar]
- Fu SL, Lucy CA. Prediction of electrophoretic mobilities. 1. Monoamines. Anal. Chem. 1998;70:173–181. doi: 10.1021/ac9706638. [DOI] [PubMed] [Google Scholar]
- Harvey D. Modern Analytical Chemistry. McGraw Hill; 2000. [Google Scholar]
- Oliver JD, Gaborieau M, Hilder EF, Castignolles P. Simple and robust determination of monosaccharides in plant fibers in complex mixtures by capillary electrophoresis and high performance liquid chromatography. J. Chromatogr. A. 2013;1291:179–186. doi: 10.1016/j.chroma.2013.03.041. [DOI] [PubMed] [Google Scholar]
- Oliver JD, Sutton AT, Karu N, Phillips M, Markham J, Peiris P, Hilder EF, Castignolles P. Simple and robust monitoring of ethanol fermentations by capillary electrophoresis. Biotechnology and Applied Biochemistry. 2015;62:329–342. doi: 10.1002/bab.1269. [DOI] [PubMed] [Google Scholar]
- Thevarajah JJ, Sutton AT, Maniego AR, Whitty EG, Harrisson S, Cottet H, Castignolles P, Gaborieau M. Quantifying the Heterogeneity of Chemical Structures in Complex Charged Polymers through the Dispersity of Their Distributions of Electrophoretic Mobilities or of Compositions. Anal. Chem. 2016;88:1674–1681. doi: 10.1021/acs.analchem.5b03672. [DOI] [PubMed] [Google Scholar]
- Toutounji MR, Van Leeuwen MP, Oliver JD, Shrestha AK, Castignolles P, Gaborieau M. Quantification of sugars in breakfast cereals using capillary electrophoresis. Carbohydr. Res. 2015;408:134–141. doi: 10.1016/j.carres.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Miramon H, Cavelier F, Martinez J, Cottet H. Highly Resolutive Separations of Hardly Soluble Synthetic Polypeptides by Capillary Electrophoresis. Anal. Chem. 2010;82:394–399. doi: 10.1021/ac902211f. [DOI] [PubMed] [Google Scholar]
- Mnatsakanyan M, Thevarajah JJ, Roi RS, Lauto A, Gaborieau M, Castignolles P. Separation of chitosan by degree of acetylation using simple free solution capillary electrophoresis. Anal. Bioanal. Chem. 2013;405:6873–6877. doi: 10.1007/s00216-013-7126-4. [DOI] [PubMed] [Google Scholar]
- Taylor DL, Ferris CJ, Maniego AR, Castignolles P, in het Panhuis M, Gaborieau M. Characterization of Gellan Gum by Capillary Electrophoresis. Australian Journal of Chemistry. 2012;65:1156–1164. [Google Scholar]
- Thevarajah JJ, Gaborieau M, Castignolles P. Separation and characterization of synthetic polyelectrolytes and polysaccharides with capillary electrophoresis. Adv. Chem. 2014;2014:798503. [Google Scholar]
- Sutton AT, Read E, Maniego AR, Thevarajah J, Marty J-D, Destarac M, Gaborieau M, Castignolles P. Purity of double hydrophilic block copolymers revealed by capillary electrophoresis in the critical conditions. J. Chromatogr. A. 2014;1372:187–195. doi: 10.1016/j.chroma.2014.10.105. [DOI] [PubMed] [Google Scholar]
- Righetti PG, Sebastiano R, Citterio A. Capillary electrophoresis and isoelectric focusing in peptide and protein analysis. Proteomics. 2013;13:325–340. doi: 10.1002/pmic.201200378. [DOI] [PubMed] [Google Scholar]
- Ali I, Al-Othman ZA, Al-Warthan A, Asnin L, Chudinov A. Advances in chiral separations of small peptides by capillary electrophoresis and chromatography. J. Sep. Sci. 2014;37:2447–2466. doi: 10.1002/jssc.201400587. [DOI] [PubMed] [Google Scholar]
- Kasicka V. Recent developments in capillary and microchip electroseparations of peptides (2011-2013) Electrophoresis. 2014;35:69–95. doi: 10.1002/elps.201300331. [DOI] [PubMed] [Google Scholar]
