Abstract
Although often considered “minimal” organisms, mycoplasmas show a wide range of diversity with respect to host environment, phenotypic traits, and pathogenicity. Here we report the complete genomic sequence and proteogenomic map for the piscine mycoplasma Mycoplasma mobile, noted for its robust gliding motility. For the first time, proteomic data are used in the primary annotation of a new genome, providing validation of expression for many of the predicted proteins. Several novel features were discovered including a long repeating unit of DNA of ∼2435 bp present in five complete copies that are shown to code for nearly identical yet uniquely expressed proteins. M. mobile has among the lowest DNA GC contents (24.9%) and most reduced set of tRNAs of any organism yet reported (28). Numerous instances of tandem duplication as well as lateral gene transfer are evident in the genome. The multiple available complete genome sequences for other motile and immotile mycoplasmas enabled us to use comparative genomic and phylogenetic methods to suggest several candidate genes that might be involved in motility. The results of these analyses leave open the possibility that gliding motility might have arisen independently more than once in the mycoplasma lineage.
Often considered the smallest independently self-replicating organisms, the mycoplasmas are wall-less bacteria characterized by small physical dimensions and genome sizes (Razin et al. 1998). Partly because of this latter characteristic, they are currently the best represented phylogenetic class (Mollicutes) of organisms to have complete genomic sequences (Barre et al. 2004). Seemingly having undergone reductive evolution from the Bacillus/Clostridium branch of the Gram-positive eubacteria, mycoplasmas generally have fewer than 1000 genes, perform little de novo synthesis of required precursors, and are obligate commensals or parasites (Dandekar et al. 2002; Maniloff 2002). Yet the mycoplasmas display a wide range of phenotypic characteristics with regard to pathogenesis, host preference, morphology, and growth requirements (Razin et al. 1998).
Mycoplasma mobile is one of the flask-shaped mycoplasmas (∼1 μm × 0.3 μm) and was originally isolated from a fish, the tench (Tinca tinca; Kirchhoff and Rosengarten 1984). It belongs to the Mycoplasma hominis group of the mycoplasma phylogeny, which also includes two other sequenced species (Mycoplasma pulmonis, complete; and Mycoplasma hyopneumoniae, in progress; Chambaud et al. 2001; Maniloff 1992, 2002). It is believed to be pathogenic and grows optimally at around 20°C, somewhat lower than most other well-studied mycoplasmas, whose optima are around 37°C, yet its doubling time (∼10 h) is within the range of mycoplasmas with mammalian hosts. However, M. mobile's single most defining characteristic is its ability to glide (defined as smooth translocation while in contact with a surface) on glass or plastic surfaces at speeds up to 7 μm/sec (Rosengarten and Kirchhoff 1987). It exerts sufficient force to tow an erythrocyte (∼1000 times its mass) without substantial loss of gliding speed, and is believed to have its motility apparatus located in the head or neck region of its flask-shaped cell body (Rosengarten et al. 1988; Miyata and Uenoyama 2002; Miyata et al. 2002). Gliding is not unique to M. mobile among the mycoplasmas, but its locomotion is certainly the most pronounced of species studied thus far (Kirchhoff 1992). No genes similar to other effectors of cell motility in bacteria (e.g., flagellae, pili) or eukaryotes (e.g., myosin, kinesin, actin) have been revealed in other gliding mycoplasmas' genome sequences (Fraser et al. 1995; Himmelreich et al. 1996; Chambaud et al. 2001; Papazisi et al. 2003). It was demonstrated recently that gliding is an ATP-powered process and that M. mobile contains several large ultrastructural proteins that play a role in surface adhesion (Jaffe et al. 2004b; Uenoyama et al. 2004). However, little else is known about the genes involved in gliding motility. Therefore, we thought that it would be useful to know the complete genomic sequence of M. mobile to help elucidate the mechanism of gliding, especially in comparison with the sequences of other mycoplasmas that are either immotile or less motile than M. mobile.
We also sought to use a new combined genomic/proteomic approach to genome sequencing and annotation called proteogenomic mapping (Jaffe et al. 2004a). By this method, proteomic data are collected concurrently with primary DNA sequence data, and the two sets are combined to yield an annotation that is informed by knowledge of the proteins that are produced by the organism under study. This also affords an opportunity to incorporate posttranslational modification information into the primary annotation when such events can be found. Here we report the genomic sequence and proteogenomic map for M. mobile.
RESULTS
Primary Genome and Proteome Features
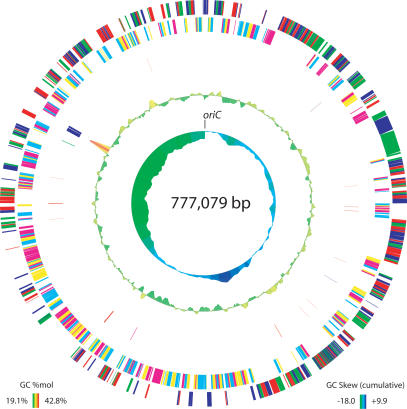
Our assembly of the complete genome (Fig. 1) consists of a single circular chromosome of 777,079 bp with a GC content of 24.9 mole%, low even among the GC-poor mycoplasmas (range 24%–40%). We have proposed 635 protein coding sequences (CDSs), of which 557 (88%) have been validated as expressed proteins by proteogenomic mapping. Initially, 612 genes were computationally predicted, but four of these were manually removed and one other gene was added prior to incorporation of proteomic evidence based on homology searches. Subsequently, 26 additional genes were added through proteomics to reach the total of 635. Of these proteins, 463 could be placed into clusters of orthologous groups (COGs) via homology search (Tatusov et al. 2000). M. mobile contains a single copy each of the 16S–23S–5S ribosomal DNAs. The 5S rDNA is not located within the 16S–23S rDNA operon, as is the case in M. pulmonis (Chambaud et al. 2001). Interestingly, it is located ∼180° away (with respect to the circular chromosome) from the conserved 16S–23S rDNA, suggesting a major genome rearrangement may have taken place about this axis (see Fig. 1). We have found evidence for 28 tRNA genes, the fewest of any organism yet reported (see Supplemental tables of RNA and Codon Usage; Chambaud et al. 2001). We have also located one copy each of the genes for the 4.5S SRP-RNA, the ribonuclease P RNA component, and the tmRNA in the genomic sequence. No insertion sequences (ISs), transposons, or endogenous plasmids were found in the genome, although the presence of these elements has been reported in several other mycoplasma genomes (Sasaki et al. 2002; Papazisi et al. 2003; Westberg et al. 2004).
Figure 1.
Circular genome plot. Ring definitions, from outermost to innermost: Predicted proteins colored by reading frame on the plus strand (red, green, blue are forward frames); predicted proteins colored by reading frame on the minus strand (cyan, magenta, and yellow are reverse frames); RNA coding features (red: tRNAs; blue: rRNAs; magenta: RNAse P RNA; green: tmRNA; cyan: 4.5S SRP RNA); GC content (calculated using a 5000-bp window sliding 100 bases at a time, see plot for range, height of trace proportional to deviation from 25% GC); cumulative GC-skew (calculated using a 5000-bp window sliding 100 bases at a time, see plot for range, height of trace proportional to deviation from 0) as in Grigoriev (1998).
We have assigned the origin of replication based on homology to other mycoplasma genomes (Cordova et al. 2002; Lartigue et al. 2003). The conserved gene order rpmH → dnaA → dnaN is present as is observed in M. pulmonis and several other mycoplasmas. In this case there also is a copy of the presumed major surface antigen in between rpmH and dnaA (see below). In following the convention of M. pulmonis, we have numbered as base 1 the first nucleotide after the stop codon of the gene immediately preceding dnaA (Chambaud et al. 2001). M. mobile lacks any clear dnaA-boxes upstream of the dnaA or dnaN genes, but this is not unusual and also is observed in Mycoplasma pneumoniae and Mycoplasma genitalium (Lartigue et al. 2003). A plot of the cumulative GC-skew (Fig. 1) also suggests that the origin should be localized to the dnaA region.
Because we were able to collect and analyze proteomic data concurrently with genome sequencing, we have good evidence for many of the proteins that we predict for this genome (Fig. 2; Supplemental Gene Table). As always with a novel genome, there are a fair number of “unknown” proteins that have no homologs or have dubious functionality based on homology searches, but now with proteomics we have verified that many of them are expressed. Hence, we have adopted a controlled vocabulary in annotating these “unknowns.” Predicted proteins for which there are no proteomic data are annotated as “hypothetical” or “conserved hypothetical” if there is supporting evidence of homology in other species. Proteins for which we do have proteomic evidence but little functional information are usually annotated as “expressed protein of unknown function.” In our annotation scheme, the word “putative” refers only to the presence of a particular functionality, and never to the existence of the protein or not. Because GenBank does not yet have a facility for integrating proteomic data into genome annotation, we have provided a summary of proteomic evidence for each protein in the “note” field of the GenBank annotation, where we specify the number of unique peptides detected for the protein and the percent coverage by amino acid sequence for the protein. Full proteomic coverage can be viewed interactively at http://www.broad.mit.edu/annotation/microbes/mycoplasma/.
