Abstract
Large chromosomal rearrangements, duplications, and inversions are relatively common in mammalian genomes. Here we report interesting features of DNA strands flanking a Multiple Sclerosis (MS) susceptibility locus on Chromosome 17q24. During the positional cloning process of this 3-Mb locus, several markers showed a radiation hybrid clone retention rate above the average (1.8-fold), suggestive for the existence of duplicated sequences in this region. FISH studies demonstrated multiple signals with three of the tested regional BACs, and 24 BACs out of 187 showed evidence for duplication in shotgun sequence comparisons of the 17q22–q24 region. Specifically, the MS haplotype region proved to be flanked by palindromic sequence stretches and by long segmental intrachromosomal duplications in which highly homologous DNA sequences (>96% identity) are present at both ends of the haplotype. Moreover, the 3-Mb DNA segment, flanked by the duplications, is inverted in the mouse genome when compared with the orientation in human and chimp. The segmental duplication architecture surrounding the MS locus raises the possibility that a nonallelic homologous recombination between duplications could affect the biological activity of the regional genes, perhaps even contributing to the genetic background of MS.
The emerging information on the structure of the human genome has provided an entirely new view to the genome landscape. Large structural repeats, inversions, and other type of rearrangements add a new level of complexity to the genome structure and set new challenges for detailed understanding of the normal and disease-associated abnormal functions of individual genes and genome regions (Subramanian et al. 2001; Eichler and Sankoff 2003).
We have applied mapping and positional cloning strategy to identify genetic loci for multiple (MS) sclerosis using the study samples collected nationwide from the isolated population of Finland. Population isolates generally possess certain advantages in genetic studies of human diseases, even of those with a complex, polygenic background (Peltonen et al. 2000). Within Finland, the Southern Ostrobothnian region represents a high-risk area for MS, showing an exceptional familial clustering of MS cases. This particular study sample thus offers several advantages for genetic studies including high incidence of disease, high number of familial cases, and high proportion of progressive-type of MS (Sumelahti et al. 2000, 2003). Our genome-wide search and previous studies have identified four main candidate regions for MS: the HLA locus on 6p, the Myelin Basic Protein (MBP) locus on 18q, and two relatively wide regions on 5p12–p14 and 17q22–q24 (Tienari et al. 1992, 1998; Kuokkanen et al. 1997). Interestingly, the locus on 17q has also been implicated for linkage to MS in other genome scans performed in study samples from more heterogeneous populations (Ebers et al. 1996; Sawcer et al. 1996; Dyment et al. 2001). The biological significance of this region is further emphasized by the fact that the 17q locus is syntenic to the mouse experimental allergic encephalomyelitis (Eae) locus on Chromosome 11 (Butterfield et al. 1998) and to a rat Eae locus on Chromosome 10 (Jagodic et al. 2001). We have more recently strengthened the evidence for linkage to this region, as well as restricted the critical MS region by haplotype and association analysis in Finnish multiplex families to 3.0 Mb, between markers D17S1792 and ATA43A10 (Saarela et al. 2002).
For fine mapping and positional cloning of the 17q MS locus, we confirmed the physical map of the region using radiation (RH) panels and observed a significant variation of the number of positive clones for each marker. We interpreted this as suggestive for duplicated sequence structures. This hypothesis stimulated more extensive physical mapping of the region, including FISH and sequence comparison of different databases. It became evident that this particular region of the genome contained an abundance of interchromosomal and intrachromosomal segmental duplications, also known as duplicons or low-copy repeat sequences (Saarela et al. 2002). Such regions often demarcate regions of genomic instability associated within recurrent chromosomal structural rearrangements and disease (Stankiewicz and Lupski 2002). Chromosome 17 is among the chromosomes known to represent segmental duplications and thus presents a special challenge for positional cloning (Bailey et al. 2002a). The critical region for MS on 17q22–24 is a large segment with >300 genes and numerous chromosomal segmental duplications. These rearrangements can potentially lead to the loss or gain of a dosage-sensitive gene or to disruption of a gene or its regulatory elements. One example of a putative role of such rearrangements in a complex disease is the region on 8p (Giglio et al. 2001). This particular region is positioned within a common submicroscopic inversion (26% of certain European descent), and the rearrangement is potentially involved in the pathogenesis of diseases like bipolar disease, mapped to this region (Giglio et al. 2001).
In this study we have used several physical and genetic mapping techniques as well as tools of computational analysis to refine more precisely the genomic structure of the 17q22–q24 region as a part of the process aiming to characterize the molecular pathogenesis of MS. The obtained data exemplify the complexity of the landscape of the human genome as well as position the critical MS region between two long-range rearrangements raising an interesting possibility that the particular chromosomal architecture may contribute to the pathogenesis of MS.
RESULTS
RH Retention Number Provides Evidence for Duplicated/Rearranged Region
The routine ordering of markers used for the fine mapping of 17q22–24 using RH panels indicated in a significant variation in the observed number of clones PCR-positive for tested markers (Table 1). On multiple occasions, two closely located STS markers had a very different number of positive clones in both Stanford G3 and TNG4 RH panels. For instance, SHGC-52535 and SHGC-32453 are two STS markers located only 200 kb apart on the same BAC clone (AC003663). The distance of 200 kb corresponds to ∼8 crays on a radiation hybrid G3 map after conversion. Two markers this close to each other should have a very similar pattern of PCR positive clones on the RH panel. However, PCR using primers for SHGC-52535 picked up 20 clones (retention rate ∼24.2%), whereas primers for SHGC-32453 detected only 13 positive clones (retention rate ∼15.6%). A likely explanation for such a difference would be that an exceptionally high number of positive clones results from signals of two genome regions, the original and the duplicated position.
Table 1.