- JoVE Science Education Database. Essentials of Analytical Chemistry. Cambridge, MA: JoVE; 2016. Capillary Electrophoresis (CE) Available from: http://www.jove.com/science-education/10226/capillary-electrophoresis-ce. [Google Scholar]
- Taylor DL, Thevarajah JJ, Narayan DK, Murphy P, Mangala MM, Lim S, Wuhrer R, Lefay C, O'Connor MD, Gaborieau M, Castignolles P. Real-time monitoring of peptide grafting onto chitosan films using capillary electrophoresis. Anal. Bioanal. Chem. 2015;407:2543–2555. doi: 10.1007/s00216-015-8483-y. [DOI] [PubMed] [Google Scholar]
- Rinaudo M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006;31:603–632. [Google Scholar]
- Li Z, Leung M, Hopper R, Ellenbogen R, Zhang M. Feeder-free self-renewal of human embryonic stem cells in 3D porous natural polymer scaffolds. Biomaterials. 2010;31:404–412. doi: 10.1016/j.biomaterials.2009.09.070. [DOI] [PubMed] [Google Scholar]
- Domard A. A perspective on 30 years research on chitin and chitosan. Carbohydr. Polym. 2011;84:696–703. [Google Scholar]
- Shekaran A, Garcia AJ. Nanoscale engineering of extracellular matrix-mimetic bioadhesive surfaces and implants for tissue engineering. Biochim. Biophys. Acta Gen. Subj. 2011;1810:350–360. doi: 10.1016/j.bbagen.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio CA, Alves CM, Reis RL, Mano JF. Immobilization of fibronectin in chitosan substrates improves cell adhesion and proliferation. J. Tissue Eng. Regen. Med. 2010;4:316–323. doi: 10.1002/term.248. [DOI] [PubMed] [Google Scholar]
- Boateng SY, Lateef SS, Mosley W, Hartman TJ, Hanley L, Russell B. RGD and YIGSR synthetic peptides facilitate cellular adhesion identical to that of laminin and fibronectin but alter the physiology of neonatal cardiac myocytes. Am. J. Physiol. Cell Physiol. 2005;288:C30–C38. doi: 10.1152/ajpcell.00199.2004. [DOI] [PubMed] [Google Scholar]
- Lefay C, Guillaneuf Y, Moreira G, Thevarajah JJ, Castignolles P, Ziarelli F, Bloch E, Major M, Charles L, Gaborieau M, Bertin D, Gigmes D. Heterogeneous modification of chitosan via nitroxide-mediated polymerization. Polym. Chem. 2013;4:322–328. [Google Scholar]
- Gartner C, Lopez BL, Sierra L, Graf R, Spiess HW, Gaborieau M. Interplay between Structure and Dynamics in Chitosan Films Investigated with Solid-State NMR, Dynamic Mechanical Analysis, and X-ray Diffraction. Biomacromolecules. 2011;12:1380–1386. doi: 10.1021/bm200193u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castignolles P, Gaborieau M, Hilder EF, Sprong E, Ferguson CJ, Gilbert RG. High resolution separation of oligo(acrylic acid) by capillary zone electrophoresis. Macromol. Rapid Commun. 2006;27:42–46. [Google Scholar]
- Chamieh J, Martin M, Cottet H. Quantitative Analysis in Capillary Electrophoresis: Transformation of Raw Electropherograms into Continuous Distributions. Anal. Chem. 2015;87:1050–1057. doi: 10.1021/ac503789s. [DOI] [PubMed] [Google Scholar]
- Maniego AR, Ang D, Guillaneuf Y, Lefay C, Gigmes D, Aldrich-Wright JR, Gaborieau M, Castignolles P. Separation of poly(acrylic acid) salts according to topology using capillary electrophoresis in the critical conditions. Anal. Bioanal. Chem. 2013;405:9009–9020. doi: 10.1007/s00216-013-7059-y. [DOI] [PubMed] [Google Scholar]
- Chung TW, Lu YF, Wang SS, Lin YS, Chu SH. Growth of human endothelial cells on photochemically grafted Gly-Arg-Gly-Asp (GRGD) chitosans. Biomaterials. 2002;23:4803–4809. doi: 10.1016/s0142-9612(02)00231-4. [DOI] [PubMed] [Google Scholar]