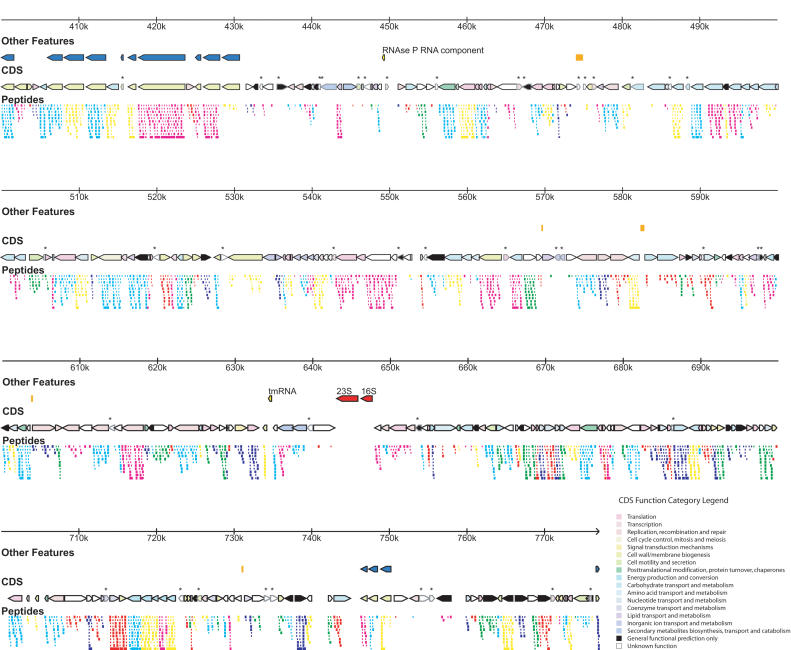
Figure 2.
Genome features and proteogenomic map. (First row) Genome ruler. (Second row) Special genome features including tRNAs (orange), mvsp genes (blue), and other features labeled on the figure. (Third row) Coding DNA sequences (CDS), color coded by COG categories automatically assigned for proteins in COGs, or manually assigned as necessary (color key in lower right corner). CDS not validated by proteomics are marked by an *. (Fourth row) Proteogenomic map showing detected peptides covering the predicted proteins color coded by reading frame, as in Figure 1.
General features of the proteome closely mirrored those in M. pneumoniae (Jaffe et al. 2004a). As mentioned earlier, at least one peptide was observed for 88% of the predicted coding regions. Two or more unique peptides were observed for 517 of the open reading frames (ORFs), and on average sequence coverage for those proteins that were detected was >40%. Some peptides were observed that were degenerate in sequence to multiple highly similar proteins, such as the lrp proteins (see below), but no protein was identified on the basis of degenerate peptides alone. Consistent with M. pneumoniae, a greater fraction of “well-annotated” proteins were detected than “poorly annotated” ones. We used the COG (clusters of orthologous groups) functional category classification where possible in deciding whether a gene was “well-annotated” or not, although 172 genes were manually assigned COG functional categories for purposes of this analysis (http://www.ncbi.nlm.nih.gov/COG/). Poorly annotated proteins were considered to be any protein automatically assigned to COG category R (general functional prediction only) or S (unknown function), or those proteins that were unmatched to COGs but were manually assigned these designations based on the lack of homologous proteins with any assigned function. The remaining proteins were considered to be well-annotated, having COGs or relatively clear functional predictions assigned to them.
We detected 94% of well-annotated proteins but only 76% of poorly annotated proteins. In fact, the proposed gene products not detected by proteomics are functionally enriched for the “unknown function” category of annotation (55% of all undetected ORFs belong to COG category S, p-value = 1 × 10–9 for functional enrichment using the hypergeometric distribution method; Sokal and Rohlf 1995). This suggests that there are probably some gene calls made in our annotation that do not represent bona fide genes. Interestingly, predicted genes not detected by proteomics tended to be shorter in amino acid length than those that were (undetected genes' mean length: 237.4 amino acids ± 157.4 SD; detected genes' mean length: 388.3 amino acids ± 332.8 SD; p-value = 0.0001 using the Student's t-test). This disparity may be related to the fact that shorter proteins have theoretically fewer peptides available for detection. We considered throwing out all gene calls without proteomic evidence but chose to err on the side of inclusiveness for several reasons. First, there are some proteins that are likely to be present in the cell (such as the RNAse P protein component) that were not detected by proteomics. In our previous study on M. pneumoniae,thisprotein was identified but only by a single peptide covering 8.5% of the protein sequence. Low coverage can be an indicator of low protein abundance. Another reason for failing to detect a protein or having low protein coverage can relate to difficulty in solubilizing a particular class of protein. We failed to detect several proteins annotated as ABC permease components and transport proteins that are often associated with the membrane, a class of protein that has been historically troublesome to detect, but we are hesitant to discard these gene calls.
Several other “named” genes were not detected by proteomics. As with M. pneumoniae, the Holliday-junction helicases ruvA and ruvB were not detected. One copy of RNAse H II was not detected, but a second copy was, thus its functionality is clearly present. As well, M. mobile's copy of ribosomal protein L36 (rpmJ) was not detected, but this particular gene has a suspicious phylogeny despite its conserved gene order with M. pulmonis and M. pneumoniae (see below). The rpmJ protein was detected in M. pneumoniae, but this protein is extremely small and missing it could have simply been an unlucky happenstance. Curiously, ATP synthase subunit C (atpE) was not detected although its presence in vivo is assumed.
Twenty-six proteins that were not computationally predicted were detected by proteomics (identification methods for each gene call can be seen in the Supplemental Table of Computational and Proteomic Predictions). There was no detectable size or functional category bias to these proteins, but eight of the 26 were <100 amino acids. Thirty-nine additional modifications were made to the start codons of computationally predicted genes based on proteomic evidence. This represents a net 10% modification to genome annotation based on incorporation of proteomics into gene modeling, and the additional 26 proteins represent 2.1% of the total genome coding capacity. Of the 608 genes originally predicted by computational methods, 531 were supported by proteomic evidence (84%). Of the 11,825 positionally unique peptides used to construct the proteogenomic map for M. mobile, 96.7% were mapped onto computationally predicted genes whereas the remainder provided the basis for the additional 26 ORFs. No detected peptides conflicted with a computationally predicted gene in terms of translational frame, however, and thus no computationally predicted genes were removed based on proteomics.
We searched for possible phosphorylation (on serine, threonine, or tyrosine), methylation (on lysine, aspartic acid, or glutamic acid), and acetylation (on lysine) of proteins by varying our SEQUEST search strategy accordingly. We did not detect any convincing posttranslational modifications by proteomics, although the data may be reanalyzed more thoroughly at a later date. This is in contrast to M. pneumoniae in which we detected phosphorylation of the HPr phosphocarrier protein using similar methods (Jaffe et al. 2004a). Other aspects of the proteomic experiments carry through the rest of the analysis of the genome.
Functional Analysis and Metabolism
A functional analysis of the proteins present in M. mobile is shown in Table 1. The basic breakdown of proteins into functional categories is similar to other mycoplasmas, with an emphasis on transport of compounds and protein synthesis (Westberg et al. 2004). As is typical of a newly sequenced genome, one-fourth to one-third of all proteins are without a well-annotated function. An additional phylogenetic breakdown of gene lineages is shown in Table 2, which highlights that 109 new proteins appear unique to M. mobile.
Table 1.
Functional Analysis of Predicted Proteins
| COG category | Number of proteins | % of total |
|---|---|---|
| Energy production and conversion | 29 | 4.6% |
| Cell division and chromosome partitioning | 2 | 0.3% |
| Amino acid transport and metabolism | 19 | 3.0% |
| Nucleotide transport and metabolism | 21 | 3.3% |
| Carbohydrate transport and metabolism | 51 | 8.0% |
| Coenzyme metabolism | 15 | 2.4% |
| Lipid metabolism | 9 | 1.4% |
| Translation, ribosomal structure, and biogenesis | 104 | 16.4% |
| Transcription | 17 | 2.7% |
| DNA replication, recombination, and repair | 58 | 9.1% |
| Cell envelope biogenesis, outer membrane | 45 | 7.1% |
| Cell motility and secretion | 10 | 1.6% |
| Posttranslational modification, protein turnover, chaperones | 19 | 3.0% |
| Inorganic ion transport and metabolism | 14 | 2.2% |
| Secondary metabolites biosynthesis, transport, and catabolism | 6 | 0.9% |
| Signal transduction mechanisms | 14 | 2.2% |
| General function prediction only | 43 | 6.8% |
| Function unknown | 159 | 25.0% |
For genes that could be placed in clusters of orthologous groups (COGs), the functional category was automatically determined. For genes that did not match to COGs, the gene was assigned to a COG category manually, with the majority of such cases being assigned to the “Function Unknown” category. For genes with more than one COG category, only the first one (in alphabetical order) was used.
Table 2.
Phylogenetic Origins of Mycoplasma mobile Proteins
| Number of genes | |
|---|---|
| Distributed in various bacterial lineages | 422 |
| Specific to mycoplasma lineages | 69 |
| Specific to M. mobile | 109 |
| Present in M. mobile and other organisms but not other mycoplasmas | 35 |
| Total | 635 |
The analysis is based on parsing the species origin of BLAST hits against the nonredundant database of proteins with an E-value <0.001.
