Summary
To understand organ function, it is important to have an inventory of its cell types and of their corresponding marker genes. This is a particularly challenging task for human tissues like the pancreas, because reliable markers are limited. Hence, transcriptome-wide studies are typically done on pooled islets of Langerhans, obscuring contributions from rare cell types and of potential subpopulations. To overcome this challenge, we developed an automated platform that uses FACS, robotics, and the CEL-Seq2 protocol to obtain the transcriptomes of thousands of single pancreatic cells from deceased organ donors, allowing in silico purification of all main pancreatic cell types. We identify cell type-specific transcription factors and a subpopulation of REG3A-positive acinar cells. We also show that CD24 and TM4SF4 expression can be used to sort live alpha and beta cells with high purity. This resource will be useful for developing a deeper understanding of pancreatic biology and pathophysiology of diabetes mellitus.
Keywords: single-cell transcriptomics, sequencing, pancreas, islets of Langerhans
Graphical Abstract
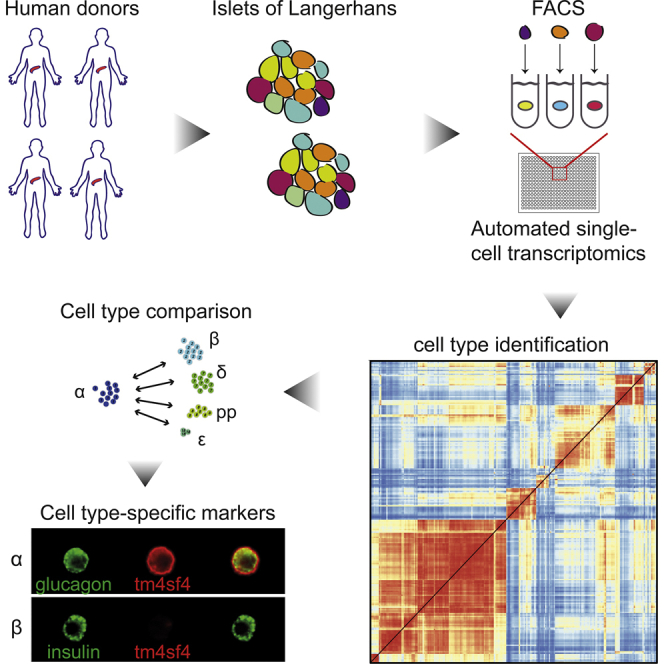
Highlights
-
•
Single-cell sequencing of human pancreas allows in silico purification of cell types
-
•
We provide cell-type-specific genes, transcription factors, and cell-surface markers
-
•
StemID finds outlier populations of acinar and beta cells
-
•
CD24 and TM4SF4 function as two markers to enrich for alpha and beta cells
Single-cell mRNA sequencing was used to describe the transcriptome of adult human pancreatic cell types. This resource was mined to find subpopulations of existing cell types and markers that can be used to purify live alpha and beta cells.
Introduction
Most organs consist of a variety of cell types with interdependent functions. To understand organ function and disease, genome-wide information on each cell type is crucial. Studies on pooled material detect global gene expression patterns but represent an average dominated by the most abundant cell types. With the advent of single-cell transcriptomics, it is possible to determine the transcriptome of individual cells, allowing the identification of cell types in an unbiased manner (Grün and van Oudenaarden, 2015, Kolodziejczyk et al., 2015, Trapnell, 2015, Wang and Navin, 2015). Initial single-cell transcriptomic studies were performed on cultured cells (Deng et al., 2014, Hashimshony et al., 2012, Islam et al., 2011, Klein et al., 2015, Shalek et al., 2013, Tang et al., 2010). Subsequent studies described cell types in the mouse lung (Treutlein et al., 2014), spleen (Jaitin et al., 2014), brain (Zeisel et al., 2015), retina (Macosko et al., 2015), small intestine (Grün et al., 2015), and pancreas (Xin et al., 2016). Studies on human tissue have so far been limited to fetal neurons (Johnson et al., 2015), glioblastomas (Patel et al., 2014), and two sets of human pancreas cells (Li et al., 2016, Wang et al., 2016). We have used single-cell sequencing of the human pancreas to reveal subpopulations of cells that show potential as progenitors (Grün et al., 2016). These studies described manually processed samples and/or low numbers of cells, which limited the number of detected genes. Here, we developed a more efficient, high-throughput method to sequence primary human cells of all pancreatic cell types.
The pancreas functions as an exocrine and endocrine gland. The exocrine compartment consists of acinar cells producing digestive enzymes and ductal cells forming channels that drain into the duodenum. The endocrine compartment consists of alpha, beta, delta, pancreatic polypeptide (PP), and epsilon cells that are found in the islets of Langerhans. Insulin-producing beta cells and glucagon-producing alpha cells play major roles in glucose homeostasis, and islet dysfunction is the hallmark of diabetes mellitus, a chronic metabolic disorder affecting approximately 9% of the adult population worldwide (WHO, 2014). Functional analysis and genetic profiling are typically performed on whole islets, masking the contribution of individual cell types to pancreas biology and disease (Bugliani et al., 2013, Cnop et al., 2014, Eizirik et al., 2012). To study heterogeneity and classify subpopulations within known cell types, single-cell resolution is essential. We developed a high-throughput approach for single-cell sequencing based on the single-cell RNA-seq by multiplexed linear amplification2 (CEL-seq2) protocol (Hashimshony et al., 2016) to create a single-cell transcriptome atlas of the human pancreas. Our method implements fluorescence-activated cell sorting (FACS), which allows the user to work with low amounts of starting material. This dataset provides an unbiased view of cell types in the human pancreas at single-cell resolution, enabling comparison of gene expression patterns among cell types and detection of subpopulations within them. This resource can be mined for genes involved in pancreatic function to define novel therapeutic targets for diseases such as diabetes mellitus.
Results
SORT-Seq Allows Deep Sequencing of Human Pancreas Cells
To assay the transcriptomes of the various human pancreatic cell types, we obtained human pancreas material from four deceased organ donors (Figure 1A). Isolation of the islets of Langerhans yielded 55%–95% islet purity (Table S1). The non-islet cells in these preparations mainly consisted of exocrine cells. After a culture period of 3–5 days, the islets were dispersed for FACS, followed by single-cell sequencing. Previously, we sorted cells from five donors, which were processed manually by single-cell RNA-seq by multiplexed linear amplification (CEL-Seq) as described (Grün et al., 2016). These yielded an average of 4,262 unique transcripts and a median of 1,958 detected genes per cell (Figures S1A and S1B) when sequencing at approximately 90.000 reads per cell. While useful to determine interesting progenitor cells and describe general differences between cell types, this dataset lacked the depth to fully describe the transcriptome of each pancreatic cell type. For example, comparing expression across endocrine cell types resulted in low numbers of differentially expressed genes (Figure S1C).
