Abstract
The major histocompatibility complex (MHC) is comprised of the class I, class II, and class III regions, including the MHC class I and class II genes that play a primary role in the immune response and serve as an important model in studies of primate evolution. Although nonhuman primates contribute significantly to comparative human studies, relatively little is known about the genetic diversity and genomics underlying nonhuman primate immunity. To address this issue, we sequenced a complete rhesus macaque MHC spanning over 5.3 Mb, and obtained an additional 2.3 Mb from a second haplotype, including class II and portions of class I and class III. A major expansion of from six class I genes in humans to as many as 22 active MHC class I genes in rhesus and levels of sequence divergence some 10-fold higher than a similar human comparison were found, averaging from 2% to 6% throughout extended portions of class I and class II. These data pose new interpretations of the evolutionary constraints operating between MHC diversity and T-cell selection by contrasting with models predicting an optimal number of antigen presenting genes. For the clinical model, these data and derivative genetic tools can be implemented in ongoing genetic and disease studies that involve the rhesus macaque.
Genetic and evolutionary studies of the immune response have often centered on the major histocompatibilty complex (MHC), comprised of the class I, class II, and class III regions, and including the MHC class I and class II genes involved in immune recognition (Parham 1999). Specifically, with the MHC class I and class II genes as a focus, comparative genetic analyses among primates have contributed significantly to both basic and clinical human studies (Bontrop 2001). An important example is the rhesus macaque, which is not only often a component of primate evolutionary comparisons, but is also one of the most widely used animal models for the study of infectious diseases, organ transplantation, and in the development of new vaccines. Altogether, relative to human, far less is known about the genetic diversity and genomics of nonhuman primate immunity.
Among the first immune loci to be carefully studied in the rhesus macaque (macaca mulatta or mamu) were the class I and class II loci (Watkins 1994; Watkins et al. 1988), respecting their fundamental roles in antigen presentation and immune recognition (Townsend and Bodmer 1989; Accolla et al. 1995; Colonna 1996). Molecular studies of class I have defined expressed orthologs to the HLA-A and HLA-B loci (Miller et al. 1991; Boyson et al. 1996), including evidence that both the mamu-A and mamu-B loci have been duplicated with as many as three B and two A loci detected in certain haplotypes (Boyson et al. 1996; Urvater et al. 2000). Homologs of the human nonclassical class I antigens HLA-E and HLA-F were described (Otting and Bontrop 1993; Knapp et al. 1998) and a functional homolog of HLA-G termed mamu-AG was characterized (Otting and Bontrop 1993; Boyson et al. 1995; Ryan et al. 2002). Humans and rhesus share most MHC class II loci including -DR, -DQ, and -DP, and an increase in the number of DRB genes in some rhesus haplotypes has been suggested (Slierendregt et al. 1994). The precise number of actively expressed class I and class II genes in an individual experimental macaque has remained largely uncertain (Mothe et al. 2002).
Beyond the preliminary characterization of class I and class II cDNAs and genomic clones, little information is available about the larger-scale genetic structure of the macaque MHC and about the diversity of MHC genes other than class I and class II. Because more than a third of MHC resident genes may be involved in the immune response (MHC Consortium 1999), such knowledge could be fundamental to basic and comparative evolutionary and clinical studies, as genomic proximity and polymorphism of other MHC resident loci can have functional consequences as well (Momburg et al. 1996).
These considerations provided motivation to better understand the genetic diversity and genomics of the immune response in rhesus macaques both to aid clinical studies, as well as to advance our understanding of the evolutionary history of primates through comparative genomic analysis. Toward these ends, we have obtained finished high-quality sequence data spanning the complete mamu MHC. Among the differences between rhesus and human observed, were a major expansion from six class I genes per haplotype in humans to as many as 22 potentially active MHC class I genes on a single haplotype in rhesus. In addition, high levels of genetic polymorphism were found over most of the region, including within many other MHC resident genes.