The genome encodes numerous transporters with a wide range of substrates and possesses some additional transporters of unknown specificity. Both ATPase Binding Cassette (ABC) and Phosphoenolpyruvate-dependent System (PTS) transporters are represented. It appears that M. mobile should be able to transport and metabolize glucose, sucrose, fructose, maltose/maltodextrin, xylose, and trehalose as energy sources. This has been shown experimentally for glucose and sucrose (Jaffe et al. 2004b). Glycerol should also be able to be used as an energy source, but no specific transporter for glycerol was found. The presence of mannose-6-phosphate isomerase (manA) suggests that mannose may also be metabolized, as is suggested elsewhere (Pollack 2002). As is common to other mycoplasmas, M. mobile should be able to produce and use glycogen and starches.
Fermentation of sugars appears to be the only method of ATP production in M. mobile. A complete glycolysis pathway is present that terminates in the formation of lactate. Most of the nonoxidative branch of the pentose phosphate pathway is present except for the reaction normally carried out by transaldolase. This is similar to the situation in M. mycoides and other mycoplasmas, and presumably this function is carried out by an as-yet-unrecognized protein (Pollack 2002; Westberg et al. 2004). No portion of the citric acid cycle is present in M. mobile. The major toxic products from central metabolism are protons that would acidify the interior of the cell. These are presumably pumped out by the F0F1-ATPase.
Other ionic homeostases are achieved through a variety of transporters. Phosphate, formate/nitrite, and cobalt transporters are specifically observed, as well as the Na+/Ca2+ antiporter ecm27 and the K+/Na+ symporter ktrAB. We also note the presence of mscL, the large conductance mechanosensitive channel for response to osmotic stress (Sukharev et al. 1994). The maintenance of a proton motive force across the membrane of M. mobile has been documented elsewhere and is presumably achieved through these genes in combination with the F0F1-ATPase and possibly other contributing proteins that have not yet been identified (Schiefer and Schummer 1982; Schummer and Schiefer 1983; Jaffe et al. 2004b).
As with other mycoplasmas, de novo amino acid and nucleotide synthesis is lacking in M. mobile (Pollack 2002). It is presumed that amino acids are obtained by ABC-type oligopeptide transporters followed by proteolytic degradation of the peptides. Extensive nucleotide salvage pathways are also present including the conversion of ribonucleotides to deoxyribonucleotides by nrdA. Only a few partial biochemical pathways novel to the mycoplasmas are present in M. mobile. One step in cobalamin (vitamin B12) synthesis is present, as is one step in biotin biosynthesis. This latter reaction is almost certainly the result of lateral gene transfer (see below). Partial pathways for phospholipid biosynthesis, riboflavin/FAD/FMN synthesis, formyl-THF biosynthesis, (deoxy)ribose phosphate utilization, pantothenate and fatty acid biosynthesis, and UDP-glucose interconversion are also detected.
A variety of DNA restriction/modification system enzymes are detected in M. mobile. This may account for their recalcitrance to genetic manipulation via transposon mutagenesis (J. Jaffe, unpubl.; M. Miyata, pers. comm.) even though it is a popular mutagenesis technique in other mycoplasma species. A similar obstacle was overcome in Mycoplasma arthritidis by using an appropriate DNA modification enzyme to circumvent that species' endogenous restriction enzymes (Voelker and Dybvig 1996). It is assumed that these systems are active, because many of them are expressed at the protein level as verified by proteomics.
Long Repeat Region
The genome contains a novel feature for bacteria that made assembly of the genome difficult. An integral unit of ∼2435 bp is repeated five times within a short span of genome sequence from coordinates 24,799 to 38,314 that range from 94% to 97% sequence identity at the DNA level (Fig. 3). The five repeats are interrupted by one abortive/divergent instance of the repeat unit after the initial repeat (from 5′ → 3′), where a new repeat begins but diverges into unrelated sequence after ∼430 nt. The divergent sequence persists for about 900 more nucleotides before a new instance or the repeat begins. Then there are four tandem repeats of the repeating sequence. We have demonstrated via proteomics that these repeats code for proteins that are expressed, and have named them lrpA–lrpE (for long repeat protein), with lrpAbor signifying the potential coding region of an abortive instance of the repeat, which may not be expressed. Moreover, slight coding differences at the protein level have allowed us to conclude that each of the five repeat proteins is uniquely expressed—that is, five slightly different isoforms of the same protein are present in the cell all at once (see Fig 3, colored boxes). BLAST searches did not yield any homologs to the lrps in other organisms (Altschul et al. 1997). Other sequence/motif analysis tools (e.g., PROSITES-can, TMPRED) similarly yield little information about these proteins (Hofmann and Stoffel 1993; Sigrist et al. 2002). They appear to be globular cytoplasmic proteins, and the degree of proteomic sequence coverage suggests that they are as abundant as the average M. mobile protein in the cell.
Figure 3.
Long repeat region. (Top) High-resolution view of the “long repeat region” of the M. mobile genome showing repeat arrangement and the lrp genes. (Bottom) Multiple sequence alignment of the lrp proteins showing all peptides detected by proteogenomic mapping (bold italic sequence) and those peptides that help to establish unique expression of that particular lrp protein (highlighted in color). Multiple alignments performed with CLUSTALX (Thompson et al. 1997).
The meaning of these repeated sequences is unclear. Because of the orientation and the site of the truncation of the abortive repeat, it appears that the repeats are generated from 5′ to 3′ on the plus strand, possibly by polymerase slippage during replication or by replication stalling with subsequent upstream repriming by the leading strand. Yet the proteins that these sequences encode are transcribed from the minus strand of the genome. At ∼20 mole%, this region of the genome is also even lower in GC-content than the rest of the genome. Southern blot experiments (data not shown) indicate that the number of repeats is stable over several passages in culture in a population. Only a single 14.9-kb band was visualized when a BbvI restriction digest designed to cut around the repeat region was probed with a sequence targeted to all of the repeat regions. Coincidentally, these experiments allowed us to fix the number of full repeats in the genome at five and complete its assembly. This finding indicates that either homologous recombination that would result in “looping out” of the repeats occurs at only a low level in M. mobile or that this region is somehow protected from that type of recombination. M. mobile should be capable of homologous recombination, as it possesses and expresses the recA gene (Cassuto et al. 1980). If these repeats were generated by simple duplication, it might have been a relatively recent event in the evolution of M. mobile. Alternatively, recombination-based DNA repair might be used to keep the sequences from diverging very much. We also note the proximity of this sequence to the putative origin of replication (only 24,799 bases from base 1), but can only speculate that it may be somehow involved in replication.
Putative Major Surface Antigen
M. mobile also contains a more familiar set of repetitive elements, namely, a presumed variable surface antigen. There are 16 copies of the element throughout the genome (marked as a special element in Fig. 2), which have been named mvspA through mvspP (for mobile variable surface protein). Eleven copies (mvspB through mvspL) are located in a tight cluster from bases 398,037 to 430,685, with only a few intervening genes (see Fig. 2). Another small cluster (mvspM through mvspO) is located from bases 746,365 to 750,245. Every copy is oriented on the minus strand of the genome except for mvspA and mvspP. They range in size from 105 amino acids for mvspG to 2002 amino acids for mvspI. The mvsp proteins contain predicted transmembrane helices, and some have an additional lipoprotein attachment motif as defined in PROSITE (Sigrist et al. 2002). The mvsps are basically unique to M. mobile, although two homologs are detected in M. pulmonis by BLAST search (mvspB is most similar to gi 15829140 [31% identity over a 207-amino-acid stretch], and mvspE is most similar to gi 15828908 [23% identity over a 478-amino-acid stretch]). The mvsp proteins are quasirepetitive and can often be aligned to each other in multiple registers. The peptide motif NGVxFxL (where x represents one of multiple possible amino acids) seems to repeat multiple times in the various mvsp copies.
Many mycoplasmas have a major antigenic protein of which there are multiple copies with variable sequences present in the genome, one of which is expressed at a given time. For example, M. pulmonis has the vsa proteins and M. pneumoniae has the P1 protein present many times each in the genome (Himmelreich et al. 1996; Chambaud et al. 2001). In the case of M. pulmonis, a promoter sequence is moved from copy to copy of vsa to effect antigenic switching (Bhugra et al. 1995; Shen et al. 2000). In M. pneumoniae, it is believed that homologous recombination transfers different variants of the P1 gene into an actively transcribed locus (Kenri et al. 1999). The net effect is that only one variant of the surface antigen is expressed at a time, and thus switching the variant from time to time can help to evade the host's immune system. However, we have proteomic evidence that almost all of the mvsp proteins are concurrently expressed in a culture. Thus, it is possible that a single cell is expressing multiple mvsp variants simultaneously or that different subpopulations in the culture each express a single variant but analysis of a batch culture results in detection of multiple variants. Our inclination is that the former case is correct because the other mycoplasmas discussed earlier do not seem to lose the ability to suppress the multiple copies of their major antigen after extended periods of culture. Although the mechanism of virulence is not well characterized in M. mobile, the genome also contains copies of the P46 antigen from Mycoplasma hyorhinis (MMOB0150), a transporter protein with known antigenic properties (MMOB0360), and an o-sialoglycoprotein endopeptidase (MMOB1300) that might be involved in antigenicity or virulence.