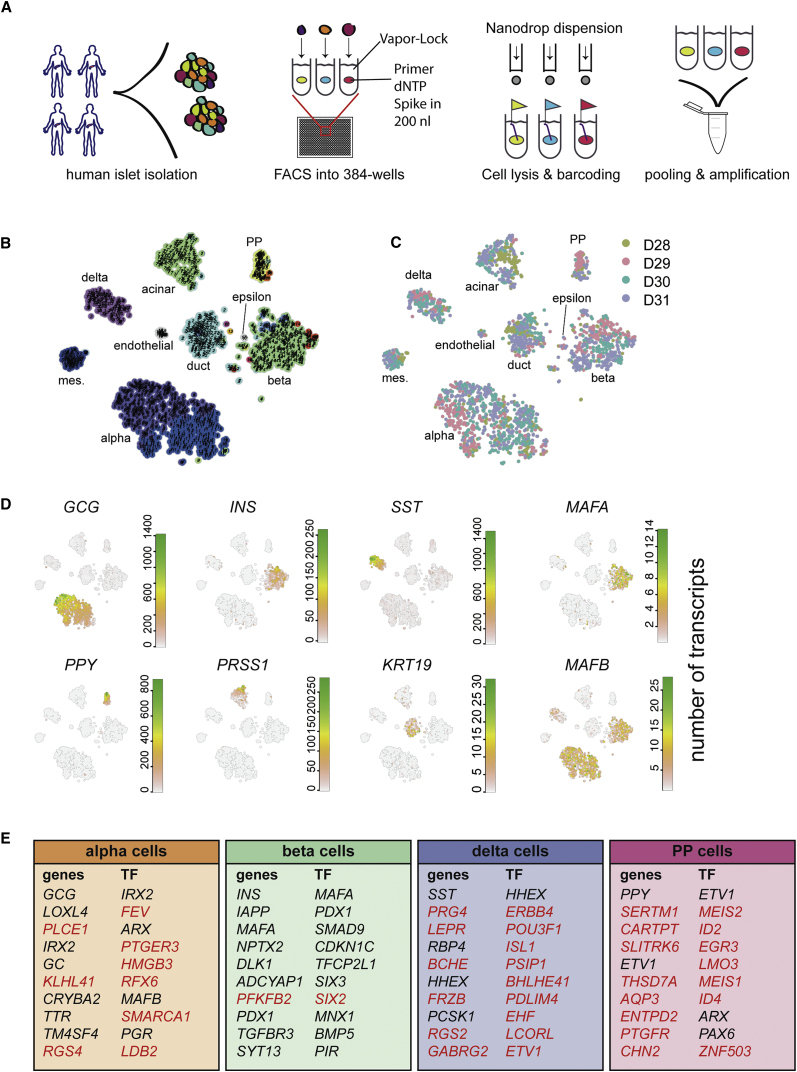
Figure 1.
SORT-Seq Allows for Deep Sequencing of Human Pancreas Cells
(A) Experimental workflow for SORT-seq. Islets were isolated from human donors. Cells were dispersed and sorted into 384-well plates with mineral oil, containing 100 nL of CEL-seq2 primers, dNTPs, and spike-ins. The RT mix was then distributed by Nanodrop II. After second-strand synthesis, material was pooled and amplified before RNA library preparation.
(B) Visualization of k-medoid clustering and cell-to-cell distances using t-SNEs. Each dot represents a single cell. Colors and numbers indicate clusters, and cell-type names are indicated with their corresponding cluster or clusters.
(C) t-SNE map highlighting donor source. Each color represents one donor.
(D) t-SNE maps highlighting the expression of marker genes for each of the six main pancreatic cell types. Transcript counts are given in a linear scale.
(E) Tables denoting the top ten differentially expressed genes and transcription factors (TFs) when comparing one cell type to all other cells in the dataset (p < 10−6). Genes whose cell-type specificity was previously unknown in the human pancreas are marked in red.
To more efficiently capture single-cell transcriptomes, we used FACS and robotics liquid handling to perform automated single-cell sequencing based on the CEL-Seq2 protocol (Hashimshony et al., 2016). We refer to this platform as SORT-seq (SOrting and Robot-assisted Transcriptome SEQuencing) (Figure 1A). Briefly, live single cells (based on DAPI and scatter properties) are sorted into 384-well plates with 5 μL of Vapor-Lock oil containing a droplet of 100 nL of CEL-seq primers, spike-ins, and dinucleotide triphosphates (dNTPs). For cDNA construction, cells are first lysed by heat, after which a robotic liquid handler dispenses reverse transcription (RT) and second-strand mix. Cells are then pooled, and the aqueous phase is extracted from the oil. The CEL-Seq2 protocol can be followed from this point onward. Compared to the manual method, the percentage of reads that could be mapped to the reference transcriptome increased from 15% to 35%. In addition, the number of unique transcripts per cell increased (median of 14,604 compared to 4,262) (Figure S1D), as did the number of genes detected per cell (median of 4,497 compared to 1,958) while sequencing an average of 41.000 reads per cell, half the depth used for the manual method (Figure S1E). This resulted in more complex single-cell libraries with more differentially expressed genes between cell types (Figure S1F).
To investigate whether we could detect the expected pancreatic cell types, we used StemID, an approach we developed for inferring the existence of stem cell populations from single-cell transcriptomic data (Grün et al., 2016). StemID calculates all pairwise cell-to-cell distances (1 − Pearson correlation) and uses this to cluster similar cells into clusters that correspond to the cell types present in the tissue (Figure S1G). This resulted in well-separated cell clusters with low intra-cluster and high inter-cluster cell-to-cell distances, as visualized in t-distributed stochastic neighbor embedding (t-SNE) maps (Figure 1B) (van der Maaten and Hinton, 2008). These maps were also used to highlight expression of specific genes across all cells (Figure 1D). To test whether the donor source influenced cluster formation, we plot donor contribution to the clusters in Figure 1C, showing that none of the clusters consist of cells from only one donor. When we compared all cells from each cell type of one donor to that of all others, we did not find major differences between donors. Most differentially expressed genes differed by less than 2-fold. As expected, XIST was upregulated in all cell types of D30 (Table S2), the only female donor of the set. The donor-independent clustering shows StemID groups cells based on cell type, rather than donor.
We found the clusters to highly express markers for all pancreatic cell types (Figure 1D). We found cluster-specific expression of GCG (alpha cells), INS (beta), SST (delta), PPY (PP), PRSS1 (acinar), KRT19 (duct), and COL1A1 (mesenchyme) (Figures 1D and S1H). Because the algorithm did not distinguish clusters with either epsilon or endothelial cells, we looked for expression of the markers GHRL or ESAM. We found two clusters of cells exclusively expressing these markers and manually annotated them as epsilon and endothelial cells (Figure 1B).
We also detected the expression of MAFA and MAFB, transcriptional regulators important for determining the identity of endocrine cell types (Nishimura et al., 2006) (Figure 1D). MAFA expression is restricted to beta cells, while MAFB expression is found in both alpha and beta cells, as previously reported in mice (Dai et al., 2012).
We next set out to generate a resource with which to compare pancreatic cell types and mine their transcriptomes for interesting genes. To this end, we compared all alpha (clusters expressing high GCG), beta, epsilon, delta, PP, duct, acinar, mesenchymal, and endothelial cells based on their distinct clustering from other cell types. Each group of cells was compared to all other cell groups, yielding a list of differentially expressed genes. The top ten of each list can be found in Figures 1E and S1I, and the full list is in Table S3. We then selected only those genes that have been reported to function as transcription factors using the TFcheckpoint database (Table S4) (Chawla et al., 2013). Several genes and transcription factors found here have never been reported as markers for specific cell types of the human pancreas (Figure 1E).
Apart from the classically known alpha cell transcription factors IRX2, ARX (Dorrell et al., 2011b), and PGR (Doglioni et al., 1990), our analysis reveals transcription factors FEV, PTGER3 (Kimple et al., 2013), SMARCA1 (Rankin and Kushner, 2010), HMGB3, and RFX6 (Piccand et al., 2014) that, to our knowledge, have not been reported to be enriched in human alpha cells and have been previously implicated in beta cell function. Some of these factors have broader expression across other endocrine cell types, such as RFX6, but they are most highly expressed in alpha cells.