Summary of Results for RH Markers on the Stanford Radiation Hybrid Map (Panel G3)
| BAC clone | UCSC July 2003 position (bp) | RH marker | Number of clones in G3 panel | Retention rate (%) | Duplication detected/not detected |
|---|---|---|---|---|---|
| AC069007.3 | chr17:44,081,749-44,286,700 | RH32232 | 37 | 44.6 | Dup |
| RH42841 | 20 | 24.1 | Dup | ||
| AC003663.1 | chr17:63,238,604-63,370,672 | D17S1557* | 17 | 20.5 | Dup |
| RH42841 | 20 | 24.1 | Dup | ||
| RH14914 | 10 | 12 | Non-dup | ||
| RH42755 | 18 | 21.7 | Dup | ||
| AC074102.2 | chr17:58,210,816-58,381,497 | RH37851 | 12 | 14.5 | Non-dup |
| AC037475.9 | chr17:58,899,542-59,015,469 | RH110597* | 11 | 13.3 | Non-dup |
| AC025048.5 | chr17:58,602,379-58,740,299 | RH38146 | 12 | 14.5 | Non-dup |
| RH84338 | 10 | 12 | Non-dup | ||
| RH94909 | 33 | 39.8 | Dupa | ||
| AC004686.1 | chr17:58,373,575-58,507,051 | RH9059 | 24 | 28.9 | Dup |
| RH31407 | 11 | 13.3 | Non-dup | ||
| RH14802 | 9 | 10.8 | Non-dup | ||
| RH83863 | 10 | 12 | Non-dup | ||
| AC005702.1 | chr17:58,470,068-58,617,753 | RH74575 | 9 | 10.8 | Non-dup |
| RH9059 | 24 | 28.9 | Dup | ||
| AC080038.3 | chr17:61,048,819-61,267,940 | RH32867 | 14 | 16.9 | Dupb |
| RH99431 | 18 | 21.7 | Dupb |
Results of RH markers on the Stanford Radiation Hybrid map. All markers were analyzed in silico, but those indicated with * have been experimentally confirmed by PCR. An additional 27 microsatellite markers used in our fine-mapping experiment were also analyzed both in silico and by PCR on the Radiation Hybrid Panel G3, and the results were comparable to the ones shown here.
Not in a duplicon, but the EST sequence maps with >90% identity to five separate locations on Chromosome 17.
Not in a duplicon, but RH markers map to both Chromosomes 7 and 17.
To further investigate the possibility of using the RH retention number as a means to detect potential genome duplication, we queried the database of all the potential RH markers positioned on Chromosome 17q22–q24. Subsequently, seven STS markers with radiation hybrid G3 data were found to map to the duplicated area as determined by in silico analysis. These markers have an average of 27.1% positive clone retention rate. In contrast, the average retention rate for STS markers with radiation hybrid G3 data is only 15.1% on Chromosome 17 (200 markers tested). The difference would imply that the RH retention number could potentially be used to detect genome duplications and that RH mapping data based on unusually high retention ratios should be approached with considerable caution.
Copy Number of Target Sequences Determined by FISH
Fluorescence in situ hybridization (FISH) was used to confirm the RH panel finding suggestive for duplications. Eight fluorescent-labeled BAC clones mapping to 17q22–q24 according to the sequence information were hybridized to metaphase chromosomes. Three of these BACs, AC003663 (D17S1557), AC015821 (D17S1825), and AC005702 (WI-6034), used as probes in FISH experiments showed at least two signals on 17q, a stronger signal on 17q24 and a weaker signal on 17q21, confirming the initial hypothesis of the existence of duplication in this chromosomal region. Additionally, the signals of two labeled BACs (AC015821 and AC005702) were exceptionally wide and strong, suggesting that multiple copies of target sequences for the BAC hybridization existed in this region. To provide higher resolution, we used these BAC clones as probes for stretched chromosome preparations, and observed two closely located fluorescence signals with each probe (Fig. 1). One of these signals was exceptionally strong, providing further evidence for multiple target sequences. FISH results would thus suggest a complex genomic structure on the 17q22–24 region involving both short distance and distant duplication patterns.
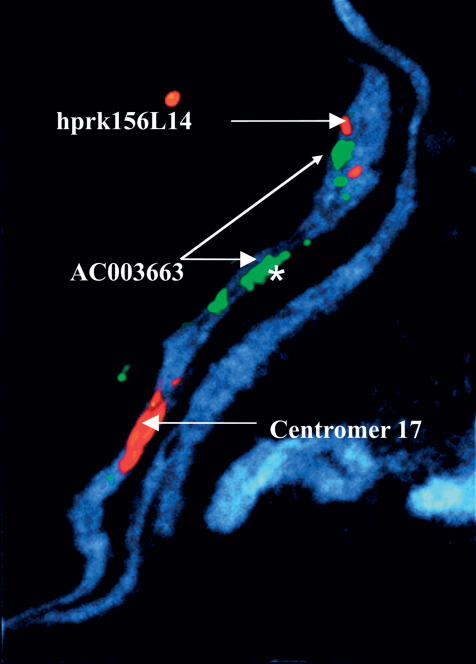
Figure 1.
A mechanically stretched, DAPI-stained chromosome demonstrating multiple hybridization signals for BAC AC003663 (green) as an indication of duplicated sequences in multiple positions of Chromosome 17q. One of these signals is exceptionally strong (labeled with an *), suggesting that this area contains multiple target sequences for BAC AC003663. BAC AC005821 (red) is detected only as two signals, one for each sister chromatin, thus suggesting that no large duplicated sequences exist in this BAC. The centromere of Chromosome 17 is seen as a large red signal in the lower part of the figure.
