Abstract
MicroRNAs (miRNAs) are an extensive class of tiny RNA molecules that regulate the expression of target genes by means of complementary base pair interactions. Although the first miRNAs were discovered in Caenorhabditis elegans, >300 miRNAs were recently documented in animals and plants, both by cloning methods and computational predictions. We present a genome-wide computational approach to detect miRNA genes in the Arabidopsis thaliana genome. Our method is based on the conservation of short sequences between the genomes of Arabidopsis and rice (Oryza sativa) and on properties of the secondary structure of the miRNA precursor. The method was fine-tuned to take into account plant-specific properties, such as the variable length of the miRNA precursor sequences. In total, 91 potential miRNA genes were identified, of which 58 had at least one nearly perfect match with an Arabidopsis mRNA, constituting the potential targets of those miRNAs. In addition to already known transcription factors involved in plant development, the targets also comprised genes involved in several other cellular processes, such as sulfur assimilation and ubiquitin-dependent protein degradation. These findings considerably broaden the scope of miRNA functions in plants.
Keywords: comparative genomics, thale cress, rice, noncoding RNA
MicroRNAs (miRNAs) are small noncoding RNA gene products ≈22 nt long that are found in a variety of organisms, including animals and plants (1-3). The first miRNAs were discovered in Caenorhabditis elegans and control developmental timing by binding to specific target mRNAs (4-9). By pairing to mRNAs, documented miRNAs initiate cleavage of the mRNA or repress active translation (1-3, 10). miRNAs have similarities with small interfering (si)RNAs, which are short (21-24 nt) molecules involved, at least in plants, in posttranscriptional gene silencing (11). In this process, long, double-stranded RNAs are cleaved into siRNA fragments by dicer, an RNase III helicase (12). These fragments bind to complementary RNA, which activates the RNA interference silencing complex to cleave the target mRNA (13). One main difference between siRNAs and miRNAs is that siRNAs mediate the silencing of the genes at the locus from which they originate, and possibly their paralogs, whereas miRNAs regulate a wide variety of genes that differ from those of which they originated (1, 14-16).
In plants, as in animals, miRNAs are processed from transcripts that can fold into a stable hairpin (17, 18). However, the size of the potential hairpin precursors of the plant miRNAs is much more variable than that of animals. For example, C. elegans miRNAs are cleaved from precursors of ≈70 nt in length, with the mature miRNA located from 2 to 10 nt from the terminal loop of the stem-loop structure (19). Although some of the Arabidopsis precursors resemble those of C. elegans, others are much larger, such as the 190-nt-long precursor of mir169 (10). In addition, the shape of the predicted secondary structure of plant miRNAs appears to be more complex, sometimes with branched structures instead of a simple hairpin. Unlike animal miRNAs, plant miRNAs generally interact with their targets through near-perfect complementarity (17-19), which greatly facilitates the computational identification of plant miRNA targets. For example, for described plant miRNAs (20, 21), 49 unique targets could already be identified. Later, many of these targets have been confirmed experimentally (15, 16, 22-24). Recent work (25, 26) also demonstrated the role of miRNAs in the control of leaf, stem, and flower development.
Different biochemical approaches were used to try to identify small RNAs in plants, but so far only a fraction of them could be verified. For example, Park et al. (21) identified 230 sequences of which only five appeared to be “true” miRNAs. With a protocol to preferentially clone dicer cleavage products, only 16 true miRNAs were isolated from a starting pool of 300 small RNAs of seedlings and flowers (18). From this set of 16, eight were also conserved in the rice (Oryza sativa) genome. Interestingly, the sequences adjacent to these miRNAs could form stem-loop structures analogous to those of Arabidopsis, with the miRNA sequence invariably on the same arm of the precursor in both species. Furthermore, although the Arabidopsis and rice sequences seemingly diverged considerably upstream and downstream of the miRNA, the miRNA itself differed in only a few base pairs. This conservation of secondary structure, despite the sequence variability observed in the precursor sequences, suggests that the secondary structure plays a major role, presumably in the processing of the mature miRNA from the precursor.
To estimate the numbers of potential miRNAs in organisms such as Caenorhabditis, Drosophila, fish, and human, different computational gene-finding strategies have been developed (27-30), all of them being based on a comparative approach. The core principle is to look for conserved sequences between different species that can fold into extended hairpins. The fact that there are much more conserved stem-loop structures than true miRNA genes stresses the importance of considering additional information to validate potential miRNA (29). Unfortunately, methods applied to detect animal miRNAs cannot be directly applied to plants. For example, the algorithms that used a fixed-length window to search for hairpins in intergenic sequences are justified in animals because most animal miRNA precursors have almost identical lengths (≈70-80 nt). On the contrary, plants show a much greater length variability of miRNA precursors, invalidating this fixed-window approach. Additionally, whereas animal miRNAs are conserved for most of the complete precursor sequence, in plants only the mature miRNA is conserved (20).