Classical beta cell markers like INS, MAFA, and PDX1 (Kulkarni, 2004) top the beta cell list, and we detect PFKFB2 (Arden et al., 2008), a gene thought to regulate insulin secretion, and the transcription factor SIX2. To our knowledge, neither PFKFB2 nor SIX2 have been reported previously in human beta cells. SIX2 is known to interact with the transcription factor TCF7L2 (Xu et al., 2014), a well-known SNP for type 2 diabetes (Grant et al., 2006). This makes it interesting for further investigations in the context of beta cell function.
Apart from the classical SST and HHEX expression in delta cells (Zhang et al., 2014), genes like LEPR and GHSR imply a possible role of leptin and ghrelin on delta cell function. PP cells have substantial expression of genes related to neuronal cells, which hints at the developmental proximity of PP and neuronal cells. This has been previously described by others in the context of beta cells (Arntfield and van der Kooy, 2011, Le Roith et al., 1982)
In summary, these gene lists confirm markers and reveal gene expression patterns in the endocrine cell types that can be further investigated for their roles in cellular identity and function.
Cluster-Restricted Gene Expression Patterns and Identification of Cell-Type-Specific Genes
We next analyzed each cluster in detail to see whether the remaining differentially expressed genes corroborated the initial identification of the six major pancreatic cell types. To investigate to what extent gene expression patterns are shared among cell types, we focused on the expression of both the top differentially expressed genes and the classical marker genes (Figure 2A). In particular, the expression of hormones was restricted to individual clusters, taking up one-fifth of the transcriptome, while being near zero in other clusters. For most clusters, the top differentially expressed genes were documented markers (Table S3). For example, INS and IAPP were co-expressed in beta cells, LOXL4 was expressed with GCG (alpha cells), and PNLIP was expressed with PRSS1. PRG4 was most highly expressed in delta cells after SST. Ductal markers SPP1 and KRT19 were relatively lowly expressed but limited to the ductal cluster. Further inspection of the top differentially expressed genes per cluster yielded new cell-type-specific genes, such as ALDH1A1, which was enriched in alpha cells and co-expressed with GCG (Figures S2C and S2D).
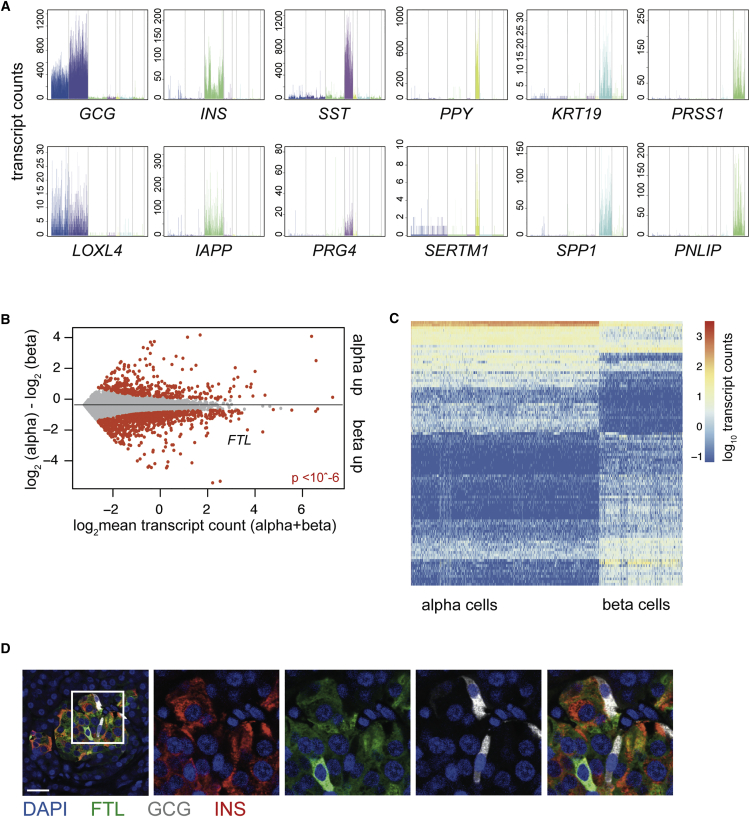
Figure 2.
Cluster-Restricted Gene Expression Patterns and Identification of Cell-Type-Specific Genes
(A) Expression of well-known marker genes (top) and the most differentially expressed gene (bottom) in each of the six main pancreatic cell types. If the most differentially expressed gene is also a canonical marker gene, the next most differentially expressed gene is shown. Gene expression values are plotted on the y axis. Each bar represents a cell, and cells are grouped by cluster with a specific color in the following order: alpha, beta, delta, PP, duct, and acinar.
(B) Differential gene expression analysis between alpha and beta cells reveals 1,376 differentially expressed genes. Gray dots indicate genes; red dots indicate significant genes (p < 10−6). The y axis indicates the log2 fold change, and the x axis shows the mean transcript count over both groups of cells.
(C) Heatmap of the top 100 differentially expressed genes between alpha and beta cells. Rows are genes, and columns are cells. Log2 expression of transcript counts for genes is plotted. Columns are ordered based on cell type (alpha on the left and beta on the right). Genes are grouped based on hierarchical clustering.
(D) Immunohistochemistry for ferritin light subunit (FTL, green) glucagon (GCG, gray), and insulin (INS, red) with counterstaining for DAPI (blue) on human pancreatic tissue sections. Scale bar represents 25 μM.
Going further down the list of differentially expressed genes continued to show cell type-restricted patterns (Figure S2A). To test whether we could use StemID clustering to compare different types of cells, we determined differentially expressed genes between all endocrine and exocrine cells. This yielded 2,858 genes that were differentially expressed (p < 10−6 after Benjamini-Hochberg correction). Clear separation of endocrine and exocrine was visible by plotting the top 100 differentially expressed genes (Figure S2B). This list consisted of many genes related to endocrine function, proving single-cell sequencing yields useful data on specific pancreatic cell types. This allowed us to continue exploring differences between more closely related cell types such as alpha and beta cells, which yielded a list of 1,376 differentially expressed genes (p < 10−6) (Figure 2B).
Plotting these differences in expression patterns showed clear cell-type-specific patterns (Figure 2C). Not surprisingly, canonical marker genes for alpha and beta cells (GCG, MAFA, IAPP, CHGB, INS, INS-IGF2, SCG2, PCSK1, and PCSK2) were in the list, as were genes found in studies that analyzed enriched populations of alpha or beta cells, such as TTR, which is specific in mouse alpha cells (Dorrell et al., 2011a); NPTX2 in beta cells (Figure S2A) (Nica et al., 2013); and group-specific component (GC) in human alpha cells (Ackermann et al., 2016). We also identified several previously unreported cell-type-specific genes for both alpha cells (CRYBA2, TM4SF4, and ALDH1A1) and beta cells (ID1, RBP4, SQSTM1, MT1X, FTL, and FTH1) (Ackermann et al., 2016, Benner et al., 2014). Many of these beta cell-specific genes have been linked to Type 2 diabetes (T2D) or to oxidative and/or endoplasmic reticulum (ER) stress responses (Åkerfeldt and Laybutt, 2011, Chen et al., 2001, Orino et al., 2001, Yang et al., 2005). To validate our results, we visualized protein levels of FTL and ALDH1A1 in tissue sections of human pancreas. FTL expression was visible in insulin-producing cells and absent from GCG-positive alpha cells in the islets of Langerhans (Figure 2D). ALDHI1A1 expression appeared to be quite similar in acinar cells and alpha cells, whereas in general, higher mRNA expression was observed in alpha cells (Figures S2C and S2D). Within the islets of Langerhans, we detected ALDH1A1 expression only in glucagon-positive alpha cells, not in other cells in the islets.