Segmental Duplication Analysis Using Sequence Representation in Different Databases
To further elucidate the genomic landscape of this region, we implemented methods developed by Bailey et al. (2002a) to interrogate both the genome assembly (April and July 2003 freezes, UCSC Human Genome Map database) sequences and the underlying BAC sequences spanning this portion of 17q22–24. According to the April 2003 freeze of the UCSC Human Genome Map, this region is represented by 187 BAC clones spanning >18.24 Mb and containing two major sequencing contigs. The whole-genome shotgun sequence detection (WSSD) method by Bailey and coworkers determines areas of potential genome duplications by analyzing each BAC clone for overrepresentation within a whole-genome shotgun sequence. A stretch of sequence within a BAC clone is defined as being duplicated if the overrepresentation of the whole-genome shotgun sequence exceeds 2 standard deviations beyond the mean coverage based on an analysis of WGS depth of coverage within unique regions of the genome. At least five overlapping windows (5 kb in size) are required before a region is scored as duplicated (Bailey et al. 2002a). Among the 187 BAC clones analyzed, 24 BACs show evidence of duplications. Our analysis shows that these correspond to the largest and most homologous segmental duplications (alignments showing >96% sequence identity and >15 kb in size). All three BACs that showed evidence for duplication by FISH were also represented by duplicated sequences by the depth-of-coverage method, whereas the five BACs with regular, single signals in the FISH hybridization showed no evidence for duplications in the bioinformatics-based analysis. We compared this pattern of segmental duplication to that based on a whole-genome assembly comparison of the finished human genome for this region (Bailey et al. 2002a). We observed an excellent correspondence between these two methods validating the enrichment of segmental duplications within this portion of 17q22–q24 (Fig. 2). Our combined analysis shows that 6.05% of the 19.57-Mb sequences are duplicated (>1 kb, >90% identity). The majority of the duplications (80.8%; 957 kb of nonoverlapping duplicated bases) belong to Chromosome 17 intrachromosomal alignments (Fig. 2). Moreover, the majority of the duplications are a large part of duplication blocks that are >10 kb in length. These properties may predispose this portion of Chromosome 17 to significant rearrangements.
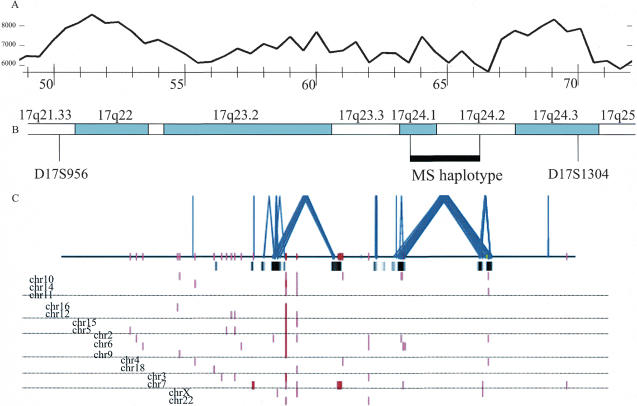
Figure 2.
Segmental duplications within 17q22–17q24 and their position in relation to chromosomal bands, MS haplotype, and palindromic sequence distribution. (A) The frequency of palindromes >6 bp. For details, see Figure 4. (B) The chromosomal banding, STS markers, and the shared MS haplotype (black horizontal bar). (C) The pattern of interchromosomal (red) and intrachromosomal segmental duplications (blue) based on the whole-genome analysis comparison method (>1 kb and >90% sequence identity) is shown for 18 Mb between markers D17S956and D17S1304. Highly homologous (>96% sequence identity) regions confirmed by whole-genome shotgun sequence detection are shown as black bars below the horizontal line.
Comparison of Physical Versus Genetics Maps
To assess if intrachromosomal segmental duplications would have an effect on the meiotic recombination, we compared the physical map of Chromosome 17q22–24 (July 2003 freeze of the UCSC genome map) with the deCODE genetic map (Fig. 3). The 19.67-Mb physical map of this region is flanked by two multiallelic markers (D17S956 and D171304) and covers a sex average distance of 25.44 cM, which translates to an overall 1.29 cM/1 Mb sex average conversion in this area, with an average intermarker distance of 0.71 cM and 0.55 Mb. This interval, covered by 166 BAC clones, contains 21 large (>10 kb) and >100 small (1–10 kb) DNA segments, which are duplicated within Chromosome 17 or in other chromosomes (Fig. 3). None of the 37 markers mapping to the interval maps to duplicated regions per se. The ratio of genetic and physical distance seemed not to be dramatically affected by the intra- or interchromosomal duplications: The average recombination rate for pairs of subsequent markers not separated by duplication was 1.09 cM/Mb (varying between 0 and 4.10, SD = 1.18), whereas the average recombination rate for marker pairs separated by duplicated sequence(s) was slightly higher, 1.24 cM/Mb (varying between 0 and 2.83, SD = 0.96).
Figure 3.
Comparison of the genetic and physical distances in the Chromosome 17q22–24 region. The black arrows indicate areas of intrachromosomal duplications and the gray arrows areas of interchromosomal duplication (long arrow: >10 kb; short arrow: <10 kb), and the hatched box in the physical map shows the shared MS haplotype.
Number of SNPs in the Duplicated and Nonduplicated Regions
While searching the databases for SNP markers mapping on the 17q region, it became evident that significantly more SNPs were mapped to the duplicated than nonduplicated regions. To pursue this observation further, we counted the total number of SNPs assigned to sequences shown to be duplicated according to the SDD database and compared that with the number of SNPs on unique regions. Currently, the SNPs in the UCSC Genome Browser are divided into two groups according to the method used for their identification. The clone overlap method of SNP discovery finds an average of 0.25 SNPs/kb on nonduplicated regions, whereas the SNP density was fourfold in the duplicated sequences, being 1.0 SNPs/kb. In contrast, there is no significant difference in the SNP density between the duplicated and unique sequences when the random clone method is used for SNP identification (0.40 SNPs in duplicated regions, 0.31 SNPs/kb in nonduplicated regions). This indicates that a large proportion of SNPs reported for the duplicated region are not true SNPs but paralogous sequence variants (PSVs; Estivill et al. 2002).
Among the 120 SNPs designed to be genotyped in our attempted fine mapping of the MS locus, six SNPs mapped to BAC clones that are shown to be duplicated in this study, but seemed unique when the SNP sequence was aligned with the publicly available human genome sequence and thus represent PSVs. None of these SNPs produced good, high-quality genotypes and were systematically excluded from the large-scale genotyping effort in the MS study sample.