Here, we propose a computational approach for detecting plant miRNAs, based on a comparison of the Arabidopsis and Oryza genomes, and considering the core features of experimentally determined plant miRNAs. By using this approach, we were able to identify many miRNA genes and their targets.
Materials and Methods
miRNA Reference Set. To derive a set of rules and parameters that describe and characterize known Arabidopsis miRNAs, we first defined a reference set of miRNA sequences, which consisted of 22 Arabidopsis miRNA sequences and their corresponding 43 precursor sequences. The difference between 22 and 43 is due to the fact that some identical miRNAs are found at different places in the genome and with different precursors. These sequences were obtained from different in vivo biochemical analyses, such as accumulation of miRNA in mutants and expression levels of miRNA sequences by northern analysis (18, 20, 21, 31), and were downloaded from the miRNA registry (32).
Arabidopsis Intergenic Sequences. Sequences and annotations were downloaded from The Institute for Genomic Research (Arabidopsis thaliana release 3, July 2002). Because most known miRNA genes have been detected in intergenic regions (IGRs) (1) the study was limited to the Arabidopsis IGR sequences that were extracted, excluding known elements, such as protein-encoding genes plus their experimentally defined UTRs, pseudogenes, ribosomal RNA, small nucleolar RNA, and tRNAs. For protein-encoding genes without experimentally defined UTR, a 300-nt region, i.e., the average length for experimentally defined UTRs with a full-length cDNA sequence, was added both upstream and downstream. A total of 23,433 sequences were thus obtained with an average length of 2 ± 1.9 kb.
O. sativa Sequences. In total, 3,601 rice bacterial artificial chromosome sequences from the International Rice Genome Sequencing Project (33) were downloaded from The Institute for Genomic Research assemblies (June 2003). The average length of the sequences was 138 ± 30 kb.
Conserved Intergenic Short Segments. Arabidopsis intergenic sequences were masked for repeat elements by using the program repeatmasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html). Conserved IGRs between Arabidopsis and rice were obtained with blast (34), by using a bit-score low-cutoff value set at 30 bits. This cutoff value allows a limited dissimilarity between sequences, which is in agreement with what is observed in the miRNAs of the reference set (18). blast hits between Arabidopsis and rice with a length between 20 and 25 nt were retained for further analysis. To consider the secondary structure of the miRNAs precursors, conserved sequences were extended with 350 nt both upstream and downstream, which is the maximum length known for Arabidopsis miRNA sequences (18). Analyses of the reference set showed that the miRNA sequence could be located on either the 5′ or 3′ arm of the precursor sequence, without any preference for strand orientation. Consequently, the reverse complement of the sequences was also considered when the miRNAs were on the reverse strand.
Removing Vector Contamination and Known Noncoding RNAs. Potential miRNA precursors were carefully checked for repeat sequences, for vector contamination with the univec database (www.ncbi.nlm.nih.gov/VecScreen/VecScreen.html), for virus sequences (35), and also for homology with well known other types of noncoding RNAs, such as tRNA, ribosomal RNA, and small nucleolar RNA. The tRNA, ribosomal RNA, and small nucleolar RNA sequences were downloaded from the GtRDB database (36), the European ribosomal RNA database (37), and the small nucleolar RNA database (38), respectively. Sequence matches with a low e-value (<0.01) were discarded.
GC Content and Low-Complexity Filtering. We analyzed blast hits from IGRs of the genome, which are well known to contain a large amount of repeat and low-complexity sequences, thus giving many false-positive conserved sequences. To reduce the number of false positives, a filter based on GC content and low complexity was applied to potential miRNA sequences. A Shannon entropy measure (39) was used as a low-complexity pattern filtering. The cutoff values were defined based on the distribution of the values obtained with the miRNA reference set. Sequences with a GC content ≥0.3 and ≤0.7 and with an entropy value ≥1.75 were retained for further analysis.
Potential miRNA Precursor and Precursor Secondary Structure. To define the potential stem-loop precursors within the extended sequences, we made use of the characteristic miRNA precursor molecules property that the mature miRNA is always excised from one side of an unbranched RNA helix (40). As a result, a sequence that corresponds to the reverse complement of the miRNA should be found on the complementary side of the mature miRNA. Therefore, for each potential miRNA, the reverse complement was aligned to the extended precursor molecule by using the local alignment algorithm implemented in the matcher program from the emboss package (41). We modified the scoring matrix to allow G·U and U·G base pairs in the precursor pairing. These parameters were chosen so that all miRNAs in our reference set were identified. Each reverse complement was checked against the cutoff values that were extracted from the reference set. After this step, all sequences flanked by the miRNA and its reverse complement were extracted as potential miRNA precursors. Potential precursor sequences were all folded with the viennarna package (42). The statistical significance of the folding of the miRNA precursor was assessed with a randomization test (see Supporting Text, which is published on the PNAS web site, for a detailed explanation).