GO-Term Analysis Reveals Cell-Type-Specific Gene Expression Patterns Relevant to Diabetes and Glucose Metabolism
We used EnrichR (Chen et al., 2013) to perform gene ontology (GO)-term analysis on the full list of genes differentially expressed in each cell type compared to all other pancreatic cell types. We determined the top 15 enriched GO terms for alpha, beta, delta, and PP cells (Figure S3A). In addition, we provide the lists of GO terms for each type, along with the genes that are involved in this GO term (Table S5). Parsing the file for alpha cell-related GO terms shows the inositol receptor ITPR1 is involved in insulin secretion. ITPR1 has previously been associated with a diabetic phenotype in mice (Figure S3C) (Ye et al., 2011). GO terms, like negative regulation of nervous system development, are highest in PP cells, indicating these cells have a more neuronal nature than other cells. The serotonin transporter SLC6A4 is found predominantly in PP cells (Figure S3C) and has well-documented roles in neurons and behavior (Murphy and Lesch, 2008). To focus on differences between cell types in more detail, we performed GO-term analysis on gene sets obtained after comparing beta cells to alpha, delta, and PP cells separately (Figure S3B). In particular, delta cells show more hits in behavior and synaptic transmission. The ghrelin receptor GHSR is involved in several of these processes. This receptor is only present in delta cells (Figure S3C), indicating a role for ghrelin in delta cell function, which has indeed been demonstrated in mice (DiGruccio et al., 2016). These results are an example of how genes obtained in this resource can be used for GO-term analysis. By zooming in on specific genes from interesting terms, we can generate hypotheses regarding cell-type-specific processes in the human pancreas.
Outlier Identification Reveals Heterogeneity within Acinar and Beta Cells
We set out to analyze cellular heterogeneity by detecting outliers within specific populations of cells using the RaceID algorithm (Grün et al., 2015). The most striking results were found in beta and acinar cells, in which we found subpopulations of cells with distinct gene expression patterns. In beta cells, the most significant genes dictating this heterogeneity were SRXN1, SQSTM1, and three ferritin subunits: FTH1P3, FTH1, and FTL (Figures 3A and S4A). All these genes were highly expressed in cluster 2 (Figure S4A) and are implicated in response to ER and oxidative stress (Orino et al., 2001, Zhou et al., 2015, Rantanen et al., 2013). The main acinar cluster split into four clusters, of which cluster 2 showed the high levels of REG3A expression (Figures 3C, 3D, and S4B). Meanwhile, the acinar marker PRSS1 was expressed in all clusters but was highest in a group of cells in clusters 3 and 4 (Figure S4C).
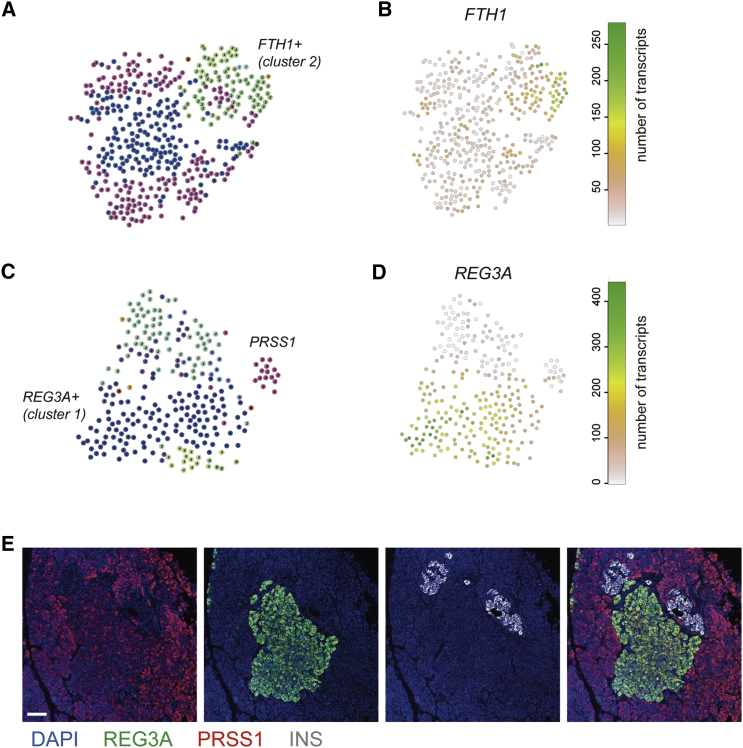
Figure 3.
Outlier Identification Reveals Heterogeneity within Acinar and Beta Cells
(A) t-SNE map of RaceID clusters after clustering of only beta cells.
(B) t-SNE map highlighting the expression of FTH1.
(C) t-SNE map of RaceID clusters after clustering of only acinar cells.
(D) t-SNE map highlighting the expression of REG3A.
(E) Immunohistochemistry for REG3A (green), trypsin (red), and insulin (gray), with counterstaining for DAPI (blue). Scale bar represents 75 μM.
To confirm the existence of subpopulations of REG3A-positive acinar cells, we stained sections of human pancreas for REG3A and PRSS1. Scattered individual REG3A/PRSS1 double-positive cells were observed (Figure S4D) in acinar tissue. Interestingly, we also detected large clusters of brightly REG3A/PRSS1-positive acinar cells close to the islets of Langerhans (Figure 3E).
To characterize subpopulations obtained in silico in more depth, we averaged the expression profiles of all single cells belonging to the different subpopulations. By averaging and pooling the transcriptomes from these groups of cells, we achieve transcriptome coverage that is similar to that of bulk sequencing experiments (Table S6).
In summary, we detected subpopulations of beta cells expressing higher levels of FTH1 and validated acinar subpopulations expressing high levels of REG3A. This subtype of acinar cell merits more investigation, because the role of REG3A in pancreas biology is unclear.
Enrichment of Alpha and Beta Cells Based on Cell-Surface Markers
We next mined our transcriptome resource for novel cell-surface markers to enrich specific pancreatic cell types using live-cell sorting. As a proof of principle, we set out to deplete the exocrine fraction from islet isolations of low purity. We found cell-surface markers CD24 and CD44 were restricted to acinar and ductal clusters (Figure 4A). Next, we prepared six FACS libraries, two with only live cells and four with negative selection for CD24 and CD44 (Figure 4B). This yielded compact clusters of cells that correspond to the main pancreatic cell types (Figure S5B). Nearly all endocrine cells were derived from the negatively selected libraries (Figure S5A), demonstrating the efficiency of the predicted cell-surface markers. Alpha cells seemed to be preferentially enriched with this strategy (Figure S5A).
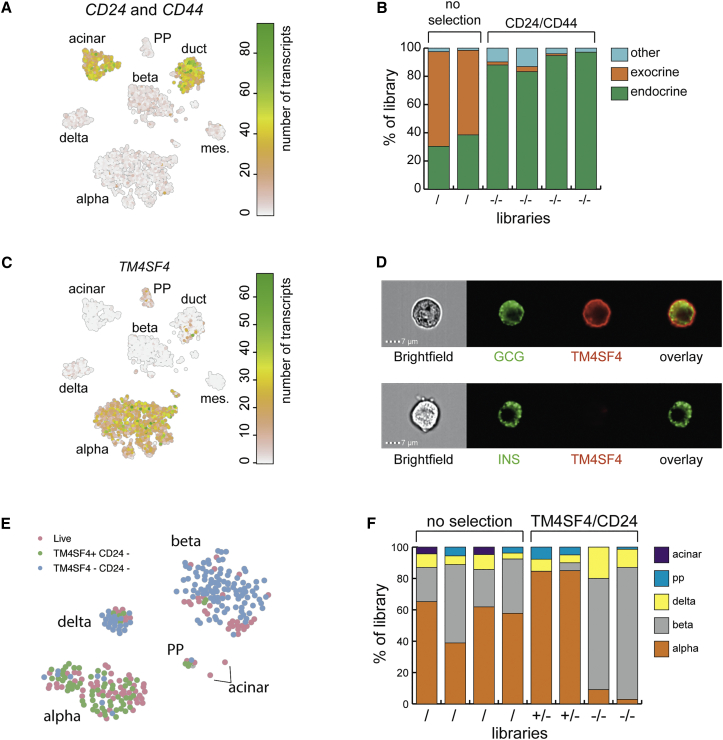
Figure 4.