Distribution of Short Palindromic Sequences
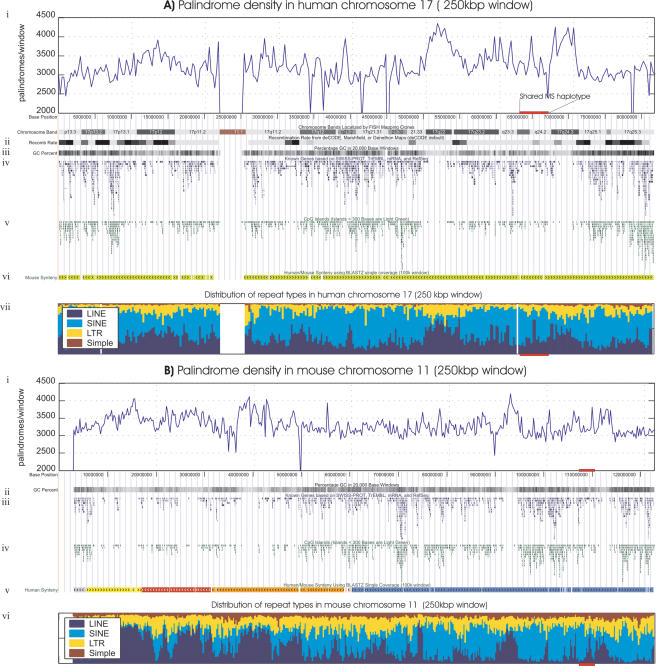
Because the palindromic sequences are often associated with the rearrangements and genetic instability in prokaryotes, we analyzed systematically the distribution of palindromes on 17q. Figure 4 shows the frequency of perfect palindromes >6 bp in length for 250,000-bp windows of human Chromosome 17 and mouse Chromosome 11. In the human Chromosome 17 (Figs. 2A and 4A), the regions 17q22 (49.0–55.8 Mb) and 17q24.3 (66.5–70.8 Mb) flanking the MS susceptibility locus show a distinct enrichment with perfect palindromes and repeats. The frequency of palindromes in these regions was significantly higher than the frequency observed in the whole Chromosome 17 (p = 1.78 × 10–15 for region 49–55.8 Mb and p = 1.22 × 10–11 for region 66.5–70.8 Mb). These landmarks seem to be conserved in the mouse Chromosome 11 (89.0–95.3 Mb and 109.8–114.0Mb corresponding to the bands 11C and 11E1) around the region syntenic to the MS susceptibility locus. Both in man and mouse, the palindrome-rich areas are also gene poor and have a high number of LINE repeats (Fig. 4B, bands 11C and 11E1).
Figure 4.
(A) Sequence landscapes of human Chromosome 17. (i) Density of perfect palindromic motifs of at least 6 bp within 250,000 base windows; (ii) recombination rate from the deCODE map; (iii) G+C percentage in 20,000 base windows; (iv) known genes based on SWISS-PROT, TrEMBL, mRNA, and RefSeq; (v) CpG islands; (vi) mouse synteny; (vii) distribution of major repeat types (LINE, SINE, LTR, and Simple) in 250,000 base windows. (B) Mouse Chromosome 11 landscapes. (i) Palindrome density; (ii) genes; (iii) G+C percent; (iv) CpG islands; (v) human synteny; (vi) distribution of major repeats. The physical location of the shared MS haplotype in human and its syntenic region in mouse are shown as red horizontal bars.
Evolution of the Duplicated 17q Region
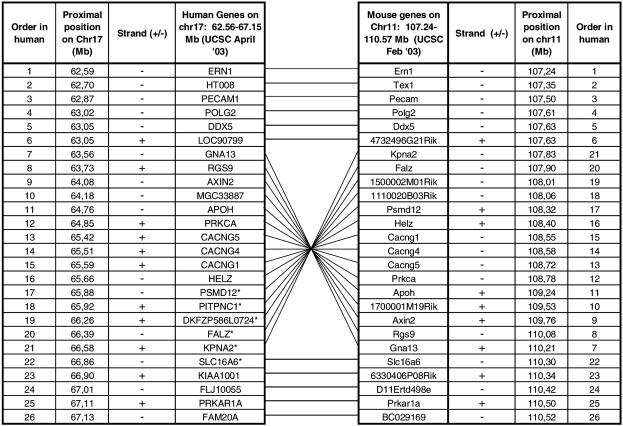
To obtain some evolutionary view to this complex chromosomal region, we compared the structure of human 17q23.3–q24.2, harboring the critical MS locus, to the syntenic region of mouse Chromosome 11 by monitoring the order and organization of the genes. The region contains 26 genes, five of which partially overlap with duplicated sequence segments: HT008, PECAM1, CACNG5, CACNG4, and PITNPC1. There are both exonic and intronic fragments overlapping with the duplications and all but one short sequence (1.4 kb) are repeated within the same locus on Chromosome 17q23–q24. The overall map order and the orientation of the genes are conserved between human and mouse chromosomes, except for a 3-Mb segment flanked by genes LOC90799 and SLC16A6, which is inverted on the human chromosome when compared with mouse (Fig. 5). This “inverted” segment contains 13 known genes and includes the critical MS locus. Interestingly, according to the UCSC July freeze, there is a predicted/hypothetical gene MGC40489, which maps with >90% homology to the sequences before and after the inverted region, as well as to 17q21.31. Two other hypothetical proteins, AK091625 and KIAA0563, map just before the inverted region and to two or more locations on 17q21.31–q21.32. The syntenic mouse region on Chromosome 11 did not contain MGC40489 and AK091625 gene sequences; neither could they be located anywhere else in the mouse genome, whereas KIAA0563 was also located on several locations on mouse Chromosome 11 (February 2003 freeze). This could, however, be caused by the more incomplete stage of the mouse sequence assembly. Interestingly, the two palindrome-rich areas identified in the human 17q are located in syntenic regions of the mouse Chromosome 11 flanking the particular region of the mouse map, which is inverted when compared with the human sequence. Potential duplications in the mouse genome have not yet been analyzed using the method by Bailey and coworkers, and it thus remains unclear if similar duplicated structures that are observed in the human chromosome would exist in mouse Chromosome 11.
Figure 5.
Comparison of the organization of the human Chromosome 17q22–24 region and the syntenic region on mouse Chromosome 11. The crossing lines indicate that the genes on mouse Chromosome 11 are in an inverted order compared with human Chromosome 17. The inverted region represents the shared haplotype region in MS. Only those human genes that have a mouse ortholog in the syntenic region of mouse Chromosome 11 are listed.