Potential Targets. Because most plant miRNA sequences bind to their target with a near-perfect complementarity (22-24), this property was exploited to predict potential targets for miRNA sequences by using a computational approach (20). By using exactly the same procedure, potential target mRNAs were searched for by the patscan software of Dsouza et al. (43). The validity was tested on the same data set as described in Rhoades et al. (20). The number of allowed mismatches varies with the length of the potential miRNA: two, three, and four mismatches for sequences of up to 21 nt, 23 nt, or more, respectively. This variable rule set took into account the possible diffuse nature of the boundaries of the miRNA sequence, which occurs more probably in longer sequences. Hits with four mismatches should be considered with caution because such hits have a great likelihood to occur by chance, although they might be genuine targets (20). The mRNA sequences (coding sequences including UTR sequences when available; version 04/17/2003) were downloaded from the Arabidopsis Information Resource database (44).
Clustering of miRNA Sequences. MiRNA sequence similarity was estimated with a procedure similar to that used by Grad et al. (30). A pairwise homology score between all of the sequences was computed with the matcher program from the emboss package (41). All pairwise comparisons were converted to Euclidean distances and a hierarchical clustering was performed with the single-linkage method. A cutoff was set to group highly similar sequences. Statistical computations were carried out with the R package (www.r-project.org).
Expression Data. Potential miRNAs were checked against EST sequences (206,678 sequences downloaded from EMBL) and Arabidopsis Massively Parallel Signature Sequencing (mpss.udel.edu/at; ref. 45) for 20-nt signatures.
All supporting information cited in the text can be accessed at www.psb.ugent.be/bioinformatics.
Results and Discussion
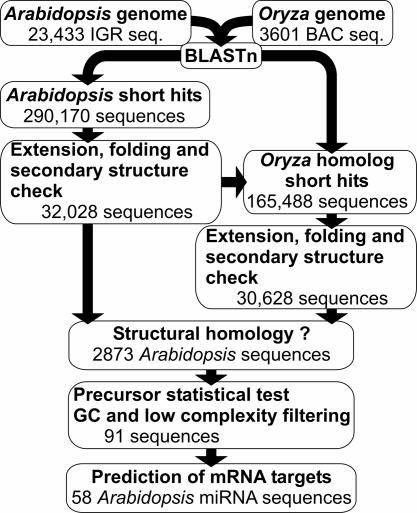
The mirfinder Computational Pipeline. A flowchart describing the general procedure of plant miRNA detection is shown in Fig. 1. The mirfinder computational pipeline was based on three major rules derived from the miRNA reference set: (i) the miRNA sequence is conserved between Arabidopsis and rice, whereas the rest of the precursor sequence has diverged; (ii) even though the precursor sequence has diverged, the ability of the precursor sequence to form a stem-loop secondary structure in both Arabidopsis and Oryza is conserved; and (iii) for two miRNA orthologs, the miRNA sequence is always located on the same arm of the stem-loop secondary structures in both species.
Fig. 1.
Overview of the mirfinder computational pipeline to detect miRNAs in plants. For details, see text.
To select valid miRNA stem-loop structures, in addition to the three general rules described above, five characteristic features of the secondary structure of miRNA and their cutoff values were also derived from the reference set, of which one qualitative and five quantitative parameters. These parameters are (see Fig. 3, which is published as supporting information on the PNAS web site): (i) the miRNA should be part of a continuous helix; (ii) the minimum free energy value should be less than -30 kcal/mol; (iii) the minimum number of paired residues in the miRNA should be 15; (iv) the maximum number of unpaired residues in both the miRNA coding and complementary strand should be 5; and (v) the maximum number of G·U pairs in the miRNA should be 5.
The mirfinder pipeline (Fig. 1) starts by identifying short highly conserved sequences between the Arabidopsis and rice genomes. The 290,170 short sequences found in Arabidopsis with at least one corresponding hit in rice were extended to see whether they could form potential stem-loop secondary structures, which were determined by looking at the possible reverse complement of the miRNA within each extended sequence. Thus, for one potential miRNA sequence conserved between Arabidopsis and Oryza, multiple potential precursor sequences could be present. A total of 1,394,939 potential precursor Arabidopsis sequences were fold with the viennarna package (42). From this pool, 32,648 sequences could be retained after filtering for typical miRNA precursor secondary structure features (see Materials and Methods). To verify whether these sequences have at least one homolog in rice, the same procedure was applied to the homologous sequences found in that genome. Every Arabidopsis stem-loop structure and its ortholog in Oryza were compared. The potential miRNA was retained only when an Arabidopsis miRNA sequence had an miRNA homolog in the rice genome that was located on the same arm (either 5′ or 3′) of the stem-loop structure in both species. A total of 2,873 Arabidopsis sequences were found to have at least one such structural homolog in the Oryza genome. Examples of such miRNAs (Fig. 2) clearly show the conservation for the miRNA sequence, the non-conservation of the precursor sequence, and the conservation of the secondary structure. The use of GC content filtering (30) further reduced the set of 2,873 Arabidopsis potential miRNA to 852 sequences. A test study showed that ≈5% of randomly selected genomic sequences from C. elegans could fold into a plausible miRNA precursor hairpin (19). To reduce the number of such false positives, all Arabidopsis precursor sequences were statistically tested with the randomization test (see Supporting Text). With this approach, the set of potential miRNA sequences was further reduced to 501. Last, an entropy-based filtering step removed miRNA sequences with a low-complexity pattern and further reduced our data set to 297 sequences.