Enrichment of Alpha and Beta Cells Based on Cell-Surface Markers
(A) t-SNE map highlighting the combined expression of CD24 and CD44.
(B) Results of FACS enrichment based on selection against CD24/CD44. Two libraries were selected for live staining (/), and four were selected against CD24 and CD44 expression (−/−). The y axis indicates the portion of the library consisting of a particular cell type. Colors indicate cell types.
(C) t-SNE map highlighting the expression of TM4SF4.
(D) Imagestream analysis of dispersed, fixed single-cells from human pancreas. The left panels show a bright field image of the cell and then immunostaining against glucagon (green) and Tm4sf4 (red). The lower panel shows insulin in green.
(E) t-SNE map highlighting libraries from a TM4SF4/CD24 sort. Cells that were not stained are in pink. Cells sorted for TM4SF4+/CD24− are in green and for TM4SF4−/CD24− are in blue.
(F) Results of FACS enrichment based on selection for TM4SF4/CD24. Four libraries were selected for live staining (/). Two libraries were TM4SF4+/CD24− (+/−), and two were TM4SF4−/CD24− (−/−). The y axis indicates the portion of the library consisting of a particular cell type. Colors indicate cell types.
To test whether we could enrich for one pancreatic cell type, we explored alpha cell-surface markers, finding TM4SF4, a tetraspanin family member that has been linked to pancreatic development (Anderson et al., 2011) and to be specifically expressed in alpha cells, with sparse expression in PP cells (Figure 4C). To verify the membrane-localized expression of TM4SF4 in alpha cells, we performed imaging flow cytometry analysis on fixed cells that were co-stained with either glucagon or insulin and TM4SF4 antibodies. We found TM4SF4 to be localized at the membrane of alpha cells but not that of beta cells (Figure 4D). To test whether this antibody can be used to enrich for alpha cells, we processed eight libraries from an endocrine-rich islet extraction (Table S1): four libraries were composed of live cells, two were CD24−/TM4SF4+, and two were CD24−/TM4SF4−. We found the main endocrine pancreatic cell types after clustering (Figure S5C). Libraries sorted for TM4SF4 consisted of >85% alpha cells. When selecting against TM4SF4 and CD24, alpha cells were depleted from the resulting population and enrichment for beta cells became possible with similar purity (Figure 4F).
In conclusion, this shows that our resource can be used to mine for genes with a specific subcellular location in a pancreatic cell type of choice. Table S7 provides a list of cell-type-enriched cell-surface markers in each of the main pancreatic cell types.
Discussion
Scarcity of material, lack of reliable cell-surface markers, and analysis of pooled populations of cells often hamper analysis of human organ cell-type composition. Most importantly, methods relying on pooled cells average gene expression profiles over thousands of cells, masking any heterogeneity to be found within one cell type and potentially missing interesting intermediate cell types. To overcome these challenges, we have sequenced single cells from donor pancreata from four donors using SORT-seq, a FACS-compatible, automated version of the CEL-Seq2 protocol. We readily detected several clusters corresponding to the canonical pancreatic cell types, allowing us to purify cell types in silico for further analysis.
Due to consideration for transplantation, the islets obtained for this study were cultured for 3–5 days before dispersion to single cells and FACS. Culture conditions might affect the varied pancreatic cell types differently (progenitor cells are more likely to be affected than terminally differentiated cell types). However, shorter culture times for human islets are difficult to achieve, and we could not detect any major biases among donors, irrespective of their culture times.
Because the efficiency of single-cell sequencing (especially when using manual TRIzol-based methods) is on the order of 10% (Grün et al., 2014), lowly expressed genes are detected only sporadically. However, sequencing many cells enabled us to detect transcription factors, rare cell types, and heterogeneity within canonical pancreatic cell types such as acinar and beta cells. To further test the predictive power of this resource, we describe a panel of cell-surface markers specifically expressed in exocrine or alpha cells. Using these markers, we were able to enrich for endocrine, alpha, and beta cells.
In conclusion, we present this dataset as a resource that can be used to study pancreas composition and function. We envision broad applicability of this single-cell transcriptome atlas of the human pancreas to improve our understanding of pancreas biology and diabetes research.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Ftl | Abcam | ab69090; RRID: AB_1523609 |
| Mouse anti-Glucagon | Abcam | ab10988; RRID: AB_297642 |
| Guinea pig anti-Insulin | Abcam | ab7842; RRID: AB_306130 |
| Mouse anti-trypsin-1 | Santacruz | sc-137077; RRID: AB_2300318 |
| Rabbit anti-Reg3a | Abcam | ab134309 |
| Rabbit anti-Aldh1a1 | Abcam | ab23375; RRID: AB_2224009 |
| TM4SF4-APC | BD | FAB7998A |
| FITC-mouse anti Human CD24 Clone ML5 | BD | 560992; RRID: AB_10562033 |
| PE-mouse anti CD44(156-3C11) | Cell Signaling | 8724S; RRID: AB_10829611 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CMRL 1066 medium | Mediatech | 99663-CV |
| Accutase | StemCell Technologies, Inc. | 07920 |
| Vapor-lock | QIAGEN | 981611 |
| Critical Commercial Assays | ||
| BD Cytofix/Cytoperm Fixation/Permeabilization Kit | BD | 554717 |
| Thermo Scientific reagents for CEL-Seq2 | Hashimshony et al., 2016 | N/A |
| Deposited Data | ||
| Single-cell mRNA sequencing of cells from the pancreas of 4 human donors | this paper | GEO: GSE85241 |
| Sequence-Based Reagents | ||
| Reagents for library prep from CEL-Seq2 | Hashimshony et al., 2016 | N/A |
| Software and Algorithms | ||
| IDEAS software | EMD Millipore | N/A |
| StemID algorithm | Grün et al., 2016 | N/A |
| Rstudio software | Version 0.99.491 | |
| Bwa | Li and Durbin, 2009 | N/A |
| Other | ||
| Nanodrop II liquid handling platform (or any other liquid handling platform that can dispense sub-microliter quantities quickly into 384-well plates | Innovadyne | N/A |
| SP8 confocal microscope | Leica | N/A |
| Amnis ImagestreamX Mark II Imaging Flow cytometer | EMD Millipore | N/A |
| 384-well plates for sorting and SORT-Seq protocol | Biorad | HSP3801 |
| FACSJazz (or any other FACS-machine that can sort into 384-well plates) | BD | N/A |
Contact for Reagent and Resource Sharing
Further information and requests may be directed to primary contact Alexander van Oudenaarden, Hubrecht Institute (a.vanoudenaarden@hubrecht.eu).
Experimental Model and Subject Details
Human cadaveric donor pancreata were procured through a multiorgan donor program. Pancreatic tissue was only used if the pancreas could not be used for clinical pancreas or islet transplantation, only if research consent was given and according to national laws. In total, 4 human donor pancreata were procured (3 male, 1 female). See Table S1 for details on donor age, sex and BMI.