Other Genes Present in Multiple Copies
There are 63 genes (∼10% of the total) present in tandem, multiple, operonic, or near-tandem duplications (this includes the lrp and mvsp clusters; see Table 3). This rate is somewhat higher than in M. pulmonis (M. mobile's closest fully sequenced relative), which has only 54 of its 782 genes (∼7%) present in regions that appear to have arisen from duplication (including the vsa genes; Chambaud et al. 2001). Interestingly, these two genomes share only three pairs of duplicated genes. Each organism has a pair of cbiO (cobalt transporter), mldB (multidrug/protein/lipid transporter), and COG0595-like genes (predicted hydrolase of the metallo-β-lactamase superfamily) in close proximity to each other. For the first two pairs, they are tandem duplications in both organisms; for the final pair, it is a tandem duplication in M. pulmonis, but two genes have been inserted between the M. mobile copies. This suggests that these particular duplications are ancient in the mycoplasma lineage. Indeed Mycoplasma penetrans and M. pneumoniae share the tandem duplications of cbiO and mldB in their genomes (Sasaki et al. 2002). Overall, the pattern of tandem duplications suggests a mechanism of rapid differentiation among the mycoplasmas. Based on phylogenetic tree branch lengths and calculated mutation rates of 16S rDNA of the mycoplasmas, M. pulmonis and M. mobile probably shared the last common ancestor (LCA) ∼190 million years ago (Mya; Maniloff 2002). It seems that both organisms have an affinity for tandem duplications in the genome, but the targets for such events were random. This affinity for tandem duplication may also suggest a mechanism for generation of the lrp cluster.
Table 3.
Tandem Duplication of Genes in Mycoplasma mobile
| Accession | Duplicated by | Gene | Function | |||
|---|---|---|---|---|---|---|
| Tandem duplications of M. mobile specific genes | ||||||
| MMOB0190 | MMOB0200 | MMOB0190 | Expressed protein of unknown function | |||
| MMOB0220 | MMOB0230, MMOB0240, MMOB0250, MMOB0260, MMOB0270 | Irp clusters | Long repeat protein | |||
| MMOB3920 | MMOB3940 | malE | Maltose-binding protein | |||
| MMOB3930 | MMOB3950 | MMOB3930 | Hypothetical protein | |||
| MMOB3980 | MMOB3990 | glgC | Glucose-1-phosphate adenylyltransferase | |||
| MMOB5080 | MMOB5090 | MMOB5080 | Expressed putative lipoprotein protein of unknown function | |||
| MMOB5940 | MMOB5950 | MMOB5940 | Expressed protein of unknown function | |||
| MMOB6040 | MMOB6050 | MMOB6040 | Expressed protein of unknown function | |||
| MMOB6250 | MMOB6260 | MMOB6250 | Expressed protein of unknown function | |||
| Tandem duplications of single genes present in Mycoplasma pulmonis | ||||||
| MMOB0420 | MMOB0430 | MMOB0420 | Unspecified transport or permease protein | |||
| MMOB0450 | MMOB0460, MMOB0470 | MMOB0450 | COF family HAD hydrolase protein | |||
| MMOB0510 | MMOB0520 | potD | Probable spermidine/putrescine or other ABC transporter substrate-binding protein | |||
| MMOB1140 | MMOB1150 | MMOB1140 | Putative amino acid permease | |||
| MMOB1420 | MMOB1430 | trxA | Thioredoxin | |||
| MMOB1780 | MMOB1790 | degV | degV homolog | |||
| MMOB2840 | MMOB2860 | hsdM | Type I restriction enzyme m protein | |||
| MMOB2850 | MMOB2870 | hsdS | Restriction-modification enzyme mpuUVIIIs subunit | |||
| MMOB3220 | MMOB3230, MMOB3280, MMOB3290, MMOB3330, MBOB3320, MMOB3330, MMOB3340, MMOB3360, MMOB3370, MMOB3380 | mvsp cluster | M. mobile variable surface protein | |||
| MMOB4890 | MMOB4910 | pcrA1 | ATP-dependent DNA helicase | |||
| MMOB5810 | MMOB5820 | pdhC | Dihydrolipoamide acetyltransferase | |||
| MMOB6070 | MMOB6080, MMOB6090 | mvsp cluster | M. mobile variable surface protein | |||
| Tandem duplications present in both M. mobile and M. pulmonis | ||||||
| MMOB4320 | MMOB4330 | cbiO | ABC transporter, ATPase component, possibly cobalt transporter | |||
| MMOB5100 | MMOB5110 | mldB1 | ABC-type multidrug/protein/lipid transport system, ATPase component | |||
| MMOB6180 | MMOB6210 | MMOB6180 | Predicted hydrolase of the metallo-β-lactamase superfamily or serine protease | |||
Duplications were identified by BLAST hits against the nonredundant database of proteins plus the 635 predicted ORFs from M. mobile with an E-value <0.001. Duplications are separated into categories by phylogenetic lineage.
Several other duplications of note are present in the M. mobile genome. There are three copies of manB scattered throughout the genome as opposed to one in M. pulmonis. There are also multiple copies of ATP synthase α and β subunits located outside the main ATP synthase cluster. This feature is shared in common with M. pulmonis and other mycoplasmas. Again, our proteomics data show that each copy of ATP synthase α and β is uniquely expressed based on detected variations at the coding level of the multiple paralogs. The genome segment containing MMOB0970 and MMOB0980 appears to have been duplicated at MMOB5690 and MMOB5700. As well, the tandem duplication of MMOB0190 and MMOB0200 is repeated at MMOB6040 and MMOB6050—a strange example of a nontandem duplication of a tandem duplication.
Potential Motility Genes
Although several genomes from motile mycoplasmas have been sequenced, no genes specifically involved in the force-generation mechanism of gliding have been revealed (Fraser et al. 1995; Himmelreich et al. 1996; Chambaud et al. 2001; Papazisi et al. 2003). Moreover, no homologs of genes traditionally involved in cell motility have been detected in these genomes, and that trend continues with M. mobile. Some groundwork has already been done in the search for motility genes. M. mobile mutants with an altered gliding phenotype have been isolated, and several proteins important for substrate adhesion (a prerequisite for gliding motility) have been recognized (Miyata et al. 2000). The proteins P123 (MMOB1020), Gli349 (gliA), and Gli521 (gliB), reside together in an operon and are implicated in glass binding and hemadsorption in M. mobile (Uenoyama et al. 2004). The correlation of pathogenicity with motility in the mycoplasmas implicates these genes in pathogenic processes as well. Gli349 and Gli521 are the two largest proteins coded by the genome, and their coding sequence accounts for >3% of the genome as a whole (the average gene accounts for 0.16%). Interestingly, we did not detect any paralogs or fragments of these proteins elsewhere in the genome, although these proteins are so huge that spurious paralogs will be detected if composition-based BLAST statistics are not used (Schaffer et al. 2001). This is contrast to the major cytadhesin of M. pneumoniae (P1) that is present in numerous copies in that organism's genome (Himmelreich et al. 1996). It has also recently been demonstrated that motility in M. mobile is powered by ATP, and therefore any proteins with ATPase functionality might be interesting as motility gene candidates (Jaffe et al. 2004b).
There are now several motile and immotile mycoplasmas that have had their genomes sequenced, and we attempted a comparative genomic analysis to search for potential motility proteins. We searched for several patterns that might indicate a gene's relevance for motility. According to the literature, M. pneumoniae, M. pulmonis, M. genitalium, and Mycoplasma gallisepticum all possess the ability to locomote, although the latter appears to do so extremely slowly (Kirchhoff 1992). At the time of this writing, we also had available annotated genome sequences for the immotile mycoplasmas Mycoplasma mycoides mycoides SC, Ureaplasma parvum (formerly U. urealyticum), Mesoplasma florum, and M. penetrans (Glass et al. 2000; Sasaki et al. 2002; Westberg et al. 2004; Me. florum courtesy of Tom Knight and Nicole Stange-Thomann). We performed BLAST searches against the nonredundant database of proteins (plus Me. florum proteins) as of March 5, 2004, and identified orthologs of M. mobile genes present in other fully sequenced mycoplasmas with E-values of 1 × 10–3 or less. The simplest case would be to find M. mobile genes that are present only in the motile mycoplasmas and absent from the immotile ones, as defined above. Unfortunately, no genes met these criteria. From these results, we conclude that this simple “present/absent” model is insufficient to identify motility genes in M. mobile.
We then reanalyzed the data under the premise that ancestral motility genes might be present throughout the mycoplasmas, but mutation might have specifically inactivated them in the immotile ones. Because we hold M. mobile as the paradigmatic motile mycoplasma, we attempted to look for M. mobile genes that were phylogenetically clustered with genes from the other motile mycoplasmas and more divergent from their orthologs in the immotile species. We used a rudimentary form of nearest-neighbor clustering termed the “Group Phylogenetic Bias” (see Methods for details). In this case, the two groups considered are the Motile Group and the Immotile Group, as defined above. We used the BLAST E-value as a surrogate for phylogenetic distance of each M. mobile gene to orthologs in the other mycoplasma species discussed above (i.e., a lower E-value implies a closer phylogenetic relationship). Table 4 shows the high-scoring Motile Group Phylogenetic Bias genes. Several interesting candidates are identified by this method.
Table 4.