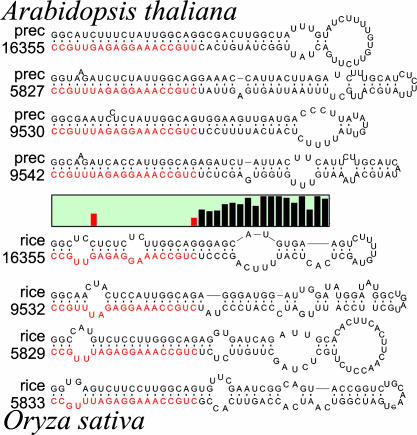
Fig. 2.
RNA secondary structure models of the eight precursor molecules (four each from Arabidopsis and rice) of the miRNAs that target the 5′ UTR of the Arabidopsis gene At2g33770.1 and its rice homolog. The precursor molecules are drawn with the miRNAs aligned and highlighted in red. (Middle) The graph represents the Shannon entropy of the nucleotide content of each corresponding position on the 3′ strand of the precursors. A low value means that all molecules have the same nucleotide at that position, whereas a high value reflects little or no nucleotide conservation. The sequence divergence outside of the mature miRNA positions is clearly shown. RNA sequences are drawn from 5′ to 3′ in clockwise orientation. All precursors are truncated at the same position. RNA secondary structure drawings were made by using rnaviz (59).
Because an exhaustive approach was used, some potential miRNAs differed only by a few base pairs on either side, or were the reverse complement of each other. To avoid redundancy and overprediction, we clustered the overlapping miRNAs, with human supervision, and kept parent only when they complied with the features discussed above, also taking into account potential targets. After this clustering process, and after removing one potential miRNA because of unexpected sequence degeneracy (see below), 91 were kept as potential miRNAs. We could not observe any close cluster of miRNA sequences as it is the case for animal miRNAS (19). miRNA fasta files are available as Data Sets 1 and 2, which are published as supporting information on the PNAS web site.
A blast search against Arabidopsis Massively Parallel Signature Sequencing 20-nt signatures did not uncover any hits. The Arabidopsis precursor sequences gave two significant hits with ESTs, namely MIR30 and MIR36 and EST sequences BX838271 and AU239920, respectively, both with very high P value (<1e-45). MIR30 (synonym of mir171) was already known to be expressed (18).
Arabidopsis miRNA Targets. We could identify 58 Arabidopsis miRNA sequences that have at least one mRNA target sequence (see Table 1). One miRNA was excluded from this set because it had 76 targets, which is very unlikely. The highly repetitive motif corresponding to this sequence (CUUCAUCUUCAUCAUCAUCAG) might explain such a ubiquity, thus revealing a potential false positive.
Table 1. List of the potential protein targets for the 91 potential miRNAs, grouped by annotated gene family and by biological function.
| Protein annotation | MiRNA | Common function/type |
|---|---|---|
| CCAAT box-binding factor | MIR13, MIR43, MIR47, MIR57, MIR81, MIR87 | |
| CCAAT box-binding factor Hap2a | MIR77 | Core transcription factor |
| CCAAT-binding factor B subunit homolog | MIR13, MIR43, MIR47, MIR57 | |
| CCAAT-binding factor B subunit-related | MIR13, MIR43, MIR57, MIR77, MIR81, MIR87 | |
| Auxin-response transcription factor (ARF) | MIR75 | |
| ARF8 | MIR54 | |
| Transcription factor B3 family similar to ARF10 | MIR40, MIR75 | |
| Transcription factor B3 family | MIR40, MIR47 | |
| NAC1/NAM protein family | MIR29, MIR82 | Auxin signaling |
| NAM-related protein | MIR29, MIR82 | |
| NAM-like protein | MIR29, MIR82 | |
| NAM-related protein | MIR82 | |
| NAM protein CUC2 | MIR82 | |
| Scarecrow transcription factor family | MIR30, MIR46, MIR58 | Asymmetric cell division |
| Scarecrow-like transcription factor 14 (SCL14) | MIR14 | Specify meristem quiescent Center and radial patterning |
| SCL6 | MIR30, MIR46, MIR58 | |
| Squamosa promoter-binding protein-related 2 | MIR17, MIR85, MIR91 | |
| Squamosa promoter-binding protein homologs | MIR17, MIR63, MIR85, MIR91 | |
| Squamosa promoter-binding protein 4 (SPL4) | MIR17, MIR43, MIR63, MIR85, MIR91 | Homeotic MADS box genes controling floral development |
| AP2 domain transcription factor RAP2.7 | MIR21, MIR84 | |