Method Details
Human Islet Isolation, Dispersion, and Sorting
Human islet isolations from pancreatic tissue were performed in the islet isolation facility of the Leiden University Medical Center according to a modified protocol originally described by Ricordi et al., (1988). Islets were cultured in CMRL 1066 medium (5.5 mM glucose) (Mediatech) supplemented with 10% human serum, 20 μg/ml ciprofloxacin, 50 μg/ml gentamycin, 2 mM L-glutamin, 0.25 μg/ml fungizone, 10 mM HEPES and 1.2 mg/ml nicotinamide for 3-6 days. Islets were maintained in culture at 37°C in a 5% CO2 humidified atmosphere. Medium was refreshed the day after isolation and every 2-3 days thereafter until cell sorting. The islets were cultured for 3.-5 days after islet isolation. Culture time depended on the decision time needed for considering islets for transplantation and FACS.
For cell sorting cultured Islets were briefly washed in cold PBS. The islet pellet was then suspended in 1 ml of Accutase (Stemcell technologies) per 5000 islet equivalents and incubated at 37 degrees with gentle intermittent shaking for 8-10 min until the islets were dispersed into single cells. The digestion process was stopped using an excess volume of cold RPMI medium containing 10% FCS. The dispersed tissue was washed briefly with cold PBS followed by filtering through a sieve to get rid of any debris and undigested material. DAPI was added to access the viability of the cells. The tissue was stored on ice until sorting using a FACS Aria II or FACSJazz (BD biosciences). Live single cells (based on DAPI exclusion and forward/side scatter properties) were sorted into 384-well hard shell plates (Biorad) with 5 μl of vapor-lock (QIAGEN) containing 100-200 nl of RT primers, dNTPs and synthetic mRNA Spike-Ins and immediately spun down and frozen to −80°C. For cells sorted on cell surface markers; filtered, dispersed cells were incubated with FITC-CD24 (BD, 560992), PE-CD44 (Cell signaling, 8724S) and/or APC-TM4SF4 (BD, FAB7998A) antibodies for 30 min post dispersion on ice, followed by brief washing and sorting as above.
Single-Cell mRNA Sequencing of Single Cells
For SORT-seq, cells were lysed by 5 min at 65°C, after which RT and second strand mixes were dispersed with the Nanodrop II liquid handling platform (GC biotech). Aqueous phase was separated from the oil phase after pooling all cells in one library, followed by IVT transcription. The CEL-Seq2 protocol was used for library prep. Primers consisted of a 24 bp polyT stretch, a 4bp random molecular barcode (UMI), a cell-specific 8bp barcode, the 5′ Illumina TruSeq small RNA kit adaptor and a T7 promoter. mRNA of each cell was then reverse transcribed, converted to double-stranded cDNA, pooled and in vitro transcribed for linear following the CEL-Seq 2 protocol (Hashimshony et al., 2016). Illumina sequencing libraries were then prepared with the TruSeq small RNA primers (Illumina) and sequenced paired-end at 75 bp read length the Illumina NextSeq.
Immunofluorescence and Imaging Flow Cytometry
Pancreatic tissue samples were fixed overnight in 4% formaldehyde (Klinipath), stored in 70% ethanol, and subsequently embedded in paraffin. Sections were deparaffinized in xylene and rehydrated in a series of ethanol, followed by heat assisted antigen retrieval in citric buffer (pH 6.0). Sections were blocked by incubating with 2% normal donkey serum and 1% lamb serum in PBS. Primary antibodies included rabbit anti-FTL (ab69090), mouse anti-Glucagon (ab10988) and guinea pig anti-Insulin (ab7842), mouse anti-trypsin-1 (sc-137077), rabbit anti-REG3a (ab134309) and rabbit anti-ALDH1A1 (ab23375). Sections were incubated in with primary antibody in PBS/1% lamb serum at 4°C overnight. Alexa Fluor 488-, 568- and 647- conjugated secondary antibodies against rabbit, mouse, and guinea pig IgG as appropiate (Life Technologies A11008, A10037 and A21450) were diluted 1:200 and incubated at room temperature for 1 hr. Nuclear counterstaining was done with DAPI and by additionally embedding with DAPI vectashield (Vector Laboratories #H-1500). Imaging was done on a Leica SP8 confocal microscope using hybrid detectors.
TM4SF4 staining on alpha versus the beta cells was performed on fixed, stained single cells from dispersed human islets. Dispersed Islet cells were fixed with 4%PFA and washed using 2% FCS/PBS, followed by permeabilization using Perm/Wash buffer from BD Cytofix/Cytoperm Fixation/Permeabilization Kit (Cat. 554717) 15 min at room temperature. The samples were incubated with antibodies diluted in Perm/Wash buffer rabbit anti glucagon (1:200) or guinea pig anti insulin (1:200) or anti TM4SF4-APC (1:50) for 30 min at room temperature. Alexa Fluor 488- conjugated secondary antibodies (directly or in biotin-streptavidin system) against rabbit, and guinea pig as appropriate (Life Technologies A11008) were diluted 1:200 and incubated at room temperature for 30 min. These samples were imaged using Amnis ImagestreamX Mark II Imaging Flow cytometer (EMD Millipore, WA USA) with 488 nm and 642 nm lasers respectively. Analysis was done using the IDEAS software.
Quantification and Statistical Analysis
Data Analysis
Paired-end reads from illumina sequencing were aligned to the human transcriptome with BWA (Li and Durbin, 2009). Read 1 was used for assigning reads to correct cells and libraries, while read 2 was mapped to gene models. Reads that mapped equally well to multiple locations were discarded. Read counts were first corrected for UMI barcode by removing duplicate reads that had identical combinations of library, cellular, and molecular barcodes and were mapped to the same gene. Transcript counts were then adjusted to the expected number of molecules based on counts, 256 possible UMI’s and poissonian counting statistics.
Samples were normalized by downsampling to a minimum number of 6000 transcripts. StemID was used to cluster cells and to perform outlier analysis. Differentially expressed genes between two subgroups of cells were identified similar to a previously published method (Anders and Huber, 2010). First, a negative binomial distribution was calculated reflecting the gene expression variability within each subgroup based on the background model for the expected transcript count variability computed by StemID (Grün et al., 2016). Using these distributions a p value for the observed difference in transcript counts between the two subgroups is computed as described in Anders and Huber (2010). These p values were corrected for multiple testing by the Benjamini-Hochberg method.
Data and Software Availability
Software
The StemID algorithm and custom scripts (Data S1) were run with RStudio, version 0.99.491.
Data Resources
The accession number for the single-cell sequencing data described in this study is GEO: GSE85241.
Author Contributions
M.J.M., G.D., E.J.P.d.K., and A.v.O. conceived the project. M.J.M. and G.D. carried out the experiments. M.A.E. supervised the human islet isolation procedure. D.G. helped with StemID. N.G., T.D., E.J., L.v.G., and F.C. aided with the experiments. M.J.M., G.D., and A.v.O analyzed the data. M.J.M., G.D., E.J.P.d.K., and A.v.O wrote the manuscript.
Acknowledgments
We thank Tamar Hashimshony and Itai Yanai for sharing CEL-Seq2. We thank USF for sequencing, Anko de Graaf for help with microscopy, and Reinier van der Linden for FACS. Many thanks to Nicola Crosetto for ideas on automation of CEL-Seq. This work was supported by an ERC Advanced grant (ERC-AdG 294325-GeneNoiseControl), a NWO VICI award, and grants from Stichting DON and the Dutch Diabetes Research Foundation. N.G. is supported by the JDRF.