Motile Group Phylogenetically Biased Genes
| Accession | Gene | Putative function | Motile group phylogenetic biasa | σs above mean |
|---|---|---|---|---|
| MMOB3970 | algA | α-Glucosidase | 98.95 | 7.3 |
| MMOB2720 | secDF | secD- and secF-like type II secretion system protein | 70.55 | 5.1 |
| MMOB2030 | glcK | Similar to sugar-related transcriptional regulator but lacks DNA-binding domain | 49.13 | 3.5 |
| MMOB2860 | hsdM | Type I restriction enzyme m protein | 39.29 | 2.7 |
| MMOB3120 | pncB | Nicotinic acid phosphoribosyltransferase | 36.87 | 2.5 |
| MMOB3310 | pgm | Phosphoglycerate mutase | 35.66 | 2.4 |
| MMOB4610 | MMOB4610 | Type III restriction modification system methylase | 35.56 | 2.4 |
| MMOB2840 | hsdM | Type I restriction enzyme m protein | 35.41 | 2.4 |
| MMOB1730 | MMOB1730 | Predicted signaling protein | 34.87 | 2.4 |
| MMOB2040 | MMOB2040 | Unspecified permease of the major facilitator superfamily | 34.2 | 2.3 |
| MMOB0670 | mgpA | mgpA-like protein | 33.76 | 2.3 |
| MMOB4580 | chrA2 | Chromate transport protein | 32.7 | 2.2 |
| MMOB2280 | MMOB2280 | Expressed protein of unknown function | 31.66 | 2.1 |
| MMOB5840 | pdhA | Pyruvate dehydrogenase E1 component, α subunit | 31.22 | 2.1 |
| MMOB4490 | pgk/oppD | Bidomainal protein: phosphoglycerate kinase/ABC transporter ATPase | 31.1 | 2.1 |
See Methods section for description of the Group Phylogenetic Bias score.
The second highest-scoring gene is secDF, part of the protein secretion apparatus. This is notable because it is likely to be localized to the membrane in the cell and, in combination with secA and secY, hydrolyzes ATP to translocate proteins across the membrane. One hypothesis might be that M. mobile has a specialized sec system that effects motility rather than secretes proteins. However, the only other member of the Motile Group to have secDF is M. pulmonis, which means that a specialized sec system would not be a universal effector of motility in the mycoplasmas. Those in the M. pneumoniae branch would require a separate mechanism. A specialized sec system might also indicate that protein extrusion plays a role in motility, analogous to the slime extrusion hypothesis for some cyanobacteria (Hoiczyk and Baumeister 1998).
Another interesting candidate is MMOB2040, which is annotated as an unspecified permease of the major facilitator superfamily. Again, this would be a membrane protein that would have the capability of ATP hydrolysis when coupled to other permease subunits. In contrast to secDF, this protein is found in the motile mycoplasmas M. genitalium, M. pneumoniae, and M. pulmonis and the immotile species M. penetrans and M. mycoides. Because its permease specificity is ambiguous based on homology to other transporters, one can again imagine that its activity might be altered to provide a motility function.
Yet another noteworthy gene is the mgpA gene. This gene was originally identified as a major antigen and cytadhesin in M. genitalium, but every sequenced mycoplasma genome so far has revealed an ortholog (Hu et al. 1987). Given its pre-existing association with surface adhesion, a prerequisite for gliding motility, we found it interesting that the sequence of this protein was more conserved among the motile mycoplasmas. In a multiple sequence alignment of mgpA genes from all nine sequenced mycoplasmas, those sequences from the motile mycoplasmas clustered together. This result lends further support for mgpA as a motility-associated gene. mgpA belongs to the DHH family (named after a characteristic amino acid sequence motif) of predicted phosphoesterases (with polyphosphatase activity), which includes processive enzymes like DnaJ (Sutera Jr. et al. 1999). Could this protein be used to generate force required for motility? DHH family phosphoesterases are not known to use ATP, but ATP might be used to build a polyphosphate substrate that is subsequently hydrolyzed by mgpA during force generation.
We attempted one other approach to search for motility genes in the mycoplasmas, which we term the “required core set” hypothesis. If the mechanism of motility is the same among the motile mycoplasmas, then they should all contain a core set of genes that potentiates motility. The immotile mycoplasmas might have one or more members of the core set, but not all of them. There are 52 genes shared by all the motile mycoplasmas where at least one of the immotile species lacks it. This list was then reduced by eliminating any genes that had an obvious functional assignment that seemed unlikely to contribute to motility. The remaining set of 12 genes is shown in Table 5, and fulfills the requirement that each of the immotile mycoplasmas could have one or more but not the complete set of these genes. It might be relatively easy to test the hypothesis that any one of these genes is required for motility by obtaining a suitable deletion in any of the mycoplasmas with amenable genetic tools, or conversely by adding back the putatively required components to any one of the immotile mycoplasmas by similar means. We note that another mgpA homolog (different from the one suggested by the previous analysis) is suggested by this method, further implicating it in motility.
Table 5.
Possible “Core Set” of Motility Genes
|
Conservation pattern
|
|||||
|---|---|---|---|---|---|
| Gene | Function | Mycoplasma penetrans | Ureaplasma parvum | Mycoplasma mycoides | Mesoplasma florum |
| MMOB0420 | Unspecified transport or permease protein | ✓ | ✓ | ||
| MMOB0430 | Unspecified transport or permease protein | ✓ | ✓ | ||
| MMOB1310 | Expressed protein of unknown function | ✓ | ✓ | ✓ | |
| MMOB1700 | Conserved expressed protein of unknown function, possibly protein prenyltransferase | ✓ | ✓ | ✓ | |
| MMOB1730 | Predicted signaling protein consisting of a modified GGDEF domain and a DHH domain (mgpA homolog) | ✓ | ✓ | ✓ | |
| MMOB1910 | Conserved expressed protein of unknown function | ✓ | ✓ | ✓ | |
| MMOB3070 | Expressed protein of unknown function | ✓ | ✓ | ✓ | |
| MMOB3080 | Expressed protein of unknown function | ✓ | ✓ | ||
| sua5 (MMOB3710) | Putative translation factor | ✓ | ✓ | ||
| mraZ (MMOB3790) | Expressed protein of unknown function | ✓ | ✓ | ✓ | |
| MMOB4550 | Expressed protein of unknown function | ✓ | |||
| thiJ (MMOB5890) | Putative intracellular protease/amidase and/or 4-methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis protein | ✓ | ✓ | ✓ | |
Each of the genes listed has an ortholog in all of the motile mycoplasmas, but one or more is missing in each of the immotile mycoplasmas.
Lateral Gene Transfer and Atypical Gene Phylogeny
M. mobile contains 35 genes that have homologs outside of the mycoplasmas but none within them (see Table 6 and Supplemental Table of Ambiguous Gene Phylogenies). These may be genes remaining from the last common Gram-positive ancestor (LCA) of the mycoplasmas before the onset of reductive evolution, or they may be genes inserted into M. mobile by lateral transfer. Of the 35 genes, 22 have a clear Gram-positive ancestry, and therefore they may be vestiges of the LCA. However, for two of them, bioF (MMOB5780) and wcaG (MMOB5790), sequence similarity to the closest Gram-positive is too good to represent a divergent copy of the gene after reductive evolution. These genes have 84% conserved residues (69% identity) over their entire sequence length to their ortholog in Enterococcus faecium (average hits to E. faecium are 45% ± 16% conserved, 28% ± 12% identical), and are adjacent in genomic sequence just as they are presumed to be in E. faecium (GenBank accessions gi 22992057 and gi 22992058). Given the promiscuous conjugative nature of the Enterococci,itis likely that these genes have been transferred to M. mobile via a direct encounter between the two organisms (Ruffin et al. 2000).
Table 6.
Lateral Transfer Candidates
| Accession | Gene name | Function | Closest homolog in | BLAST E-value | % GCa | CAIb | Comment |
|---|---|---|---|---|---|---|---|
| MMOB0940 | MMOB0940 | Conserved hypothetical protein | Helicobacter pylori | 1 × 10-15 | 24% | 0.654 | |
| MMOB0950 | MMOB0950 | Putative type II DNA methylase protein | Chlorella virus NY2A | 2 × 10-19 | 24% | 0.659 | Also strong homologs in H. pylori |
| MMOB4460 | arsR | Arsenical resistance operon repressor family protein | Dechloromonas aromatica | 6 × 10-6 | 25% | 0.661 | Stop codon is TGA in Dechloromonas ortholog, translated as Trp in M. mobile |
| MMOB5030 | MMOB5030 | Expressed protein of unknown function | Pirellula sp. | 9 × 10-32 | 24% | 0.678 | Pirellula is an aquatic bacterium |
| MMOB5780 | bioF | 8-Amino-7-oxononanoate synthase (or related protein) | Enterococcus faecium | 1 × 10-153 | 30% | 0.689 | See text |
| MMOB5790 | wcaG | NAD-dependent nucleoside-diphosphate-sugar epimerase | E. faecium | 1 × 10-129 | 30% | 0.717 | See text |
| MMOB5850 | MMOB5850 | Conserved hypothetical protein | Rickettsia rickettsii | 3 × 10-15 | 19% | 0.739 | Has some homologs in Gram-positive (including M. penetrans) but best hits are in Gram-negative |
| MMOB5990 | MMOB5990 | Hypothetical protein | Ferroplasma acidarmanus | 8 × 10-12 | 24% | 0.668 | F. acidarmanus is an archaeon |
| MMOB6010 | udgA | UDP-glucose dehydrogenase | Environmental sequence | 5 × 10-34 | 25% | 0.781 | Best named species hit is Mycobacterium avium, also hits in archaea |
The average GC content of M. mobile is 24.9%.