| AP2 domain transcription factor, potential | MIR18, MIR21, MIR84 | |
| Floral homeotic protein APETALA2 | MIR18, MIR21, MIR84 | |
| HD-Zip transcription factor Athb-15 | MIR5, MIR70 | Vascular development and leaf development |
| Phabulosa HD-Zip TF Athb-14 | MIR70 | |
| HD-Zip transcription factor Athb-9 | MIR70 | |
| PIL6 Myc-related bHLH transcription factor | MIR32 | Circadian rhythm control by light |
| Myb family transcription factor MYB33 | MIR89 | |
| Myb family transcription factor MYB30, MYB120, MYB65 | MIR88 | Myb family transcription factor |
| Zinc finger (C3HC4-type RING finger) family | MIR12, MIR44, MIR56, MIR76, MIR80, MIR86 | Zinc finger transcription factor |
| TIR1, E3 ubiquitin ligase SCF complex F-box subunit | MIR10 | |
| TIR1-like genes, E3 ubiquitin ligase F-box subunit | MIR10, MIR20 | Ubiquitination pathway |
| F-box protein GRR1-like protein 1, AtFBL 18 | MIR10 | Controling targeted |
| E2 UBC, ubiquitin-conjugating enzyme family | MIR16, MIR27, MIR67 | Protein degradation (auxin-related for many) |
| CDC48 domain-containing AAA-type ATPase/NSF | MIR2 | |
| TAZ RING BTB/POZ domain protein | MIR47 | |
| F-box protein family | MIR63 | |
| FLU, TPR-containing protein | MIR15 | Chloroplast import of PORA |
| VQ-motif containing protein | MIR60 | Control of plastid genes |
| Thioredoxin-like protein 3 | MIR59 | Redox control in chloroplast |
| ATP sulfurylase | MIR64 | Sulfur metabolism |
| Sulfate transporter | MIR64 | |
| Acyl transferase-containing multifunctional enzyme | MIR64 | |
| Copper/zinc superoxidase dismutase (CSD1) | MIR1 | Cell death |
| Potential cytosine/deoxycytidine deaminase | MIR55 | Purine salvage |
| Amine oxidase-related | MIR83 | Overlaps auxin gene NAC1 |
| Ypt/Rab GTPase-activating protein | MIR4 | Intracellular trafficking |
| WASP domain containing-protein | MIR11 | Signal transduction to actin |
| SAG101, leaf senescence-associated acyl hydrolase | MIR73 | Senescence |
| Target of rapamycin, phosphatidylinositol 3-kinase | MIR34 | Embryo development |
| Potential (glycerol) phosphate acyltransferase | MIR31, MIR72 | Phospholipid synthesis |
| Laccase (diphenol oxidase), potential | MIR9 | |
| Lucine-rich repeat receptor kinase, potential | MIR55 | |
| ABC transporter family protein | MIR51 | Specific function unknown |
| Proline-rich protein family | MIR53 | |
| Expressed protein | MIR21 | |
| Expressed protein | MIR25 | |
| Hypothetical protein | MIR6, MIR15 | |
| Gypsy family retrotransposon gag protein | MIR88 | Retrotransposon |
An extended version of this table, which includes the gene loci (At codes) for each protein, appears as Table 2, which is published as supporting information on the PNAS web site.
The overall sensitivity of our procedure is demonstrated by the fact that we could identify six of the eight described miRNAs that are known to be conserved between Arabidopsis and rice, namely MIR156, MIR160, MIR164, MIR166, MIR167, and MIR171 (18, 46). The reason why we missed the two other miRNAs will be discussed below. If a hierarchical clustering approach were applied, the 91 Arabidopsis miRNAs would be clustered in 51 families (dendrogram and list are available as Fig. 4, which is published as supporting information on the PNAS web site).
The different mRNA targets were grouped into families (Table 1), of which 56% of all potential targets represent transcription factor (TF)-related proteins. Many of the TF targets listed were already known to be targets of miRNAs (1, 10). For example, we successfully identified both the previously described Arabidopsis mir171 (MIR30 in Table 1) and its targets, the Scarecrow TF family (At2g45160, At3g60630, and At4g00150). The function of mir171 is supported by both prediction and experimental evidence (17, 18, 20, 31). A few other TF targets, to our knowledge, are new, such as the Zn-finger protein family (At1g54150 in Table 1).