Published: September 29, 2016
Footnotes
Supplemental Information includes five figures, seven tables, and one data file and can be found with this article online at http://dx.doi.org/10.1016/j.cels.2016.09.002.
Contributor Information
Eelco J.P. de Koning, Email: e.koning@hubrecht.eu.
Alexander van Oudenaarden, Email: a.vanoudenaarden@hubrecht.eu.
Supplemental Information
List of differentially expressed genes in each cell type between donor D30 and the other three donors.
Differentially expressed genes between each cell type compared to all others (across all donors).
Differentially expressed transcription factors between each cell type compared to all others (across all donors).
GO-term analysis for alpha, beta, delta, and PP cells compared to all others (across all donors).
Average of gene expression across all cells of acinar and beta subpopulations.
Differentially expressed cell-surface markers factors between each cell type compared to all others (across all donors).
Data analysis script detailing StemID parameters and differential gene expression analysis between one cell type and all others.
References
- Ackermann A.M., Wang Z., Schug J., Naji A., Kaestner K.H. Integration of ATAC-seq and RNA-seq identifies human alpha cell and beta cell signature genes. Mol. Metab. 2016;5:233–244. doi: 10.1016/j.molmet.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åkerfeldt M.C., Laybutt D.R. Inhibition of Id1 augments insulin secretion and protects against high-fat diet-induced glucose intolerance. Diabetes. 2011;60:2506–2514. doi: 10.2337/db11-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. Differential expression analysis for sequence count data. (2010) Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K.R., Singer R.A., Balderes D.A., Hernandez-Lagunas L., Johnson C.W., Artinger K.B., Sussel L. The L6 domain tetraspanin Tm4sf4 regulates endocrine pancreas differentiation and directed cell migration. Development. 2011;138:3213–3224. doi: 10.1242/dev.058693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden C., Hampson L.J., Huang G.C., Shaw J.A.M., Aldibbiat A., Holliman G., Manas D., Khan S., Lange A.J., Agius L. A role for PFK-2/FBPase-2, as distinct from fructose 2,6-bisphosphate, in regulation of insulin secretion in pancreatic beta-cells. Biochem. J. 2008;411:41–51. doi: 10.1042/BJ20070962. [DOI] [PubMed] [Google Scholar]
- Arntfield M.E., van der Kooy D. β-Cell evolution: how the pancreas borrowed from the brain: The shared toolbox of genes expressed by neural and pancreatic endocrine cells may reflect their evolutionary relationship. BioEssays. 2011;33:582–587. doi: 10.1002/bies.201100015. [DOI] [PubMed] [Google Scholar]
- Benner C., van der Meulen T., Cacéres E., Tigyi K., Donaldson C.J., Huising M.O. The transcriptional landscape of mouse beta cells compared to human beta cells reveals notable species differences in long non-coding RNA and protein-coding gene expression. BMC Genomics. 2014;15:620. doi: 10.1186/1471-2164-15-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugliani M., Liechti R., Cheon H., Suleiman M., Marselli L., Kirkpatrick C., Filipponi F., Boggi U., Xenarios I., Syed F. Microarray analysis of isolated human islet transcriptome in type 2 diabetes and the role of the ubiquitin-proteasome system in pancreatic beta cell dysfunction. Mol. Cell. Endocrinol. 2013;367:1–10. doi: 10.1016/j.mce.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Chawla K., Tripathi S., Thommesen L., Lægreid A., Kuiper M. TFcheckpoint: a curated compendium of specific DNA-binding RNA polymerase II transcription factors. Bioinformatics. 2013;29:2519–2520. doi: 10.1093/bioinformatics/btt432. [DOI] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Carlson E.C., Pellet L., Moritz J.T., Epstein P.N. Overexpression of metallothionein in pancreatic beta-cells reduces streptozotocin-induced DNA damage and diabetes. Diabetes. 2001;50:2040–2046. doi: 10.2337/diabetes.50.9.2040. [DOI] [PubMed] [Google Scholar]
- Cnop M., Abdulkarim B., Bottu G., Cunha D.A., Igoillo-Esteve M., Masini M., Turatsinze J.V., Griebel T., Villate O., Santin I. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes. 2014;63:1978–1993. doi: 10.2337/db13-1383. [DOI] [PubMed] [Google Scholar]
- Dai C., Brissova M., Hang Y., Thompson C., Poffenberger G., Shostak A., Chen Z., Stein R., Powers A.C. Islet-enriched gene expression and glucose-induced insulin secretion in human and mouse islets. Diabetologia. 2012;55:707–718. doi: 10.1007/s00125-011-2369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Q., Ramsköld D., Reinius B., Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- DiGruccio M.R., Mawla A.M., Donaldson C.J., Noguchi G.M., Vaughan J., Cowing-Zitron C., van der Meulen T., Huising M.O. Comprehensive alpha, beta and delta cell transcriptomes reveal that ghrelin selectively activates delta cells and promotes somatostatin release from pancreatic islets. Mol. Metab. 2016;5:449–458. doi: 10.1016/j.molmet.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doglioni C., Gambacorta M., Zamboni G., Coggi G., Viale G. Immunocytochemical localization of progesterone receptors in endocrine cells of the human pancreas. Am. J. Pathol. 1990;137:999–1005. [PMC free article] [PubMed] [Google Scholar]
- Dorrell C., Grompe M.T., Pan F.C., Zhong Y., Canaday P.S., Shultz L.D., Greiner D.L., Wright C.V., Streeter P.R., Grompe M. Isolation of mouse pancreatic alpha, beta, duct and acinar populations with cell surface markers. Mol. Cell. Endocrinol. 2011;339:144–150. doi: 10.1016/j.mce.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorrell C., Schug J., Lin C.F., Canaday P.S., Fox A.J., Smirnova O., Bonnah R., Streeter P.R., Stoeckert C.J., Jr., Kaestner K.H., Grompe M. Transcriptomes of the major human pancreatic cell types. Diabetologia. 2011;54:2832–2844. doi: 10.1007/s00125-011-2283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eizirik D.L., Sammeth M., Bouckenooghe T., Bottu G., Sisino G., Igoillo-Esteve M., Ortis F., Santin I., Colli M.L., Barthson J. The human pancreatic islet transcriptome: expression of candidate genes for type 1 diabetes and the impact of pro-inflammatory cytokines. PLoS Genet. 2012;8:e1002552. doi: 10.1371/journal.pgen.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant S.F.A., Thorleifsson G., Reynisdottir I., Benediktsson R., Manolescu A., Sainz J., Helgason A., Stefansson H., Emilsson V., Helgadottir A. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- Grün D., van Oudenaarden A. Design and analysis of single-cell sequencing experiments. Cell. 2015;163:799–810. doi: 10.1016/j.cell.2015.10.039. [DOI] [PubMed] [Google Scholar]