RESULTS
Isolation and Sequencing of the Rhesus MHC
To isolate the rhesus MHC region, we utilized PCR amplicons derived from the finished human MHC sequence (MHC Consortium 1999) to generate macaque probes for screening the BAC resource (Geraghty et al. 2002). Using this strategy, it was possible to map clones for sequencing and to assign each BAC to one of the two haplotypes represented in the library. Subsequent clone isolations required the use of sequence data from the ends of BACs bordering gaps to generate new PCR probes for additional library screening. A contiguous sequence assembly derived from 54 BACs and five short gaps spanning the mamu MHC is presented in Figure 1. The sequence extends from the telomeric end near mamu-F to the centromeric end following mamu-DP over a distance of 5,291,973 bp. The telomeric class I sequence extending into the approximate middle of the mamu-B region, is derived from a single haplotype excepting ∼29 kb derived from BAC 337E2. The sequence including the entire B region extending to the end of mamu class II was obtained from a second haplotype without gaps. An additional 2,131,545 bp have been derived from overlapping sequence from the alternate chromosome. All of the sequences were high-quality finished data, essential to distinguish functional genes from nonfunctional genes, especially in the mamu-A and mamu-B regions, where single nucleotide substitutions were responsible for inactivating some of the class I genes.
The Rhesus MHC Compared With the Human
The first striking observation apparent between the rhesus and human was the relatively large increase in size of the rhesus MHC. Where the human sequence extends over ∼3.7 Mb, the rhesus spans about 5.3 Mb, given the same start and end positions. Virtually all of this size difference is the result of significant internal expansions within the mamu class I A and class I B gene regions (Fig. 1). In other regions, the overall gene content of rhesus largely parallels that of human. In total, 10 genes or pseudogenes are distinct between human and rhesus, four found in human, but not rhesus, and six in rhesus, but not human, and an additional eight that are common to both MHCs, but which are altered relative to one another by changes in the protein-coding start or stop positions (color coded in Fig. 1). The remainder of the gene content of some 140 genes appears unaltered, with expected coding-sequence divergence. All comparisons with the human MHC are largely paralleled with any of the chimpanzee, as a recent analysis confirmed the existence of a high degree of sequence similarity between the human and chimpanzee class I regions (Anzai et al. 2003).
The Mamu-A and Mamu-B Regions
The mamu-A region has diverged from human in two respects, both through an expansion of the number of class I genes and through the alteration in the class I gene and pseudogene content of the region. The telomeric portion of the mamu-A region has apparently arisen as a result of a tandem duplication of copies of an ∼100-kb segment in the haplotype depicted in Figure 2. The first two of these segments contain remarkable similarity in having only eight nucleotide substitutions over a span of 80 kb. Immediately downstream, two additional tandem copies of this segment occur, each including one additional partial class I gene fragment. Whereas all four copies of the mamu-G genes contain inactivating alterations, previous work has suggested that mamu-AG is a functional homolog, acting as a replacement for mamu-G (Ryan et al. 2002). All four mamu-AG genes have functional gene structures, indicating that they may all be active in the placental environment. Two mamu-A genes with complete identity to mamu-A cDNA sequences are located centromeric in a region that appears to be part of an older duplication event (Fig. 2B,C). Of the full-length class I pseudogenes, it was possible to identify homologs to the human pseudogenes HLA-59 (HLA-J), HLA-70 (HLA-L), and HLA-92, but not HLA-54 (HLA-H; Geraghty et al. 1992a,b). This may be relevant when considering the origin of the rhesus monkey A locus alleles, which are more closely related to HLA-H than to HLA-A (Adams and Parham 2001). Overall, the similarities between human and rhesus in this region include similar functional genes, whereas the differences include an increase in the number of genes and pseudogenes (Table 1).
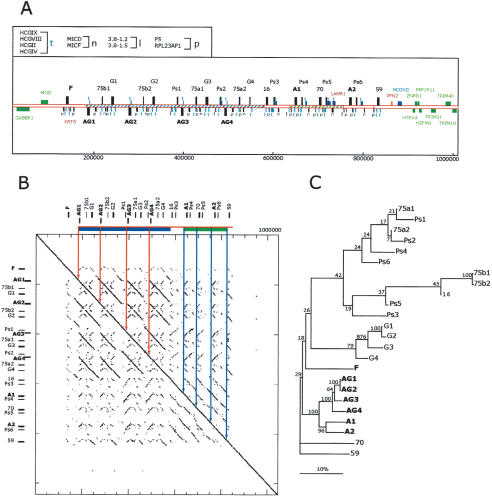
Figure 2.