Interestingly, we discovered that almost half of the predicted targets are non-TF mRNAs. As shown in Table 1, the function of most of these targets is known, at least to some extent, and some can be clustered according to this function and/or their corresponding miRNAs into larger functional groups. The largest one, with TIR1 as a typical member, comprises some of the many genes of the ubiquitination pathway, involved in proteasome-dependent degradation of targeted proteins (47), which plays an important role in plant development (48). TIR1 is a component of the E3 ubiquitin ligase SCFTIR1 complex that mediates auxin response through the Aux/IAA proteins, which act as negative regulators. TIR1 itself is responsible for the specificity of the complex and binds the Aux/IAA proteins that need to be destroyed (47, 48). This interaction is promoted by auxin (49). The fact that TIR1 mRNA is targeted by the potential miRNAs MIR10 and MIR20 adds a further layer of negative regulation to the auxin pathway. Also, besides the genes in the ubiquitination pathway, a large group of auxin-related TFs, including auxin-response transcription factors and no apical meristems (NAMs; refs. 50 and 51), are present that are also potential targets of miRNAs (Table 1). Altogether, there is strong suggestive evidence that miRNAs play a major role in auxin signaling. To validate prediction of MIR20, we found homologs with perfectly conserved miRNA sequences with a valid stem-loop precursor structure in three plant genomes (Fig. 5, which is published as supporting information on the PNAS web site), namely Medicago truncatula (working draft sequences downloaded from the EMBL database), Populus trichocarpa (public reads downloaded from http://genome.jgi-psf.org), and Lotus corniculatus (sequences downloaded from EMBL).
Another interesting example is the potential miRNA MIR64 that targets three different gene families (Table 1) involved in sulfur metabolism. The first target encodes ATP sulfurylase (APS), the first enzyme in the sulfate assimilation pathway, which is present both in the cytosol and the chloroplast. Of the four APS proteins present in Arabidopsis (52), MIR64 can pair with the mRNAs of only three of them, and its target sequence is indeed missing from the fourth, APS2 (At1g19990). MIR64 also targets one of the three Arabidopsis sulfate transporters, namely that with low affinity induced in plant roots by sulfate starvation (53, 54). Altogether, MIR64 probably plays a specific role in the control of sulfur assimilation. The potential MIR1 targets a Cu/Zn superoxidase dismutase (CSD1; ref. 55), which is one of the main markers of programmed cell death in Arabidopsis (56). AtTOR, an ortholog of the yeast target of rapamycin involved in cell-cycle regulation during embryo development (57, 58) is also a miRNA target of interest.
False-Positives and -Negatives. We have tried to reduce the number of false-positives by combining multiple procedures and an overall conservative approach. First, the number of potential miRNAs was strongly reduced by considering the miRNA secondary structure parameters based on the reference set. By comparing this data set with the rice genome, this number could be further lowered. The combination of a statistical test on the precursor sequences and GC and low-complexity filtering on the miRNA sequences further reduced the number of candidates by one order of magnitude.
The apparently missing targets for part of our candidates (58 of 91) could be explained by the less perfect than previously thought complementarity with mRNA target sequences for some plant miRNAs, as is the case for metazoan miRNAs (1-3). Recently, 20 miRNAs were experimentally identified in rice (46), and only seven had targets with near-perfect complementarity, which suggests that pairing between miRNAs and their target mRNAs can be less stringent in plants.
In contrast, a fraction of the true miRNA sequences might have been discarded although they are true miRNAs. A sequence that is conserved outside the strict limit of the miRNA sequence might have a hit length of more than the used upper threshold value (25 nt). As mentioned, we identified six of the eight miRNAs previously known to be conserved between Arabidopsis and rice (18). For these two missing miRNAs (mir162 and mir169), the conserved regions seem indeed longer than 25 nt and, as a result, are not reported in our approach. We hope that refinements of the computational pipeline used here for the prediction of miRNA may cope with this difficulty in the future.
In conclusion, the stringent search conditions that we applied have, in addition to most of the previously described miRNA genes conserved between Arabidopsis and Oryza, uncovered a considerable number of unknown miRNAs and their targets. If these miRNAs could be confirmed by experimental evidence, the number of different processes and pathways regulated by miRNAs in Arabidopsis would significantly increase. We would also like to stress that we used a deliberately conservative approach that retained only those potential miRNAs that complied with every feature of experimentally confirmed plant miRNAs. Many of the more recently reported rice miRNAs (46) do not fulfill all these criteria, suggesting that miRNAs in the Arabidopsis genome are probably more abundant than reported here and that additional experimental and in silico studies are needed.
Note Added in Proof. Upon acceptance of this paper, Jones-Rhoades and Bartel (60) reported several miRNAs identical to those we found, thereby confirming our approach. In addition, 34 miRNAs from our list matched perfectly with expressed small RNAs deposited in the Arabidopsis Small RNA Project Database, which can be accessed at http://cgrb.orst.edu/smallRNA.
Supplementary Material
Acknowledgments
We thank Ivo Hofacker for sharing parts of his software code and many members of our research group for helpful discussions. This work was supported by an Instituut voor de aanmoediging van Innovatie door Wetenschap en Technologie in Vlaanderen postdoctoral fellowship (to J.W.).
Abbreviations: IGR, intergenic region; miRNA, microRNA; TF, transcription factor; NAM, no apical meristem.