To study whether entire coding regions of genes located in this region are duplicated, mRNA sequences of these 24 genes were BLASTed to human, mouse, and chimp genomes. Five genes (psmd12, pitpnc1, dkfzp586l0724, KPNA2, SLC16A6) had at least two hits in the human genome (overlap >1 kb of the gene, >85% identity; Fig. 5). Two of these genes (SLC16A6 and KPNA2) had two hits on Chromosome 17, and the other three genes had homologous sequences on Chromosomes 1, 3, and X. Interestingly, none of these five coding sequences showed evidence for duplication in the mouse genome; only one hit on the mouse Chromosome 11 was detected. However, all of these five genes had more than one hit also in the chimp genome sequence (UCSC November 2003 assembly).
The Position of the MS Locus
The two-point and multipoint linkage data combined with monitoring for shared haplotypes in 28 Finnish MS families position the critical MS locus to a chromosomal position 17q24, flanked by segmental duplications on the centromeric side. Furthermore, the telomeric half of the locus contains two long and several short duplicons and is bordered by the palindrome-rich area at 66.5 Mb (UCSC Genome Browser, July 2003). The two large duplicated segments (35.7 kb and 16.6 kb in size) map to two distinct positions within this locus (at 63.5 Mb and 66.5 Mb), whereas the shorter segments map to other chromosomes in addition to separate locations on Chromosome 17.
We wanted to explore whether the complex structure of Chromosome 17 and its possible instability could result in a false-positive transmission distortion. A potential transmission distortion to unaffected MS family members was analyzed in 22 families where genotypes for both affected and nonaffected family members were available. In two-point linkage analysis, no hint of linkage was observed with any of the 18 multiallelic markers located in the region to nonaffected family members (Max lod score ranging from 0.00–0.98). In the corresponding test, the linkage to MS was clearly significant (Max lod score 4.48, with marker D17S1825; 14 markers gave an lod score >1.0). The position of the MS locus is indicated in Figures 2, 3, 4 and is flanked by markers D17S1792 and ATA43A10 and covered by a minimal tailing path of 23 unique BACs.
DISCUSSION
The comprehensive DNA sequence information has provided new tools to study the geography of the complete human genome. One of the intriguing questions is the association of large chromosomal rearrangements with disease susceptibility. Such rearrangements are well demonstrated in somatic mutations associated with malignant transformation and monogenic chromosome anomalies (Kolomietz et al. 2002). Their potential role in the susceptibility of complex traits has only lately been suggested for regions like 8p inversion, polymorphic at the population level (Stankiewicz and Lupski 2002). Here we demonstrate that both experimental and biocomputational tools provide comparable information on the large-scale rearrangements of 17q. This genome region is of special interest because it contains a locus linked to the susceptibility of MS, and its complexity severely hampered the efforts of locus restriction. The structural complexity results in several technical challenges, for example, PSVs and the high retention rate of clones in the radiation hybrid analysis. Interestingly, none of the polymorphic multisatellite markers in the region resulted in ambiguous results, which is likely to reflect a selection bias of the markers: the problematic ones have been excluded from the currently available marker sets.
It has previously been shown that 17q is rich in both inter- and intrachromosomal duplications (Dorr et al. 2001; Bailey et al. 2002a; Saarela et al. 2002; Armengol et al. 2003; Cheung et al. 2003); according to Bailey et al. (2002a,b); it is among the chromosomes showing most duplications after Y, 22, and 16. Here we have further demonstrated the structural complexity of the region surrounding one MS susceptibility locus on Chromosome 17q22–24. It is noteworthy that duplications are not within the shared MS haplotype but flanking them and they are present in healthy alleles. We do not know if the structure of duplications is different in MS alleles and control alleles. Most of the current methods are not readily applicable to study specifically the MS alleles. It could, however, be speculated that such flanking regions, rich in duplications and palindromic sequences, might contribute to genetic plasticity during somatic cell divisions, but the precise effect remains to be clarified. Sequence structures on 17q22–24 are represented also on 17q11 and 17q21 as indicated by multiple techniques. Recently a t17q11;22q11 translocation associated with palindromic repeat structures in an NF1 patient (the NF1 gene is located in the 17q11 region) was reported demonstrating that at least occasionally chromosomal rearrangements can be detected in areas enriched for palindromic sequences (Kurahashi et al. 2003). Aberrations of the Chromosome 17q22–24 area is frequently involved with both solid and hematological neoplasias; translocations in hematological, deletions in solid tumors (Phelan et al. 1998; Bertherat et al. 2003), and specifically amplifications in ∼20% of breast tumors (Monni et al. 2001). It remains to be resolved if there is a possible functional background or connection of these aberrations with the complex structure (e.g., palindromic sequences or duplications) of 17q22–24.
One could speculate that this complex genomic structure of the 17q22–24 area would affect the rate of meiotic recombination and thus affect the observed genetic distances in the region. However, no striking differences between the genetic and physical maps were observed between duplicated and nonduplicated regions. We observed a trend toward meiotic recombinations potentially being slightly more frequent in the areas containing duplicated sequences, but the observed difference is so small that it does not merit any definitive conclusions. However, it is interesting to speculate that accurate recombination might be different in an area containing multiple copies of the same sequence compared with a regular two-copy situation (Selker 1999).
Here we show an interesting trend between breakpoints of conserved synteny between man and mouse and the position of segmental duplications. Associations between segmental duplications and chromosomal rearrangement breakpoints have been noted before (Valero et al. 2000; Armengol et al. 2003; Locke et al. 2003; Bailey et al. 2004). Based on the degree of sequence identity (>90%), the segmental duplications in our study likely arose specifically during hominoid evolution (<35 million years ago). In contrast, most breakpoints in conserved synteny regions between man and mouse are much more ancient in nature—many more occurring within the rodent lineage of evolution (Waterston et al. 2002). Global analyses of mouse–human synteny have shown that orthologous breakpoints and duplications are biased to reoccur within the same locations of the genome (Armengol et al. 2003; Bailey et al. 2004). In this context, it is interesting that the MS locus is flanked by an unusual abundance of palindromic sequences. This raises an interesting possibility of the relationship of these two structures. The functional implications remain speculative but stimulate the idea that a high number of certain palindromes might be associated with genomic plasticity also in eukaryotes. These structures are located in gene-poor regions of Chromosome 17, and it further seems that in this region the palindromic structures colocalize with an increased number of LINE elements. However, it is unlikely that the higher number of these palindromic sequences would be explained only by repeat structures, as the differences in the frequency distribution remain even after repeat masking. It is also intriguing that the palindrome-rich sequence areas (49–55 Mb and 66.5–71 Mb) are flanking but not within the region rich in duplicated sequences. This finding might represent pure chance, and more thorough genome-wide analysis of the presence of short palindromes, segmental duplications, and chromosomal breakpoints is necessary to correlate the positional correlation of palindrome clusters and segmental duplications.