Description of the expanded, relative to human, mamu-A class I gene region. (A) The 1.1 mb segment extending from GABBR1 to TRIM10 from mamu haplotype 2 is depicted. The positions of class I genes and pseudogenes are indicated by alphanumerical descriptions and bars with direction of transcription indicated by positioning above and below the line according to convention (Table 1 contains a summary description). The positions of homologs to non-class I human genes and pseudogenes are indicated with bars and symbols that are explained in the top, left box. Flanking genes outside of this duplicated area and homologous to those found in the human MHC with conserved structures are indicated by green bars and text. Sequences not present in the corresponding human class I region are indicated by red bars and text. Other non-class I genes are colored in blue. Segments of the sequence with significant similarity indicative of duplication events are indicated with similar hatching bounded by solid lines. The scale below is in base pairs. (B) Dot matrix analysis of the mamu-A region against itself to provide a visualization of the repetitive nature of this segment. The dot plot was generated by a custom python script that divides the query sequence into 100-kb fragments and compares them using cross_match (Ewing and Green 1998; Ewing et al. 1998) against the target sequence. The minmatch score was set at 30 and masklevel was set at 101. All other cross_match parameters were set to default values. The start and end positions for each cross_match hits were plotted using the Biggles python module (biggles.sourceforge.net). Tick marks on the vertical indicate the scale in 200-kb segments and on the horizontal in 50-kb segments. The locations of the class I genes are indicated in relative positions at top and to the left.(C) Phylogenetic analysis of the mamu-A region genes and pseudogenes. Included in this analysis are all mamu class I-like genes contained in this segment (Table 1). Conceptual translations were produced by FASTY (Pearson et al. 1997) comparison of the genomic sequence of each gene or pseudogene with the human HLA-A24 gene (SwissProt 1A24_HUMAN), correcting frameshifts and stop codons of pseudogenes and partial genes. The reconstructed amino acid sequences were aligned using ClustalW with default parameters. Phylogenetic reconstruction was generated using the Neighbor-Joining algorithm (Saitou and Nei 1987), and tested using 1000 rounds of bootstrap analysis. The tree is unrooted. The horizontal bar indicates 10% divergence along each branch.
Table 1.
Mamu-A Region Class I Genes and Related Sequences
| A gene homologuea | BAC location | Gene structure | Human equivalent |
|---|---|---|---|
| MamuF | 268P23 | functional | HLA-F |
| mamuAG1 | 151L13 | functional | HLA-Ab |
| mamu75b1 | 151L13 | partial gene | HLA-75 |
| mamuG1 | 151L13 | pseudogene | HLA-G |
| mamuAG2 | 151L13, 227F07 | functional | HLA-Ab |
| mamu75b2 | 227F07 | partial gene | HLA-75 |
| mamuG2 | 227F07 | pseduogene | HLA-G |
| mamuPs1 | 227F07 | partial gene | HLA-80/90 |
| mamuAG3 | 227F07, 139P19 | functional AG0211c | HLA-Ab |
| mamu75a1 | 227F07, 139P19 | partial gene | HLA-75 |
| mamuG3 | 139P19 | pseduogene | HLA-G |
| mamuPs2 | 179H14, 139P19 | partial gene | HLA-80/90 |
| mamuAG4 | 179H14, 139P19 | functional | HLA-Ab |
| mamu75a2 | 179H14, 139P19 | partial gene | HLA-75 |
| mamuG4 | 179H14, 139P19 | partial gene | HLA-G |
| mamu16 | 179H14 | pseudogene | HLA-16 |
| mamuPs3 | 179H14 | partial gene | HLA-80/90 |
| mamuA1 | 63G23 | functional, A1401c | HLA-A |
| mamuPs4 | 63G23 | partial gene | HLA-80/90 |
| mamu70 | 63G23 | pseudogene | HLA-70 |
| mamuPs5 | 63G23 | partial gene | HLA-80/90 |
| mamuA2 | 63G23 | functional, A0402c | HLA-A |
| mamuPs6 | 63G23 | partial gene | HLA-80/90 |
| mamu59 | 67K14, 25A04 | pseudogene | HLA-59 |
| mamuX1 | 25A04 | gene fragment | |
| mamu92 | 25A04 | pseudogene | HLA-92 |
| mamuE | 25A04 | functional | HLA-E |
Pseudogene designations are given numerical reference as in (Geraghty 1993; Geraghty et al. 1992a,b).
Mamu-AG has sequence similarity to HLA-A but functional similarity to HLA-G (Boyson et al. 1997; Ryan et al. 2002).
Mamu-A genes are identical to indicated mamu cDNAs.