CAI is the codon adaptation index as calculated by the method of Sharp and Li (1987). The average CAI for a gene in M. mobile is 0.720 ± 0.049. See also the Supplementary Table of Ambiguous Gene Phylogenies.
Another convincing example of lateral gene transfer is the gene cluster consisting of MMOB0940 and MMOB0950. Both of these genes have orthologs in Helicobacter pylori, a member of the ε-proteobacteria, although they do not appear as adjacent genes in H. pylori. They may be derived from an as-yet-unsequenced Helicobacter species in which this gene order is present. As well, the latter gene is annotated as a DNA or RNA methylase whose closest BLAST hit is to a virus of Chlorella, a green algae. The homology to a viral gene may suggest a mechanism of transfer from one species to another.
Other candidates for lateral gene transfer and genes with atypical phylogenies are shown in Table 6 and in the Supplemental Ambiguous Gene Phylogeny Table. One particularly interesting instance is a gene (MMOB5030) whose closest homolog is found in a Pirellula species, a marine bacterium. This may reflect an environmental opportunity of M. mobile to acquire new genes based on its natural aquatic habitat. Another gene's (MMOB5990) closest homolog is found in an archaeon, Ferroplasma acidarmanus.
DISCUSSION
The mycoplasmas are the most deeply sequenced genus to date. Here, we add not only the genome but also the proteome of M. mobile. When sequencing a new genome, many new and unexpected coding sequences are discovered. It is useful to be able to attribute these to real protein products at the outset of annotation rather than afterward. We discovered 26 genes only through proteomics and not through other gene-calling methods. Some of these genes were small, which suggests a reason that they may have been missed by gene-calling programs. One example is an expressed open reading frame (ORF) of 40 amino acids located between the genes atpG and atpA in the ATP synthase operon. In light of our proteomic results, we have used a controlled vocabulary and added notes to our GenBank annotation to indicate which coding sequences are validated as expressed at the protein level. M. mobile now stands as the organism with the greatest degree of proteomic coverage (88% of all predicted genes, 40% average sequence coverage of those detected) of any organism to date. This represents a new standard in genome sequencing and annotation and, we hope, will encourage future microbial genome consortia to include proteomics as an integral part of their efforts.
The importance of proteomics is also demonstrated in the detection of the multiple unique isoforms of the lrp proteins. Even with coding identities >90%, proteomic techniques can make useful distinctions when the data sets are sufficiently comprehensive. Proteomics also helped to establish that multiple isoforms of the putative major surface antigen are simultaneously expressed in culture, a key difference from other mycoplasma species studied so far. We were disappointed not to detect any posttranslational modifications of proteins, but one advantage of proteomic data sets is that they can be augmented by exploring different environmental conditions for cell growth or targeted experiments designed to capture various protein classes at a later date (Ficarro et al. 2002). The current data as well as proteogenomic and metabolic model updates will be available at http://www.broad.mit.edu/annotation/microbes/mycoplasma/ and http://arep.med.harvard.edu/mycoplasma/.
We hoped that the abundance of genomic sequences for both motile and immotile mycoplasmas would shed light on the protein components involved in motility in the mycoplasmas. However, no obvious pattern of genes was detected that separated the motile and the immotile groups. We used two strategies to suggest candidate genes that may be involved in motility. First, we looked for a phylogenetic bias toward the M. mobile ortholog of a gene in the motile mycoplasmas and against it in the immotile species (the Group Phylogenetic Bias). Second, we looked for a minimal set of genes that were all present in the motile mycoplasmas but had one or more members missing in each of the immotile mycoplasmas (the Required Core Set). Using these methods, we have suggested several genes that might be involved in motility in the mycoplasmas. We hope that these predictions can be tested experimentally in the future. To that end, genomic sequencing has now revealed several DNA restriction/modification systems that might aid in the development of genetics tools for M. mobile, and has suggested that Enterococcus might make a good conjugation partner for M. mobile.
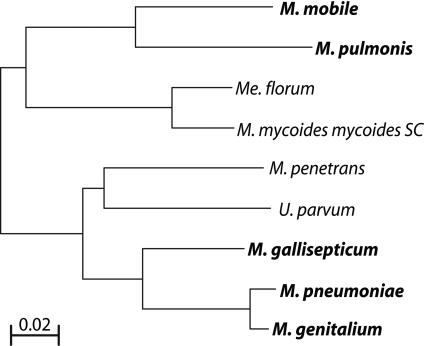
However, given the difficulty thus far of identifying motility genes by comparative genomic methods, we must raise the question of whether gliding motility was a common feature in an ancestral mycoplasma or if it arose more than once in the mycoplasma phylogeny. The motile mycoplasmas fall distinctly in two branches of the phylogeny (Fig. 4; Maniloff 2002). The first group of M. mobile and M. pulmonis (LCA ∼ 190 Mya) falls roughly into the M. hominis cluster, whereas the members of the second group of M. genitalium, M. pneumoniae, and M. gallisepticum (LCA ∼ 90 Mya) are close relatives in the M. pneumoniae cluster. The LCA of the first and second groups was probably >400 Mya. It is surprising that motility has not been observed in other members of each cluster. This might be because of lack of adequate observation of other species, selective pressure to lose the motility apparatus from the genome, or relatively recent independent development of motility in the two phylogenetic branches that contain motile species. Phenotypic differences support the latter hypothesis. For instance, both M. mobile and M. pulmonis glide continuously with little or no pausing during motility (Bredt and Radestock 1977; Rosengarten and Kirchhoff 1987). M. pneumoniae gliding is characterized by short bursts of motility followed by long periods of stationary adhesion to glass (Radestock and Bredt 1977). In fact, M. pneumoniae gliding is most readily observed during cytokinesis, and it has been hypothesized elsewhere that motility might play a role in this process (Bredt 1968; Miyata 2002). Here, the genetic evidence also seems to indicate wide enough differences in motility genes between the motile species in the two branches that it must be seriously considered that gliding motility has evolved twice independently in the mycoplasmas.
Figure 4.
16S rRNA phylogeny of sequenced mycoplasmas. Motile species are shown in bold. The Onion Yellows Phytoplasma is omitted because it was not used in the comparative genomics analysis targeted toward motility genes. The scale bar represents substitutions per site.
Although the search for motility genes is ongoing, we did discover a surprising and novel proteogenomic element in M. mobile that is unprecedented in other bacteria. The long repeat region initially complicated the assembly of this genomic sequence, and we were surprised that the repeats, in fact, code for highly similar yet uniquely expressed proteins. Deciphering the function of these repeats and their proteins should prove an interesting challenge for the future. This discovery underscores the reality that there are no “simple genomes” and demonstrates the value of continuing to sequence the genomes of even the smallest organisms.
METHODS
Growth Conditions, DNA Isolation, and Protein Isolation
M. mobile 163K (ATCC 43663) was kindly provided by Makoto Miyata of Osaka City University, who provided bacterial cultures and expertise. Cultures of M. mobile were grown in 150-mL plastic flasks without shaking in Aluotto medium at 22°C (Aluotto et al. 1970). For DNA sequencing, cultures were grown to OD600 0.15, and DNA was extracted using the Qiagen Genomic Tip system (QIAGEN). The average size of the isolated DNA was 50 kb as determined by pulse-field gel electrophoresis (PFGE). For protein extracts, cultures were grown to OD600 0.075 and treated as described (Jaffe et al. 2004a).
DNA Sequencing, Assembly, and Finishing
The M. mobile 163K genome sequence was determined by using a whole-genome shotgun sequencing approach. Random small and large insert plasmid libraries (2-kb, 4-kb, 6-kb, 8-kb, and 10-kb inserts) were constructed as described in http://www.broad.mit.edu/annotation/microbes/mycoplasma/.
Sequence reads were derived from both ends of the inserts to generate paired-end reads as previously described (Lander et al. 2001; Waterston et al. 2002). Assemblies were generated with a variety of combinations of read types and assembly parameters using the ARACHNE software package (Batzoglou et al. 2002; Jaffe et al. 2003), and the optimal assembly was selected for finishing.
The optimal assembly was generated with a total of 16,376 reads derived from the 2-kb, 4-kb, 6-kb, 8-kb, and 10-kb inserts and yielded 12-fold sequence coverage of the M. mobile genome with a PHRED quality score of ≥20. After this assembly, there were 10 gaps spanned by plasmid clones and eight unspanned gaps. To supplement regions of low coverage and to obtain sequence for the unspanned gaps, additional paired-end reads from all five plasmid libraries were incorporated into the genome assembly. A combination of standard finishing methods including transposon-mediated sequencing and PCR were applied to close gaps and to resolve regions of low sequence quality as described previously (Galagan et al. 2002). The finished M. mobile genome assembly was validated by PCR, PFGE of genomic DNA singly digested with the restriction enzymes AvrII, BamHI, BsmBI, and BstEII (New England Biolabs), and by comparison of virtual restriction enzyme digests (ApaI, BamHI, MluI, and NruI) of the assembly to a previously generated physical map of the M. mobile genome (Bautsch 1988).