References
- 1.Bartel, D. P. (2004) Cell 116, 281-297. [DOI] [PubMed] [Google Scholar]
- 2.Carrington, J. C. & Ambros, V. (2003) Science 301, 336-338. [DOI] [PubMed] [Google Scholar]
- 3.Lai, E. C. (2003) Curr. Biol. 13, R925-R936. [DOI] [PubMed] [Google Scholar]
- 4.Lee, R. C., Feinbaum, R. L. & Ambros, V. (1993) Cell 75, 843-854. [DOI] [PubMed] [Google Scholar]
- 5.Wightman, B., Ha, I. & Ruvkun, G. (1993) Cell 75, 855-862. [DOI] [PubMed] [Google Scholar]
- 6.Moss, E. G., Lee, R. C. & Ambros, V. (1997) Cell 88, 637-646. [DOI] [PubMed] [Google Scholar]
- 7.Reinhart, B. J., Slack, F. J., Basson, M., Pasquinelli, A. E., Bettinger, J. C., Rougvie, A. E., Horvitz, H. R. & Ruvkun, G. (2000) Nature 403, 901-906. [DOI] [PubMed] [Google Scholar]
- 8.Abrahante, J. E., Daul, A. L., Li, M., Volk, M. L., Tennessen, J. M., Miller, E. A. & Rougvie, A. E. (2003) Dev. Cell 4, 625-637. [DOI] [PubMed] [Google Scholar]
- 9.Lin, S. Y., Johnson, S. M., Abraham, M., Vella, M. C., Pasquinelli, A., Gamberi, C., Gottlieb, E. & Slack, F. J. (2003) Dev. Cell 4, 639-650. [DOI] [PubMed] [Google Scholar]
- 10.Bartel, B. & Bartel, D. P. (2003) Plant Physiol. 132, 709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton, A. J. & Baulcombe, D. C. (1999) Science 286, 950-952. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363-366. [DOI] [PubMed] [Google Scholar]
- 13.Hannon, G. J. (2002) Nature 418, 244-251. [DOI] [PubMed] [Google Scholar]
- 14.Mallory, A. C. & Vaucheret, H. (2004) Curr. Opin. Plant Biol. 7, 120-125. [DOI] [PubMed] [Google Scholar]
- 15.Vazquez, F., Gasciolli, V., Crété, P. & Vaucheret, H. (2004) Curr. Biol. 14, 346-351. [DOI] [PubMed] [Google Scholar]
- 16.Boutet, S., Vazquez, F., Liu, J., Beclin, C., Fagard, M., Gratias, A., Morel, J. B., Crete, P., Chen, X. & Vaucheret, H. (2003) Curr. Biol. 13, 843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llave, C., Kasschau, K. D., Rector, M. A. & Carrington, J. C. (2002) Plant Cell 14, 1605-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B. & Bartel, D. P. (2002) Genes Dev. 16, 1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau, N. C., Lim, L. P., Weinstein, E. G. & Bartel, D. P. (2001) Science 294, 858-862. [DOI] [PubMed] [Google Scholar]
- 20.Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B. & Bartel, D. P. (2002) Cell 110, 513-520. [DOI] [PubMed] [Google Scholar]
- 21.Park, W., Li, J., Song, R., Messing, J. & Chen, X. (2002) Curr. Biol. 12, 1484-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasschau, K. D., Xie, Z., Allen, E., Llave, C., Chapman, E. J., Krizan, K. A. & Carrington, J. C. (2003) Dev. Cell 4, 205-217. [DOI] [PubMed] [Google Scholar]
- 23.Tang, G., Reinhart, B. J., Bartel, D. P. & Zamore, P. D. (2003) Genes Dev. 17, 49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llave, C., Xie, Z., Kasschau, K. D. & Carrington, J. C. (2002) Science 297, 2053-2056. [DOI] [PubMed] [Google Scholar]
- 25.Emery, J. F., Floyd, S. K., Alvarez, J., Eshed, Y., Hawker, N. P., Izhaki, A., Baum, S. F. & Bowman, J. L. (2003) Curr. Biol. 13, 1768-1774. [DOI] [PubMed] [Google Scholar]
- 26.Aukerman, M. J. & Sakai, H. (2003) Plant Cell 15, 2730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim, L. P., Glasner, M. E., Yekta, S., Burge, C. B. & Bartel, D. P. (2003) Science 299, 1540. [DOI] [PubMed] [Google Scholar]
- 28.Lim, L. P., Lau, N. C., Weinstein, E. G., Abdelhakim, A., Yekta, S., Rhoades, M. W., Burge, C. B. & Bartel, D. P. (2003) Genes Dev. 17, 991-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lai, E. C., Tomancak, P., Williams, R. W. & Rubin, G. M. (2003) Genome Biol. 4, R42.1-R42.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grad, Y., Aach, J., Hayes, G. D., Reinhart, B. J., Church, G. M., Ruvkun, G. & Kim, J. (2003) Mol. Cell 11, 1253-1263. [DOI] [PubMed] [Google Scholar]