Segmental duplication has also occurred in the mamu-B region in one of the most remarkable findings of this work. In humans, the two HLA-B and HLA-C genes lie within a 100-kb segment, whereas the largely equivalent region containing the mamu-B genes has swelled to about 1.3 Mb. This increase has resulted from a complex series of segmental duplications and a single transposition of some 43 kb of sequences from the mamu-A region (including mamu-75a3). In the complete haplotype 1 sequence presented in Figure 3, 19 mamu-B-like genes were found arrayed in a tandem arrangement. Similar to the mamu-A region, the mamu-B region can be grossly subdivided into two segments. The telomeric portion extending from mamu-B1, interrupted by the mamu-A transposed segment, and continuing to mamu-B10 has arisen in part from six tandem duplications of a segment some 60 kb in length, whereas the centromeric portion from mamu-B11 to mamu-B19 has arisen in part from a single larger tandem duplication of about 150 kb (Fig. 3B). Such a historical relationship between mamu-B1 through mamu-B10 and the mamu-B11 to mamu-B19 genes is reflected in the evolutionary distances among them (Fig. 3C).
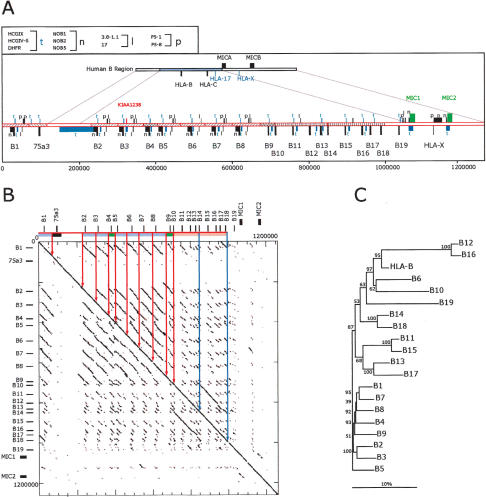
Figure 3.
Analysis of a complete mamu class I B region haplotype. (A) Comparison of human and mamu-B region segments. Top cartoon depicts the homologous human segment extending to scale over the HLA-B and HLA-C region, including the surrounding class I loci as indicated. Immediately beneath is depicted the mamu-B region extending 1.3 Mb and including 19 mamu-B-like genes (see Table 2 for gene summary). Segments of similarity between rhesus and human are bounded by lines connecting the two cartoons. The positions of homologs to human genes and pseudogenes are indicated with bars and symbols that are explained in the top, left box. Orthologous genes with conserved functional structure are indicated by green bars and text. Segments with significant similarity indicative of past duplication events are indicated with similar hatching bounded by solid lines. The placement of bars above or below the line indicates direction of transcription according to convention. The scale below is in base pairs. (B) Dot matrix analysis of the mamu-B region haplotype 2 against itself. The dot plot was generated by a custom python script that divides the query sequence into 100-kb fragments and compares them using cross_match (Ewing and Green 1998; Ewing et al. 1998) against the target sequence. The minmatch score was set at 30 and the masklevel was set at 101. All other cross_match parameters were set to default values. The start and end positions for each cross_match hit were plotted using the Biggles python module. Tick marks indicate the scale in 200-kb segments on the vertical and 50 kb on the horizontal. The locations of the class I genes are indicated in relative positions at top and to the left. Similarity among segments is indicated by similar coloring within the bar on the top and further bounded by vertical lines within the box. (C) Phylogenetic analysis of all mamu-B-like genes in the region based on 5-kb of genomic sequence surrounding each gene. Included in this analysis are all of those genes from haplotype 1 listed in Table 2. A multiple alignment of the repeat-masked genomic sequences was performed using ClustalW with default parameters. Phylogenetic reconstruction was generated using the Neighbor-Joining algorithm (Saitou and Nei 1987) and tested using 1000 rounds of bootstrap analysis. The tree is unrooted. The horizontal bar indicates 10% divergence along each branch.
Although this finding is fascinating evolutionarily, another important issue raised is whether the expansion in gene number could significantly alter the progress or development of an acquired or innate immune response of the rhesus macaque relative to human. To answer this first question requires knowledge of which of these genes are transcribed and translated into functional class I products. Of the 19 B-like genes on haplotype 1, seven have identical matches to cDNAs (derived from macaque peripheral blood), and seven have typical class I protein-encoding capacity, including apparently functional exons and highly similar promoter regions. Of the remaining five genes, four have stop codons within exons 2–4, presumably inactivating the gene, and a single gene encodes a protein without a leader peptide (Table 2).
Table 2.