- Grün D., Kester L., van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat. Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- Grün D., Lyubimova A., Kester L., Wiebrands K., Basak O., Sasaki N., Clevers H., van Oudenaarden A. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- Grün D., Muraro M.J., Boisset J.-C., Wiebrands K., Lyubimova A., Dharmadhikari G., van den Born M., van Es J., Jansen E., Clevers H. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell. 2016;19:266–277. doi: 10.1016/j.stem.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimshony T., Wagner F., Sher N., Yanai I. CEL-seq: single-cell RNA-seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Hashimshony T., Senderovich N., Avital G., Klochendler A., de Leeuw Y., Anavy L., Gennert D., Li S., Livak K.J., Rozenblatt-Rosen O. CEL-seq2: sensitive highly-multiplexed single-cell RNA-seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S., Kjällquist U., Moliner A., Zajac P., Fan J.B., Lönnerberg P., Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res. 2011;21:1160–1167. doi: 10.1101/gr.110882.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin D.A., Kenigsberg E., Keren-Shaul H., Elefant N., Paul F., Zaretsky I., Mildner A., Cohen N., Jung S., Tanay A., Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.B., Wang P.P., Atabay K.D., Murphy E.A., Doan R.N., Hecht J.L., Walsh C.A. Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat. Neurosci. 2015;18:637–646. doi: 10.1038/nn.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimple M.E., Keller M.P., Rabaglia M.R., Pasker R.L., Neuman J.C., Truchan N.A., Brar H.K., Attie A.D. Prostaglandin E2 receptor, EP3, is induced in diabetic islets and negatively regulates glucose- and hormone-stimulated insulin secretion. Diabetes. 2013;62:1904–1912. doi: 10.2337/db12-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A.M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D.A., Kirschner M.W. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161:1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodziejczyk A.A., Kim J.K., Svensson V., Marioni J.C., Teichmann S.A. The technology and biology of single-cell RNA sequencing. Mol. Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- Kulkarni R.N. The islet beta-cell. Int. J. Biochem. Cell Biol. 2004;36:365–371. doi: 10.1016/j.biocel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Le Roith D., Shiloach J., Roth J. Is there an earlier phylogenetic precursor that is common to both the nervous and endocrine systems? Peptides. 1982;3:211–215. doi: 10.1016/0196-9781(82)90080-8. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Klughammer J., Farlik M., Penz T., Spittler A., Barbieux C., Berishvili E., Bock C., Kubicek S. Single-cell transcriptomes reveal characteristic features of human pancreatic islet cell types. EMBO Rep. 2016;17:178–187. doi: 10.15252/embr.201540946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D.L., Lesch K.-P. Targeting the murine serotonin transporter: insights into human neurobiology. Nat. Rev. Neurosci. 2008;9:85–96. doi: 10.1038/nrn2284. [DOI] [PubMed] [Google Scholar]
- Nica A.C., Ongen H., Irminger J., Bosco D., Berney T., Antonarakis S.E., Halban P.A., Dermitzakis E.T. Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome. Genome Res. 2013;23:1554–1562. doi: 10.1101/gr.150706.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura W., Kondo T., Salameh T., El Khattabi I., Dodge R., Bonner-Weir S., Sharma A. A switch from MafB to MafA expression accompanies differentiation to pancreatic beta-cells. Dev Biol. 2006;293:526–539. doi: 10.1016/j.ydbio.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orino K., Lehman L., Tsuji Y., Ayaki H., Torti S.V., Torti F.M. Ferritin and the response to oxidative stress. Biochem. J. 2001;357:241–247. doi: 10.1042/0264-6021:3570241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccand J., Strasser P., Hodson D.J., Meunier A., Ye T., Keime C., Birling M.-C., Rutter G.A., Gradwohl G. Rfx6 maintains the functional identity of adult pancreatic β cells. Cell Rep. 2014;9:2219–2232. doi: 10.1016/j.celrep.2014.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin M.M., Kushner J.A. Aging induces a distinct gene expression program in mouse islets. Islets. 2010;2:345–352. doi: 10.4161/isl.2.6.13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantanen K., Pursiheimo J.-P., Högel H., Miikkulainen P., Sundström J., Jaakkola P.M. p62/SQSTM1 regulates cellular oxygen sensing by attenuating PHD3 activity through aggregate sequestration and enhanced degradation. J. Cell Sci. 2013;126:1144–1154. doi: 10.1242/jcs.115667. [DOI] [PubMed] [Google Scholar]
- Ricordi C., Lacy P.E., Finke E.H., Olack B.J., Scharp D.W. Automated method for isolation of human pancreatic islets. Diabetes. 1988;37:413–420. doi: 10.2337/diab.37.4.413. [DOI] [PubMed] [Google Scholar]
- Shalek A.K., Satija R., Adiconis X., Gertner R.S., Gaublomme J.T., Raychowdhury R., Schwartz S., Yosef N., Malboeuf C., Lu D. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Bao S., Lee C., Nordman E., Wang X., Lao K., Surani M.A. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-seq analysis. Cell Stem Cell. 2010;6:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C. Defining cell types and states with single-cell genomics. Genome Res. 2015;25:1491–1498. doi: 10.1101/gr.190595.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., Quake S.R. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L., Hinton G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- Wang Y., Navin N.E. Advances and applications of single-cell sequencing technologies. Mol. Cell. 2015;58:598–609. doi: 10.1016/j.molcel.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.J., Schug J., Won K.J., Liu C., Naji A., Avrahami D., Golson M.L., Kaestner K.H. Single cell transcriptomics of the human endocrine pancreas. Diabetes. 2016:db160405. doi: 10.2337/db16-0405. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2014). Global status report on noncommunicable diseases 2014. http://www.wsmi.org/wp-content/uploads/2015/01/Global-Status-Report-NCDs-2014.pdf.
- Xin Y., Kim J., Ni M., Wei Y., Okamoto H., Lee J., Adler C., Cavino K., Murphy A.J., Yancopoulos G.D. Use of the Fluidigm C1 platform for RNA sequencing of single mouse pancreatic islet cells. Proc. Natl. Acad. Sci. USA. 2016;113:3293–3298. doi: 10.1073/pnas.1602306113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Liu H., Park J.-S., Lan Y., Jiang R. Osr1 acts downstream of and interacts synergistically with Six2 to maintain nephron progenitor cells during kidney organogenesis. Development. 2014;141:1442–1452. doi: 10.1242/dev.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Graham T.E., Mody N., Preitner F., Peroni O.D., Zabolotny J.M., Kotani K., Quadro L., Kahn B.B. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature. 2005;436:356–362. doi: 10.1038/nature03711. [DOI] [PubMed] [Google Scholar]
- Ye R., Ni M., Wang M., Luo S., Zhu G., Chow R.H., Lee A.S. Inositol 1,4,5-trisphosphate receptor 1 mutation perturbs glucose homeostasis and enhances susceptibility to diet-induced diabetes. J. Endocrinol. 2011;210:209–217. doi: 10.1530/JOE-11-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., McKenna L.B., Bogue C.W., Kaestner K.H. The diabetes gene Hhex maintains δ-cell differentiation and islet function. Genes Dev. 2014;28:829–834. doi: 10.1101/gad.235499.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Duan S., Zhou Y., Yu S., Wu J., Wu X., Zhao J., Zhao Y. Sulfiredoxin-1 attenuates oxidative stress via Nrf2/ARE pathway and 2-Cys Prdxs after oxygen-glucose deprivation in astrocytes. J. Mol. Neurosci. 2015;55:941–950. doi: 10.1007/s12031-014-0449-6. [DOI] [PubMed] [Google Scholar]
- Zeisel A., Munoz-Manchado A.B., Codeluppi S., Lonnerberg P., La Manno G., Jureus A., Marques S., Munguba H., He L., Betsholtz C. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of differentially expressed genes in each cell type between donor D30 and the other three donors.
Differentially expressed genes between each cell type compared to all others (across all donors).
Differentially expressed transcription factors between each cell type compared to all others (across all donors).
GO-term analysis for alpha, beta, delta, and PP cells compared to all others (across all donors).
Average of gene expression across all cells of acinar and beta subpopulations.
Differentially expressed cell-surface markers factors between each cell type compared to all others (across all donors).
Data analysis script detailing StemID parameters and differential gene expression analysis between one cell type and all others.