Proteogenomic Mapping
Proteogenomic mapping by mass spectrometry was performed as previously described (Jaffe et al. 2004a), with the following modifications. For these experiments, two complete biological repeats of the proteogenomic mapping protocol were performed (i.e., two separate cultures were extracted for protein and processed). For each repeat, 1 mg of total cell protein was fractionated by strong cation exchange chromatography (SCX). LCQ DecaXP Plus mass spectrometers (ThermoFinnigan) configured for nanoelectrospray ionization were used to collect tandem mass spectra from subsequent reversed-phase separations of each of 80 SCX fractions in each repeat. These spectra were searched via SEQUEST-PVM using 61 processors of a computing cluster against a six-frame in silico translation of the primary DNA sequence determined for M. mobile (Sadygov et al. 2002). Results from the two repeats were pooled to form a final proteogenomic map.
Annotation and Analysis
Automated gene prediction of the M. mobile genome was performed using the Calhoun annotation system developed at the Broad Institute (Galagan et al. 2002, 2003). GLIMMER (Delcher et al. 1999) was run on the whole genome using protein translation Table 4 to generate an initial set of ORFs. GLIMMER ORFs longer than 200 bp were annotated as genes if they did not overlap with an adjacent ORF by more than 30 bp. These ORFs were refined based on homology to the closest known protein by searching against the GenBank NR database using BLASTX with threshold E < 1 × 10–10 (Altschul et al. 1997). Subsequently, proteogenomic information was incorporated into the annotation to validate the expression of the predicted proteins where possible. Several new ORFs that had proteomic evidence but were not identified in the initial round of automated annotation were added at this stage. Start codon differences between the proteomic and sequenced-based models for ORFs were resolved with the aid of RBSFINDER and multiple sequence alignments from orthologous genes when possible (Suzek et al. 2001). Every ORF was then Gapped-BLAST-searched against the nonredundant database of protein sequences available at NCBI using a cutoff E-value of 0.001 and sequence composition-based statistics (Altschul et al. 1997; Schaffer et al. 2001). RPS-BLAST searches against the COG database were additionally used to assign COGs to the proteins where possible (Tatusov et al. 2000; Marchler-Bauer et al. 2003). E.C. numbers were assigned where applicable by comparison of the proteins to the KEGG database (Ogata et al. 1999). Results were inspected manually, and final protein annotations were selected. Multiple alignments (when necessary) were performed with CLUSTALX (Thompson et al. 1997). A controlled vocabulary was used to reflect the degree of certainty about a predicted ORF with unknown function (see Results).
tRNA genes were detected by the tRNAscan-SE program (Lowe and Eddy 1997). rDNA genes were detected by homology search using BLASTN and other tools (Altschul et al. 1997; Wuyts et al. 2004). tmRNA, 4.5S SRP-RNA, and RNAse P RNA sequences were also detected via homology search and subsequent comparison to known secondary structure features of these molecules (Brown 1999; Laslett et al. 2002).
BIOPERL was used in parsing of homology search results and generation of some figures for this paper (Stajich et al. 2002). PATHWAY TOOLS and METACYC were used to automatically generate and visualize a metabolic network map from the annotation that was subsequently refined manually (Karp et al. 2002; Krieger et al. 2004). Annotation details, analysis results, and the complete genome sequence are available at http://www.broad.mit.edu/annotation/microbes/mycoplasma/.
Phylogenetic-Distance Search for Motility Genes
A matrix was constructed in which each row represented one M. mobile gene and each column was assigned to one of the fully sequenced mycoplasmas. This matrix was filled with the base-10 logarithm of the E-values obtained in the search described above where hits were obtained. For each gene, the average of these log-transformed E-values was taken for orthologs found in motile and immotile species. The average from the motile orthologs was subtracted from the average of the immotile orthologs to yield a surrogate phylogenetic distance score such that genes more highly conserved among the motile mycoplasmas would have a positive score (note that the BLAST E-values all had negative exponents with the stringency criteria that we used). This score is termed the Motile Group Phylogenetic Bias. The scores were approximately normally distributed around an average of 4.07 with a standard deviation of 12.9. We then identified genes >2 (95% confidence) standard deviations above the mean as candidates for motility genes.
Acknowledgments
We thank Makoto Miyata for his gift of M. mobile and for sharing his expertise in cultivation of this organism and Jeremy Zucker for assistance in our METACYC analysis. We gratefully acknowledge Eric S. Lander for his critical support of this project. We also thank all members of the genome sequencing platform at the Broad Institute. Funding of this work was provided by support from the Broad Institute and by grants to H.C.B. (NIH Grant # AI16478), and G.M.C. (DOE GTL). There are no known competing financial interests involved in this work.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2674004.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession no. AE017308. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: M. Miyata, T. Knight, and N. Stange-Thomann.]
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluotto, B.B., Wittler, R.G., Williams, C.O., and Faber, J.E. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Intl. J. Syst. Bacteriol. 20: 35–58. [Google Scholar]
- Barre, A., de Daruvar, A., and Blanchard, A. 2004. MolliGen, a database dedicated to the comparative genomics of Mollicutes. Nucleic Acids Res. 32 Database issue: D307–D310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzoglou, S., Jaffe, D.B., Stanley, K., Butler, J., Gnerre, S., Mauceli, E., Berger, B., Mesirov, J.P., and Lander, E.S. 2002. ARACHNE: A whole-genome shotgun assembler. Genome Res. 12: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautsch, W. 1988. Rapid physical mapping of the Mycoplasma mobile genome by two-dimensional field inversion gel electrophoresis techniques. Nucleic Acids Res. 16: 11461–11467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhugra, B., Voelker, L.L., Zou, N., Yu, H., and Dybvig, K. 1995. Mechanism of antigenic variation in Mycoplasma pulmonis: Interwoven, site-specific DNA inversions. Mol. Microbiol. 18: 703–714. [DOI] [PubMed] [Google Scholar]
- Bredt, W. 1968. Growth morphology of Mycoplasma pneumoniae strain FH on glass surface. Proc. Soc. Exp. Biol. Med. 128: 338–340. [DOI] [PubMed] [Google Scholar]
- Bredt, W. and Radestock, U. 1977. Gliding motility of Mycoplasma pulmonis. J. Bacteriol. 130: 937–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, J.W. 1999. The Ribonuclease P Database. Nucleic Acids Res. 27: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassuto, E., West, S.C., Mursalim, J., Conlon, S., and Howard-Flanders, P. 1980. Initiation of genetic recombination: Homologous pairing between duplex DNA molecules promoted by recA protein. Proc. Natl. Acad. Sci. 77: 3962–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambaud, I., Heilig, R., Ferris, S., Barbe, V., Samson, D., Galisson, F., Moszer, I., Dybvig, K., Wroblewski, H., Viari, A., et al. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29: 2145–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordova, C.M., Lartigue, C., Sirand-Pugnet, P., Renaudin, J., Cunha, R.A., and Blanchard, A. 2002. Identification of the origin of replication of the Mycoplasma pulmonis chromosome and its use in oriC replicative plasmids. J. Bacteriol. 184: 5426–5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar, T., Snel, B., Schmidt, S., Lathe, W., Suyama, M., Huynen, M., and Bork, P. 2002. Comparative genome analysis of Mollicutes. In Molecular biology and pathogenicity of mycoplasmas (eds. S. Razin and R. Herrmann), pp. 255–278. Kluwer Academic/Plenum, New York.
- Delcher, A.L., Harmon, D., Kasif, S., White, O., and Salzberg, S.L. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27: 4636–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficarro, S.B., McCleland, M.L., Stukenberg, P.T., Burke, D.J., Ross, M.M., Shabanowitz, J., Hunt, D.F., and White, F.M. 2002. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20: 301–305. [DOI] [PubMed] [Google Scholar]
- Fraser, C.M., Gocayne, J.D., White, O., Adams, M.D., Clayton, R.A., Fleischmann, R.D., Bult, C.J., Kerlavage, A.R., Sutton, G., Kelley, J.M., et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270: 397–403. [DOI] [PubMed] [Google Scholar]
- Galagan, J.E., Nusbaum, C., Roy, A., Endrizzi, M.G., Macdonald, P., FitzHugh, W., Calvo, S., Engels, R., Smirnov, S., Atnoor, D., et al. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12: 532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan, J.E., Calvo, S.E., Borkovich, K.A., Selker, E.U., Read, N.D., Jaffe, D., FitzHugh, W., Ma, L.J., Smirnov, S., Purcell, S., et al. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868. [DOI] [PubMed] [Google Scholar]
- Glass, J.I., Lefkowitz, E.J., Glass, J.S., Heiner, C.R., Chen, E.Y., and Cassell, G.H. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407: 757–762. [DOI] [PubMed] [Google Scholar]
- Grigoriev, A. 1998. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26: 2286–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelreich, R., Hilbert, H., Plagens, H., Pirkl, E., Li, B.C., and Herrmann, R. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24: 4420–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, K. and Stoffel, W. 1993. TMbase—A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 364: 166. [Google Scholar]
- Hoiczyk, E. and Baumeister, W. 1998. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr. Biol. 8: 1161–1168. [DOI] [PubMed] [Google Scholar]
- Hu, P.C., Schaper, U., Collier, A.M., Clyde Jr., W.A., Horikawa, M., Huang, Y.S., and Barile, M.F. 1987. A Mycoplasma genitalium protein resembling the Mycoplasma pneumoniae attachment protein. Infect. Immun. 55: 1126–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe, D.B., Butler, J., Gnerre, S., Mauceli, E., Lindblad-Toh, K., Mesirov, J.P., Zody, M.C., and Lander, E.S. 2003. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 13: 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe, J.D., Berg, H.C., and Church, G.M. 2004a. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4: 59–77. [DOI] [PubMed] [Google Scholar]
- Jaffe, J.D., Miyata, M., and Berg, H.C. 2004b. The energetics of gliding motility in Mycoplasma mobile. J. Bacteriol. 186: 4254–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp, P.D., Paley, S., and Romero, P. 2002. The Pathway Tools software. Bioinformatics 18 Suppl 1: S225–S232. [DOI] [PubMed] [Google Scholar]
- Kenri, T., Taniguchi, R., Sasaki, Y., Okazaki, N., Narita, M., Izumikawa, K., Umetsu, M., and Sasaki, T. 1999. Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: Evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect. Immun. 67: 4557–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff, H. 1992. Motility. In Mycoplasmas: Molecular biology and pathogenesis (ed. J. Maniloff), pp. 289–306. American Society for Microbiology, Washington, DC.