Mamu-B Genes
| B Locus | BAC location | Gene structure |
|---|---|---|
| MamuB01 | 244P03 | BJa |
| Mamu75a3 | 391L07, 244P03 | gene fragment |
| MamuB02 | 274L13 | B30-likeb |
| MamuB03 | 274L13 | B30-likeb |
| MamuB04 | 274L13, 429H11 | Stop Exon 3b |
| MamuB05 | 429H11 | B30 |
| MamuB06 | 429H11, 192G09 | B57b |
| MamuB07 | 192G09 | B56-likea |
| MamuB08 | 192G09 | B12b |
| MamuB09 | 23M02 | B3802b |
| MamuB10 | 23M02 | Stop Exon 3a |
| MamuB11 | 23M02 | Stop Exon 2a |
| MamuB12 | 23M02, 297I07 | B01-likec |
| MamuB13 | 297I07, 145B12 | B56 |
| MamuB14 | 297I07, 145B12 | Stop Exon 4a |
| MamuB15 | 297I07, 145B12 | B56-likea |
| MamuB16 | 145B12, 134C21 | B01-likec |
| MamuB17 | 145B12, 134C21 | B56-likea |
| MamuB18 | 145B12, 134C21, 24N14 | B4602a |
| MamuB19 | 134C21, 24N14 | Cytoplasmic (?), no signal sequence |
| Mamu-h2-B02 | 326H24 | B30-likeb |
| Mamu-h2-B03 | 326H24, 457F04 | B30-likeb |
| Mamu-h2-B04 | 457F04, 125A15 | B30-likeb |
| Mamu-h2-B05 | 125A15 | B26/B57b |
| Mamu-h2-B06 | 487N12 | B56-likea |
| Mamu-h2-B07 | 487N12 | B30b |
| Mamu-h2-B17 | 188J04 | B56-likea |
| Mamu-h2-B18 | 188J04 | B60a |
| Mamu-h2-B19 | 188J04 | Cyto, no signal sequence, several stops |
Designates cDNA contains an ATG start codon within the sequence CGAGATGCGG-TCATGGCGGC, similar to that found in mamuB56 cDNA (among others), requiring that the second ATG be used as the start codon to yield a functional protein.
Designates cDNA encoding a more typical class I sequence CGA GATGCGGGTCATGGCGCT using the first ATG start. Similar to mamu B30.
Designates cDNA contains only the second (21 amino acid leader peptide) ATG start codon. Similar to mamu B01. Bold type indicates identical match with indicated cDNA sequence.
One unique and relevant feature of mamu-B genes may be the structure of exon 1 and the placement of the ATG start codon. There are three arrangements found; the first is typical of MHC class I in having two ATG start codons with the downstream ATG following in frame at amino acid position 4 (ATGC GGGTCATG). A second arrangement contains only the downstream ATG yielding a signal peptide with 21 amino acids. A third coding sequence has the first ATG encountered out of frame with the remainder of the protein-coding sequence (AT GCGG-TCATG, typified by mamu-B*17), suggesting that the second ATG is used as the start codon (Table 2). All three types are transcribed as examples of representative cDNAs that have been found for each. In addition, peptide-binding motifs and SIV cytotoxic T-lymphocyte epitopes have been deciphered for each type (Evans et al. 2000; Mothe et al. 2002), suggesting that they may all function as antigen-presenting molecules. Thus, there may be as many as 28 functional mamu-B antigen-presenting molecules expressed in an individual macaque.
Our initial attempts to unravel the mamu-B region encountered difficulty because it was not possible to assign a BAC to one haplotype unambiguously, due to the underlying complex repetitive structure of the mamu-B sequences. Further, the complex arrangement of genes heightened interest in a comparison of the sequence from both haplotypes. Therefore, we combined the sequence from haplotype 1 with data from over half of haplotype 2, the latter including nine additional mamu-B genes. Of these, three had identical matches to cDNA sequences and five genes were structurally intact with a typical class I coding capacity (Table 2). One gene appeared to be an allele of mamu-B19, but contained several in frame stop codons not found in the allelic sequence. It was possible to align the two haplotypes at the 5′ and 3′ ends of the regions using the single occurrence of the HCGIX sequence at the 5′ end and MIC1 at the 3′ end. Missing at the 5′ end of haplotype 2 was the transposed mamu-A region sequences and mamu-B1 (Fig. 4). However, despite having anchors at the ends, it was apparent that any notion of alleles between mamu-B genes at corresponding positions between the two sequences was ambiguous. Thus, mamu-h2B5 was more closely related to mamu-B6, whereas mamu-h2B6 and mamu-h2B7 aligned most significantly with mamu-B3 and mamu-B4, respectively. Further, mamu-h2B17 and mamu-h2B18 both appeared to be allelic to mamu-B18 and only distantly related to mamu-B17 (Fig. 4B,C).