- 31.Mette, M. F., van der Winden, J., Matzke, M. & Matzke, A. J. (2002) Plant Physiol. 130, 6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffiths-Jones, S. (2004) Nucleic Acids Res. 32, D109-D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki, T. & Burr, B. (2000) Curr. Opin. Plant Biol. 3, 138-141. [DOI] [PubMed] [Google Scholar]
- 34.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol., 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 35.Peterson-Burch, B. D. & Voytas, D. F. (2002) Mol. Biol. Evol. 19, 1832-1845. [DOI] [PubMed] [Google Scholar]
- 36.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wuyts, J., Perrière, G. & Van de Peer, Y. (2004) Nucleic Acids Res. 32, D101-D103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown, J. W., Echeverria, M., Qu, L. H., Lowe, T. M., Bachellerie, J. P., Huttenhofer, A., Kastenmayer, J. P., Green, P. J., Shaw, P. & Marshall, D. F. (2003) Nucleic Acids Res. 31, 432-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon, C. E. (1948) Bell Syst. Tech. J. 27, 379-423. [Google Scholar]
- 40.Ambros, V., Bartel, B., Bartel, D. P., Burge, C. B., Carrington, J. C., Chen, X., Dreyfuss, G., Eddy, S. R., Griffiths-Jones, S., Marshall, M., et al. (2003) RNA 9, 277-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rice, P., Longden, I. & Bleasby, A. (2000) Trends Genet. 16, 276-277. [DOI] [PubMed] [Google Scholar]
- 42.Hofacker, I. L., Fontana, W., Stadler, P. F., Bonhoeffer, L. S., Tacker, M. & Schuster, P. (1994) Monatsh. Chem. 125, 167-188. [Google Scholar]
- 43.Dsouza, M., Larsen, N. & Overbeek, R. (1997) Trends Genet. 13, 497-498. [DOI] [PubMed] [Google Scholar]
- 44.Huala, E., Dickerman, A. W., Garcia-Hernandez, M., Weems, D., Reiser, L., LaFond, F., Hanley, D., Kiphart, D., Zhuang, M., Huang, W., et al. (2001) Nucleic Acids Res. 29, 102-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brenner, S., Williams, S. R., Vermaas, E. H., Storck, T., Moon, K., McCollum, C., Mao, J. I., Luo, S., Kirchner, J. J., Eletr, S., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 1665-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, J. F., Zhou, H., Chen, Y. Q., Luo, Q. J. & Qu, L. H. (2004) Nucleic Acids Res. 32, 1688-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vierstra, R. D. (2003) Trends Plant Sci. 8, 135-142. [DOI] [PubMed] [Google Scholar]
- 48.Hellmann, H. & Estelle, M. (2002) Science 297, 793-797. [DOI] [PubMed] [Google Scholar]
- 49.Dharmasiri, N., Dharmasiri, S., Jones, A. M. & Estelle, M. (2003) Curr. Biol. 13, 1418-1422. [DOI] [PubMed] [Google Scholar]
- 50.Xie, Q., Frugis, G., Colgan, D. & Chua, N. H. (2000) Genes Dev. 14, 3024-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hagen, G. & Guilfoyle, T. (2002) Plant Mol. Biol. 49, 373-385. [PubMed] [Google Scholar]
- 52.Hatzfeld, Y., Lee, S., Lee, M., Leustek, T. & Saito, K. (2000) Gene 248, 51-58. [DOI] [PubMed] [Google Scholar]
- 53.Lappartient, A. G., Vidmar, J. J., Leustek, T., Glass, A. D. & Touraine, B. (1999) Plant J. 18, 89-95. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi, H., Yamazaki, M., Sasakura, N., Watanabe, A., Leustek, T., Engler, J. A., Engler, G., Van Montagu, M. & Saito, K. (1997) Proc. Natl. Acad. Sci. USA 94, 11102-11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kliebenstein, D. J., Monde, R. A. & Last, R. L. (1998) Plant Physiol. 118, 637-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swidzinski, J. A., Sweetlove, L. J. & Leaver, C. J. (2002) Plant J. 30, 431-446. [DOI] [PubMed] [Google Scholar]
- 57.Vespa, L., Vachon, G., Berger, F., Perazza, D., Faure, J.-D. & Herzog, M. (2004) Plant Physiol. 134, 1283-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Menand, B., Desnos, T., Nussaume, L., Berger, F., Bouchez, D., Meyer, C. & Robaglia, C. (2002) Proc. Natl. Acad. Sci. USA 99, 6422-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Rijk, P., Wuyts, J. & De Wachter, R. (2003) Bioinformatics 19, 299-300. [DOI] [PubMed] [Google Scholar]
- 60.Jones-Rhoades, M. W. & Bartel, D. P. (2004) Mol. Cell 14, 787-799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.