The association of palindromes to genomic plasticity has been well described in prokaryotes (Leach 1994). The genetic instability seems to correlate with the length and structure of the palindromes; in stimulation studies of yeast, the frequency of deletions and recombinations has been shown to be directly related to the size of the repeat and inversely proportional to the size of an intervening spacer (Lobachev et al. 1998). The palindromes of the human genome can roughly be divided in short palindromes (<20 bp) and longer structures (>60 bp). Short palindromes are much more frequent, and several of them are positioned in 5′-areas of genes, where they typically serve as protein-binding sites (Sanchez et al. 2002; e.g., estrogen receptors). Larger palindromes are less characterized and have been mostly associated to noncoding areas. The Y-chromosome contains palindromes of massive sizes, the largest palindrome spanning 3 Mb (Kuroda-Kawaguchi et al. 2001). In accordance with the genetic plasticity hypothesis, the large palindromes on the Y-chromosome have been associated with deletions and spermatogenetic failure. It remains entirely speculative if relatively short palindromes (20 bp to a few hundred base pairs) have a role in rearrangement events in the genome of individual cells, for example, over time. Accordingly, the relation of palindromes, duplications, and their possible role with autoimmune disorders such as MS need further experimentation. However, the structural features reflecting the plasticity of the genome might guide us to the DNA elements important for the molecular pathogenesis of MS, for example, by a potential for gene-silencing mechanisms, potentially even affecting sex-specific expression of alleles (Selker 1999). The complex structure of the region might also add another level of complexity to expression studies of genes located in this region as similar (or extremely homologous) transcripts are coded by more than one gene locus. However, as conclusive data about the causative relationship of the genetic susceptibility to MS and the complex structure of the Chromosome 17 are lacking, more detailed analyses of the local genome architecture of the other genetic loci linked to MS or other autoimmune diseases might be justified to evaluate this hypothesis.
METHODS
Radiation Hybrid Mapping
For radiation hybrid mapping, 29 STS (sequence tagged site) markers on Chromosome 17q23–24 were selected. PCR primers for each STS marker were either obtained directly from the Stanford Radiation Hybrid Web site (http://www-shgc.stanford.edu/Mapping/rh/RH_poster/) or designed using the MIT Primer3 software (http://www.broad.mit.edu/cgi-bin/primer/primer3.cgi). Conditions for individual STSs were applied as described in http://wwwshgc.stanford.edu/Mapping/rh/procedure/rhassaynew.html. The PCR reactions were performed on 83 radiation hybrid (RH) clones from the Stanford RH G3 panel (Research Genetics). The retention number of each RH marker is the percentage of the total positive clones over all 83 clones. To determine the average retention number in Chromosome 17, we used all the G3 markers mapped on the Stanford Radiation Hybrid backbone. However, G3 markers close to thymidine kinase gene (TK1) on Chromosome 17q25.1 were omitted from this calculation because it is the selection marker for the radiation hybrid fusion cells and every clone contains the gene.
Fluorescence in Situ Hybridization (FISH)
BACs AC005821, AC006440, AC003663, AC005702, AC015821, and AC069134 were chosen as probes for metaphase and stretched chromosome FISH. The BAC clones were purchased, cultured, and prepared according to the QIAGEN large-construct purification protocol (QIAGEN). The subsequent metaphase FISH was performed as previously described (Laan et al. 1995). FISH images were captured using a Leica DMR fluorescence microscope workstation. A minimum of 10 representative chromosome spreads were evaluated for each hybridization.
Segmental Duplication Analysis Using Sequence Representation in Different Databases
The presence of recent homologous duplications was assessed using two independent computational methods. The first method identified all duplications where the individual pairwise alignments were >90% and >1 kb in length. DNA sequence between D17S956 and D17S1304 was extracted from the finished genome sequence (coordinates chr17:50388877 to 70058334; July 2003 freeze) and compared against the entire human genome using the whole-genome analysis comparison method (Bailey et al. 2001). Both intrachromosomal and interchromosomal patterns of duplications were characterized. As a second approach, we examined the underlying BAC sequences from this portion of the genome assembly for an excess of whole-genome shotgun sequence reads as previously described (Bailey et al. 2002b). Regions of duplication characteristically show increased WGS depth of coverage within the alignments because of the recruitment of both allelic and paralogous sequence reads. This approach has the ability to identify large duplications >15 kb with >96% sequence identity and to provide an assembly-independent method to assess duplication content allowing potential false positives and negatives to be excluded. For all duplications >15 kb and >96% sequence identity, nearly perfect correspondence between these two methods was observed, validating the segmental duplication architecture of this region.
HGP and Celera Genome Map Comparison at Chromosome 17q23–24
To determine the positions of individual BACs in the HGP and Celera genome maps at Chromosome 17q22–24, the accession numbers of all the BACs between the two anchors were recorded separately from the public UCSC HGP (http://genome.ucsc.edu) and Celera (http://www.celera.com) bases, and the BACs were ordered according to their 5′ position on a spreadsheet. STSs (sequence tagged sites) were determined through ePCR (http://www.ncbi.nlm.nih.gov/genome/sts/epcr.cgi) to identify corresponding BACs in the two databases.
Comparison of Genetic Versus Physical Map
The deCODE sex average genetic map for the 17q22–24 (bordering markers D17S956 and D17S1304) region was retrieved from http://www.nature.com/ng/journal/v31/n3/suppinfo/ng917_S1.html. For the physical map, the UCSC Human Genome Map (July 2003 freeze) was used. Corresponding genetic markers from deCODE were electronically mapped to the UCSC physical map.
SNP Position
SNPs from both the clone overlapping track and random clones in the November 2002 UCSC Human genome map were used. SNPs from both tracks that mapped in the region on Chromosome 17q22–24 were further separated into those that mapped to the duplicated region and those that do not map to the duplicated region.