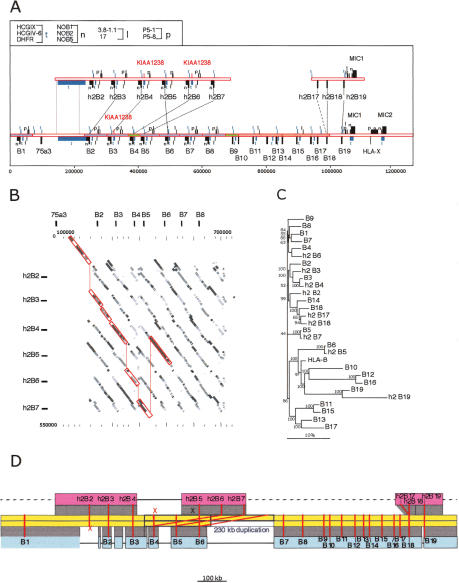
Figure 4.
Comparison of two mamu-B haplotype sequences. (A) Two segments from haplotype 2 spanning 564 and 181 kb from the telomeric and centromeric ends of the region, respectively, are aligned above the corresponding complete haplotype 1 segment. Genes and other sequences are indicated as described in the legend to Figure 3. Lines between the top and bottom mamu-B genes indicate the most closely related sequences between the two haplotypes (alleles). (B) Dot matrix similarity between the segments from haplotype 2 containing h2-B2 though h2-B7 and the B2 though B8 segment from haplotype 1. Shading of dots indicates percent identity in equal integer gradations from 5% gray (90% identity) to black (100% identity). Red lines enclose and connect the matrix at positions that have the highest percent similarity between loci from the respective haplotypes. The comparisons were done using cross_match as described in the legend to Figure 2. Tick marks indicate the scale in 100-kb segments on the vertical and 20 kb on the horizontal. The locations of the class I genes from haplotype 2 are indicated in relative positions at top and to the left. (C) Phylogenetic analysis of all mamu-B-like genes, based on 5 kb of genomic sequence surrounding each gene. Included in this analysis are all B genes listed in Table 2. A multiple alignment of the repeat-masked genomic sequences was performed using ClustalW with default parameters. Phylogenetic reconstruction was generated using the Neighbor-Joining algorithm (Saitou and Nei 1987), and tested using 1000 rounds of bootstrap analysis. The tree is unrooted. The horizontal bar indicates 10% divergence along each branch. (D) Reconstruction of the evolution of haplotype 1 (blue) and haplotype 2 (red) from the ancestral sequence (yellow), by way of a 230-kb duplication (between the blue boxes), followed by deletions on both sides (black horizontal lines). Dashed gray lines indicate yet unavailable sequence. Red lines connect alleles of mamu-B genes; (x) deleted alleles.
To begin to unravel the complex relationships between the two mamu-B haplotypes, we took advantage of the presence within the repeat units of large numbers of retroposed elements (Alus, LINEs, etc.), which produced unique repeat patterns. The simplest reconstruction of the events that might have occurred to generate these divergent haplotype structures is depicted in Figure 4D. The pivotal event in this reconstruction is the duplication of 230 kb of ancestral sequence, including genes B4, B5, and B6. Haplotype 1 evolved from the ancestral, unduplicated sequence by a series of deletions, one of which erased the allele of h2B2. Haplotype 2 evolved, after the 230-kb duplication, by deleting the alleles of B4 and B5, and by a further duplication that generated h2B17 and h2B18. Thirteen additional mamu-B genes are predicted to be located in the yet-unsequenced regions of this haplotype, but some may have also been lost by deletions.
Mamu-DR, Mamu-DQ, and Mamu-DP Loci
The DRB gene complex in humans consists of from one to three active DRB genes on different haplotypes. In rhesus, this region is organized similarly. Haplotype 2 contains two genes that appear structurally intact and match cDNA sequences consistent with functionality. The haplotype 1 sequence also contains two potentially functional genes. In contrast to the human locus, additional pseudogenes exist on both rhesus chromosomes. In humans, from one to five copies of DRB appear on different haplotypes, with two of these nonfunctional. On the two rhesus chromosomes, there are as many as seven pseudogenes or related fragments on haplotype 2 and haplotype 4 on haplotype 1. Because there is a gap between BACs covering this chromosome, the total number of DRB genes on this haplotype may be larger.