- Kirchhoff, H. and Rosengarten, R. 1984. Isolation of a motile mycoplasma from fish. J. Gen. Microbiol. 130: 2439–2445. [DOI] [PubMed] [Google Scholar]
- Krieger, C.J., Zhang, P., Mueller, L.A., Wang, A., Paley, S., Arnaud, M., Pick, J., Rhee, S.Y., and Karp, P.D. 2004. MetaCyc: A multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 32 Database issue: D438–D442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. [DOI] [PubMed] [Google Scholar]
- Lartigue, C., Blanchard, A., Renaudin, J., Thiaucourt, F., and Sirand-Pugnet, P. 2003. Host specificity of mollicutes oriC plasmids: Functional analysis of replication origin. Nucleic Acids Res. 31: 6610–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslett, D., Canback, B., and Andersson, S. 2002. BRUCE: A program for the detection of transfer-messenger RNA genes in nucleotide sequences. Nucleic Acids Res. 30: 3449–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniloff, J. 1992. Phylogeny of mycoplasmas. In Mycoplasmas: Molecular biology and pathogenesis (ed. J. Maniloff), pp. 549–560. American Society for Microbiology, Washington, DC.
- Maniloff, J. 2002. Phylogeny and evolution. In Molecular biology and pathogenicity of mycoplasmas (eds. S. Razin and R. Herrmann), pp. 31–44. Kluwer Academic/Plenum, New York.
- Marchler-Bauer, A., Anderson, J.B., DeWeese-Scott, C., Fedorova, N.D., Geer, L.Y., He, S., Hurwitz, D.I., Jackson, J.D., Jacobs, A.R., Lanczycki, C.J., et al. 2003. CDD: A curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31: 383–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata, M. 2002. Cell division. In Molecular biology and pathogenicity of mycoplasmas (eds. S. Razin and R. Herrmann), pp. 117–130. Kluwer Academic/Plenum, New York.
- Miyata, M. and Uenoyama, A. 2002. Movement on the cell surface of the gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215: 285–289. [DOI] [PubMed] [Google Scholar]
- Miyata, M., Yamamoto, H., Shimizu, T., Uenoyama, A., Citti, C., and Rosengarten, R. 2000. Gliding mutants of Mycoplasma mobile: Relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146: 1311–1320. [DOI] [PubMed] [Google Scholar]
- Miyata, M., Ryu, W.S., and Berg, H.C. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184: 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., and Kanehisa, M. 1999. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 27: 29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papazisi, L., Gorton, T.S., Kutish, G., Markham, P.F., Browning, G.F., Nguyen, D.K., Swartzell, S., Madan, A., Mahairas, G., and Geary, S.J. 2003. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain R(low). Microbiol. 149: 2307–2316. [DOI] [PubMed] [Google Scholar]
- Pollack, J.D. 2002. Central carbohydrate pathways: Metabolic flexibility and the extra role of some “housekeeping” enzymes. In Molecular biology and pathogenicity of mycoplasmas (eds. S. Razin and R. Herrmann), pp. 163–200. Kluwer Academic/Plenum, New York.
- Radestock, U. and Bredt, W. 1977. Motility of Mycoplasma pneumoniae. J. Bacteriol. 129: 1495–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin, S., Yogev, D., and Naot, Y. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62: 1094–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengarten, R. and Kirchhoff, H. 1987. Gliding motility of Mycoplasma sp. nov. strain 163K. J. Bacteriol. 169: 1891–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengarten, R., Fischer, M., Kirchhoff, H., Kerlen, G., and Seack, K.-H. 1988. Transport of erythrocytes by gliding cells of Mycoplasma mobile 163K. Curr. Microbiol. 16: 253–257. [Google Scholar]
- Ruffin, D.C., van Santen, V.L., Zhang, Y., Voelker, L.L., Panangala, V.S., and Dybvig, K. 2000. Transposon mutagenesis of Mycoplasma gallisepticum by conjugation with Enterococcus faecalis and determination of insertion site by direct genomic sequencing. Plasmid 44: 191–195. [DOI] [PubMed] [Google Scholar]
- Sadygov, R.G., Eng, J., Durr, E., Saraf, A., McDonald, H., MacCoss, M.J., and Yates III, J.R. 2002. Code developments to improve the efficiency of automated MS/MS spectra interpretation. J. Proteome Res. 1: 211–215. [DOI] [PubMed] [Google Scholar]
- Sasaki, Y., Ishikawa, J., Yamashita, A., Oshima, K., Kenri, T., Furuya, K., Yoshino, C., Horino, A., Shiba, T., Sasaki, T., et al. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 30: 5293–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer, A.A., Aravind, L., Madden, T.L., Shavirin, S., Spouge, J.L., Wolf, Y.I., Koonin, E.V., and Altschul, S.F. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29: 2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefer, H.G. and Schummer, U. 1982. The electrochemical potential across mycoplasmal membranes. Rev. Infect. Dis. 4 Suppl: S65–S70. [DOI] [PubMed] [Google Scholar]
- Schummer, U. and Schiefer, H.G. 1983. Electrophysiology of mycoplasma membranes. Yale J. Biol. Med. 56: 413–418. [PMC free article] [PubMed] [Google Scholar]
- Sharp, P.M. and Li, W.H. 1987. The Codon Adaptation Index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X., Gumulak, J., Yu, H., French, C.T., Zou, N., and Dybvig, K. 2000. Gene rearrangements in the vsa locus of Mycoplasma pulmonis J. Bacteriol. 182: 2900–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist, C.J., Cerutti, L., Hulo, N., Gattiker, A., Falquet, L., Pagni, M., Bairoch, A., and Bucher, P. 2002. PROSITE: A documented database using patterns and profiles as motif descriptors. Brief. Bioinform. 3: 265–274. [DOI] [PubMed] [Google Scholar]
- Sokal, R.R. and Rohlf, J.F. 1995. Biometry: The principles and practice of statistics in biological research. W.H. Freeman, New York.
- Stajich, J.E., Block, D., Boulez, K., Brenner, S.E., Chervitz, S.A., Dagdigian, C., Fuellen, G., Gilbert, J.G., Korf, I., Lapp, H., et al. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12: 1611–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev, S.I., Blount, P., Martinac, B., Blattner, F.R., and Kung, C. 1994. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature 368: 265–268. [DOI] [PubMed] [Google Scholar]
- Sutera Jr., V.A., Han, E.S., Rajman, L.A., and Lovett, S.T. 1999. Mutational analysis of the RecJ exonuclease of Escherichia coli: Identification of phosphoesterase motifs. J. Bacteriol. 181: 6098–6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzek, B.E., Ermolaeva, M.D., Schreiber, M., and Salzberg, S.L. 2001. A probabilistic method for identifying start codons in bacterial genomes. Bioinformatics 17: 1123–1130. [DOI] [PubMed] [Google Scholar]
- Tatusov, R.L., Galperin, M.Y., Natale, D.A., and Koonin, E.V. 2000. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28: 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uenoyama, A., Kusumoto, A., and Miyata, M. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 186: 1537–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker, L.L. and Dybvig, K. 1996. Gene transfer in Mycoplasma arthritidis: Transformation, conjugal transfer of Tn916, and evidence for a restriction system recognizing AGCT. J. Bacteriol. 178: 6078–6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Westberg, J., Persson, A., Holmberg, A., Goesmann, A., Lundeberg, J., Johansson, K.E., Pettersson, B., and Uhlen, M. 2004. The genome sequence of Mycoplasma mycoides subsp. mycoides SC type strain PG1T, the causative agent of contagious bovine pleuropneumonia (CBPP). Genome Res. 14: 221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuyts, J., Perriere, G., and Van De Peer, Y. 2004. The European ribosomal RNA database. Nucleic Acids Res. 32 Database issue: D101–D103. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://arep.med.harvard.edu/mycoplasma/; G.M. Church, Harvard University Medical School.
- http://www.broad.mit.edu/annotation/microbes/mycoplasma/; Mycoplasma mobile genome project.
- http://www.ncbi.nlm.nih.gov/COG/; NCBI, National Institues of Health.