Linkage Calculations
Two-point linkage analyses were performed using the MLINK program of the LINKAGE package, FASTLINK version 2.2 (Cottingham Jr. et al. 1993) and as described in more detail in Saarela et al. (2002).
Analysis of Palindromic Sequences
We compared the distribution of perfect palindromic patterns between human Chromosome 17 and mouse Chromosome 11, which has large syntenic regions with the human Chromosome 17. Locations of all perfect nonoverlapping DNA palindromes longer than 6 bp were identified from the DNA sequence of the human and mouse chromosomes using a custom pattern searching software (T. Miettinen, unpubl.). The latest NCBI mouse sequence assembly m30 and the NCBI human assembly version 31 were used to retrieve the sequences. To compare the sequence landscapes between the two species, we measured the density (mean distance between detected) of the palindromic patterns in 250-kb segments. The significance of the differences of the palindrome density in selected regions was estimated by a randomization test in which the p-value of the segment is measured from the empirical distribution of the density of same-length segments resampled from the same chromosome (Chromosome 17).
Acknowledgments
The work was supported in part by grants from NIH, RO1 NS 43559; the National MS Foundation NMSS: RG 3050-B-2; Helsinki University Central Hospital Research Funds, TYH 2313; and the Finnish National Multiple Sclerosis Society. Marlena Schoenberg Fejo and Mervi Eeva are acknowledged for technical assistance in FISH experiments. York Marahrens is acknowledged for valuable comments in preparing the manuscript. Our special thanks go to the multiple sclerosis families and to the collaborating physicians.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2340804. Article published online ahead of print in July 2004.
References
- Armengol, L., Pujana, M.A., Cheung, J., Scherer, S.W., and Estivill, X. 2003. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum. Mol. Genet. 12: 2201–2208. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Yavor, A.M., Massa, H.F., Trask, B.J., and Eichler, E.E. 2001. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 11: 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002a. Recent segmental duplications in the human genome. Science 297: 1003. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Yavor, A.M., Viggiano, L., Misceo, D., Horvath, J.E., Archidiacono, N., Schwartz, S., Rocchi, M., and Eichler, E.E. 2002b. Human-specific duplication and mosaic transcripts: the recent paralogous structure of chromosome 22. Am. J. Hum. Genet. 70: 83–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J.A., Baertsch, R., Kent, W.J., Haussler, D., and Eichler, E.E. 2004. Hotpots of mammalian chromosomal evolution. Genome Biol. (in press). [DOI] [PMC free article] [PubMed]
- Bertherat, J., Groussin, L., Sandrini, F., Matyakhina, L., Bei, T., Stergiopoulos, S., Papageorgiou, T., Bourdeau, I., Kirschner, L.S., Vincent-Dejean, C., et al. 2003. Molecular and functional analysis of PRKAR1A and its locus (17q22–24) in sporadic adrenocortical tumors: 17q losses, somatic mutations, and protein kinase A expression and activity. Cancer Res. 63: 5308–5319. [PubMed] [Google Scholar]
- Butterfield, R.J., Sudweeks, J.D., Blankenhorn, E.P., Korngold, R., Marini, J.C., Todd, J.A., Roper, R.J., and Teuscher, C. 1998. New genetic loci that control susceptibility and symptoms of experimental allergic encephalomyelitis in inbred mice. J. Immunol. 161: 1860–1867. [PubMed] [Google Scholar]
- Cheung, J., Estivill, X., Khaja, R., MacDonald, J.R., Lau, K., Tsui, L.C., and Scherer, S.W. 2003. Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol. 4: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham Jr., R.W., Idury, R.M., and Schäffer, A.A. 1993. Faster sequential genetic linkage computations. Am. J. Hum. Genet. 53: 252–263. [PMC free article] [PubMed] [Google Scholar]
- Dorr, S., Midro, A.T., Farber, C., Giannakudis, J., and Hansmann, I. 2001. Construction of a detailed physical and transcript map of the candidate region for Russell-Silver syndrome on chromosome 17q23–q24. Genomics 71: 174–181. [DOI] [PubMed] [Google Scholar]
- Dyment, D.A., Willer, C.J., Scott, B., Armstrong, H., Ligers, A., Hillert, J., Paty, D.W., Hashimoto, S., Devonshire, V., Hooge, J., et al. 2001. Genetic susceptibility to MS: A second stage analysis in Canadian MS families. Neurogenetics 3: 145–151. [DOI] [PubMed] [Google Scholar]
- Ebers, G.C., Kukay, K., Bulman, D.E., Sadovnick, A.D., Rice, G., Anderson, C., Armstrong, H., Cousin, K., Bell, R.B., Hader, W., et al. 1996. A full genome search in multiple sclerosis. Nat. Genet. 13: 472–476. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E. and Sankoff, D. 2003. Structural dynamics of eukaryotic chromosome evolution. Science 301: 793–797. [DOI] [PubMed] [Google Scholar]
- Estivill, X., Cheung, J., Pujana, M.A., Nakabayashi, K., Scherer, S.W., and Tsui, L.C. 2002. Chromosomal regions containing high-density and ambiguously mapped putative single nucleotide polymorphisms (SNPs) correlate with segmental duplications in the human genome. Hum. Mol. Genet. 11: 1987–1995. [DOI] [PubMed] [Google Scholar]
- Giglio, S., Broman, K.W., Matsumoto, N., Calvari, V., Gimelli, G., Neumann, T., Ohashi, H., Voullaire, L., Larizza, D., Giorda, R., et al. 2001. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 68: 874–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagodic, M., Kornek, B., Weissert, R., Lassmann, H., Olsson, T., and Dahlman, I. 2001. Congenic mapping confirms a locus on rat chromosome 10 conferring strong protection against myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis. Immunogenetics 53: 410–415. [DOI] [PubMed] [Google Scholar]