The first obvious difference between human and rhesus DQ genes is that both monkey chromosomes have only one copy of the DQA and DQB genes, whereas the human locus has two DQA loci and two copies of DQB and one DQB pseudogene (MHC Consortium 1999). Further, the arrangement of the DQA and DQB genes in the rhesus provides one of the more interesting structural variations in the class II region. On haplotype 2, represented by BAC 7H18, these genes are arranged in opposite transcriptional orientations, consistent with that found in the human MHC (MHC Consortium 1999). However, on haplotype 1, represented by BAC 281E18, an inversion of the region containing the DQB gene has taken place to yield an arrangement in which DQB is in the same transcriptional orientation as DQA. In total, 15.4 kb of clone 218E18 has been inverted relative to clone 7H18, and of this, 13.3 kb of the sequence is shared, with only minor substitution and indel variations between the sequences. Outside of the shared sequence, but within the inversion, the two allelic regions differ mostly in their interspersed repeat content. At both ends of the shared sequence in the inversion is found an Alu Y element. Both structural arrangements have been confirmed by analysis of genomic DNA and additional overlapping BACs (data not shown), removing the possibility that the rearrangement is an artifact of BAC cloning.
Sequence Divergence Spanning the Mamu MHC
Our preliminary analysis of amplicon sequences derived from mamu-B genes had indicated that the mamu MHC overall had significantly more sequence divergence than found in human (Geraghty et al. 2002). To analyze this more completely and to establish a detailed structural comparison, we obtained contiguous sequence from one mamu class II haplotype and nearly contiguous data from the second (Fig. 1). In addition to this and the mamu-B region haplotypes discussed above, we sequenced BACs from both haplotypes extending over two regions in class I and over 200 kb in class III. Further, from our initial library screening, we sampled sequence data from 300 amplicons spread over the entire MHC from three animals (data not shown). In general, the pattern of sequence divergence was both surprising in its magnitude and in its extent. In contrast to high levels of localized divergence in humans (Guillaudeux et al. 1998; Horton et al. 1998), divergence in the rhesus class I region remains at 2%–6% over most of the length of the sequence obtained.
We compared 470 kb of contiguous sequences from the mamu class II region from each of the two haplotypes (Fig. 5) to highlight sequence divergence in class II between mamu chromosomes and human. For comparison with the orthologous region in human, we used contiguous sequence data from two human chromosomes sequenced by the Sanger Institute MHC haplotype project (http://www.sanger.ac.uk/HGP/Chr6/MHC/). Sequences were aligned as described in the Methods section, and the numbers of insertions, deletions, and substitutions were plotted versus position for the rhesus versus rhesus, rhesus versus human, and human versus human comparisons (Fig. 5). From this simple comparison, it was evident that the rhesus haplotypes have diverged significantly more than in the human. In immune-related genes other than MHC class I or class II, the divergence found was about 10-fold higher than human (e.g., the transporter associated with antigen processing subunits, TAP1 and TAP2, and DMB) and near class II genes, the divergence between rhesus chromosomes was as high or higher than that found between the orthologous rhesus and human chromosomes (e.g., near DPA1). Some regions with high %GC and containing closely packed genes correlated with lower levels of divergence, implying conservation of coding sequences. This extent of divergence constitutes a second major distinguishing feature of the mamu MHC that may relate directly to the immune response. Of the ∼60 genes in the MHC with immune-related function, >40 examined appear to be more highly polymorphic than their human counterparts.
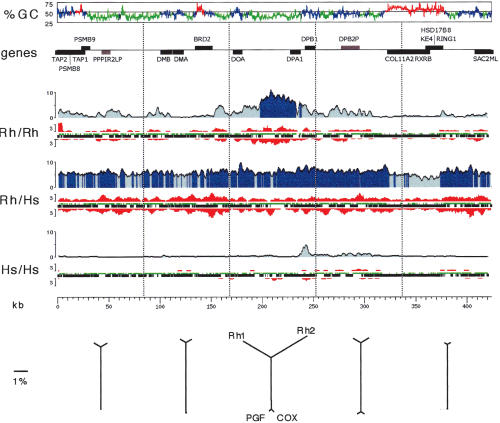
Figure 5.