- Kolomietz, E., Meyn, M.S., Pandita, A., and Squire, J.A. 2002. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer 35: 97–112. [DOI] [PubMed] [Google Scholar]
- Kuokkanen, S., Gschwend, M., Rioux, J.D., Daly, M.J., Terwilliger, J.D., Tienari, P.J., Wikstrom, J., Palo, J., Stein, L.D., Hudson, T.J., et al. 1997. Genomewide scan of multiple sclerosis in Finnish multiplex families. Am. J. Hum. Genet. 61: 1379–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi, H., Shaikh, T., Takata, M., Toda, T., and Emanuel, B.S. 2003. The constitutional t(17;22): Another translocation mediated by palindromic AT-rich repeats. Am. J. Hum. Genet. 72: 733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi, T., Skaletsky, H., Brown, L.G., Minx, P.J., Cordum, H.S., Waterston, R.H., Wilson, R.K., Silber, S., Oates, R., Rozen, S., et al. 2001. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat. Genet. 29: 279–286. [DOI] [PubMed] [Google Scholar]
- Laan, M., Kallioniemi, O.P., Hellsten, E., Alitalo, K., Peltonen, L., and Palotie, A. 1995. Mechanically stretched chromosomes as targets for high-resolution FISH mapping. Genome Res. 5: 13–20. [DOI] [PubMed] [Google Scholar]
- Leach, D.R. 1994. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 16: 893–900. [DOI] [PubMed] [Google Scholar]
- Lobachev, K.S., Shor, B.M., Tran, H.T., Taylor, W., Keen, J.D., Resnick, M.A., and Gordenin, D.A. 1998. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 148: 1507–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke, D.P., Archidiacono, N., Misceo, D., Cardone, M.F., Deschamps, S., Roe, B., Rocchi, M., and Eichler, E.E. 2003. Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster. Genome Biol. 4: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monni, O., Barlund, M., Mousses, S., Kononen, J., Sauter, G., Heiskanen, M., Paavola, P., Avela, K., Chen, Y., Bittner, M.L., et al. 2001. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc. Natl. Acad. Sci. 98: 5711–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen, L., Palotie, A., and Lange, K. 2000. Use of population isolates for mapping complex traits. Nat. Rev. Genet. 1: 182–190. [DOI] [PubMed] [Google Scholar]
- Phelan, C.M., Borg, A., Cuny, M., Crichton, D.N., Baldersson, T., Andersen, T.I., Caligo, M.A., Lidereau, R., Lindblom, A., Seitz, S., et al. 1998. Consortium study on 1280 breast carcinomas: Allelic loss on chromosome 17 targets subregions associated with family history and clinical parameters. Cancer Res. 58: 1004–1012. [PubMed] [Google Scholar]
- Saarela, J., Schoenberg Fejzo, M., Chen, D., Finnila, S., Parkkonen, M., Kuokkanen, S., Sobel, E., Tienari, P.J., Sumelahti, M.L., Wikstrom, J., et al. 2002. Fine mapping of a multiple sclerosis locus to 2.5 Mb on chromosome 17q22–q24. Hum. Mol. Genet. 11: 2257–2267. [DOI] [PubMed] [Google Scholar]
- Sanchez, R., Nguyen, D., Rocha, W., White, J.H., and Mader, S. 2002. Diversity in the mechanisms of gene regulation by estrogen receptors. Bioessays 24: 244–254. [DOI] [PubMed] [Google Scholar]
- Sawcer, S., Jones, H.B., Feakes, R., Gray, J., Smaldon, N., Chataway, J., Robertson, N., Clayton, D., Goodfellow, P.N., and Compston, A. 1996. A genome screen in multiple sclerosis reveals susceptibility loci on chromosome 6p21 and 17q22. Nat. Genet. 13: 464–468. [DOI] [PubMed] [Google Scholar]
- Selker, E. 1999. Gene silencing: Repeats that count. Cell 97: 157–160. [DOI] [PubMed] [Google Scholar]
- Stankiewicz, P. and Lupski, J.R. 2002. Molecular-evolutionary mechanisms for genomic disorders. Curr. Opin. Genet. Dev. 12: 312–319. [DOI] [PubMed] [Google Scholar]
- Subramanian, G., Adams, M.D., Venter, J.C., and Broder, S. 2001. Implications of the human genome for understanding human biology and medicine. JAMA 286: 2296–2307. [DOI] [PubMed] [Google Scholar]
- Sumelahti, M.L., Tienari, P.J., Wikstrom, J., Palo, J., and Hakama, M. 2000. Regional and temporal variation in the incidence of multiple sclerosis in Finland 1979–1993. Neuroepidemiology 19: 67–75. [DOI] [PubMed] [Google Scholar]
- Sumelahti, M.L., Tienari, P.J., Hakama, M., and Wikstrom, J. 2003. Multiple sclerosis in Finland: Incidence trends and differences in relapsing remitting and primary progressive disease courses. J. Neurol. Neurosurg. Psychiatry 74: 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tienari, P.J., Wikstrom, J., Sajantila, A., Palo, J., and Peltonen, L. 1992. Genetic susceptibility to multiple sclerosis linked to myelin basic protein gene. Lancet 340: 987–991. [DOI] [PubMed] [Google Scholar]
- Tienari, P.J., Kuokkanen, S., Pastinen, T., Wikstrom, J., Sajantila, A., Sandberg-Wollheim, M., Palo, J., and Peltonen, L. 1998. Golli-MBP gene in multiple sclerosis susceptibility. J. Neuroimmunol. 81: 158–167. [DOI] [PubMed] [Google Scholar]
- Valero, M.C., de Luis, O., Cruces, J., and Perez Jurado, L.A. 2000. Fine-scale comparative mapping of the human 7q11.23 region and the orthologous region on mouse chromosome 5G: The low-copy repeats that flank the Williams-Beuren syndrome deletion arose at breakpoint sites of an evolutionary inversion(s). Genomics 69: 1–13. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu; UCSC Genome Browser.
- http://www.broad.mit.edu/cgi-bin/primer/primer3.cgi; Whitehead/MIT Genome Center Primer 3.
- http://www.celera.com; Celera Genomic.
- http://www.nature.com/ng/journal/v31/n3/suppinfo/ng917_S1.html; Nature.
- http://www.ncbi.nlm.nih.gov/genome/sts/epcr.cgi; NCBI.
- http://wwwshgc.stanford.edu/Mapping/rh/procedure/rhassaynew.html; Stanford.
- http://www-shgc.stanford.edu/Mapping/rh/RH_poster/; Stanford Radiation Hybrid Web site.