Distribution of mutational events over a 470-kb segment of the MHC class II region. Pairwise comparisons of the two rhesus haplotypes, the two human haplotypes, and one haplotype of each species is indicated immediately to the left of each graph. The graphs reflect the analysis of the aligned sequences only, excluding insertions or deletions. (%GC) G+C content graph in 500-bp windows. Green, blue, and red indicate isochore regions of low, medium, and high G+C content, respectively (window size is 30 kb long, cutoffs are 43% and 50% G+C). (Genes) Location, strand, and identity of genes (black) and pseudogenes (gray). (Rh/Rh) Comparison of the two rhesus haplotypes. The percent divergence is shown in blue (averaged over 2.5-kb windows), with dark blue indicating regions over 5% divergence; the number of insertion/deletion events (at least 3 bp long, averaged over 4.5-kb windows) is shown in red, with upward peaks indicating insertions in Rh1 relative to Rh2, and downward peaks indicating insertions in Rh2 relative to Rh1; aligned regions deriving from interspersed repeats or from unique sequences are shown as green or black horizontal bars, respectively. (Rh/Hs) Comparison of Rh1 with the human COX haplotype. (Hs/Hs) Comparison between the two human haplotypes, COX and PGF (http://www.sanger.ac.uk/HGP/Chr6/MHC/). (kb) Kilobase scale. (Bottom) Distance relationships between the four sequences studied. A multiple alignment computed by the MAP program (see Methods) of the two human and the two rhesus haplotypes was split into five regions of equal lengths (approximate boundaries indicated by vertical dotted lines immediately above). Neighbor-joining trees were reconstructed on the basis of the calculated distances, excluding all gaps and correcting for multiple substitutions.
DISCUSSION
Immunological Divergence Between Rhesus and Human
It is reasonable to suppose that selective pressures have been responsible for the enormous expansion of the number of mamu-B genes in the rhesus. The ability of the rhesus MHC to expand is illustrated not only by an increase in the number of mamu-A and mamu-B-loci, but also by the transposition of sequences, such as the mamu-A segment transposed into the B region in haplotype 1 and by the inversion of the DQB gene on haplotype 2. These structures reflect events that are ongoing in the rhesus population, as they are haplotype specific and may reflect evolutionary mechanisms that have not been observed in human populations to date.
Among a plethora of distinguishing details, our studies have revealed two fundamental differences in MHC structure between rhesus macaques and humans that relate directly to the immune response. The expansion in the number of class I-B, and to a lesser extent, the class I-A and class II-DRB genes may be an important issue when attempting to dissect a T cell-mediated immune response, as it may not be clear how many of these loci are involved in a specific response. The high polymorphism found throughout the mamu MHC introduces a second major consideration. Polymorphisms in MHC loci with immune-related function, other than the class I and class II genes, can have a direct effect on antigen presentation (e.g., TAP, or the proteasome subunits, LMP-2 and LMP-7), on cytokine levels and efficacy, and on interactions with other components of both the innate and acquired immune response.
An essential step in resolving some of these issues is clearly to determine which of the mamu-B genes are functional. Of the 14 intact genes in haplotype 1, all are predicted to produce functional proteins on the basis of cDNA and transfection analysis (Evans et al. 2000; Mothe et al. 2002). For each of the three types of cDNAs, divided by ATG start codon position (Table 2), at least one representative has been shown to be transcribed, translated, expressed on the cell surface, bind peptide, and evoke a CTL response. Considering that as many as 28 mamu-B loci and four mamu-A loci may be expressed in a heterozygous animal raises questions not only of which class I loci are most important to a particular CTL response, but also concerning the interaction of class I with the innate immune response (Colonna 1996). An essential component of the latter response is the interaction between the killer cell Ig-like receptors (KIR) and their MHC class I ligands. In humans, specific class I antigens interact with specific KIR proteins yield both inhibitory and activatory responses from NK cells. However, the expansion of class I in the macaque is apparently not concomitant with an expansion of the number of KIR gene products (Hershberger et al. 2001; LaBonte et al. 2001). This raises the question of whether an increase in the number of mamu-B proteins has affected the associated inhibitory or activatory network. Another consequence of increased numbers of class I antigens could be to alter the interaction of the CD94/NKG2 receptor and its ligand, HLA-E. The number and abundance of different nonamer peptides derived from class I signal sequences and made available for HLA-E binding is a consideration toward HLA-E function, as the availability of nonamer peptide in a cell and HLA-E surface levels are correlated (Lee et al. 1998a,b). Further, relative surface levels of HLA-E may affect the functional interaction with CD94/NKG2 ligand (Lee et al. 1998b). In that regard, the alteration in the ATG start codon common to many mamu-B genes might be of interest, as the use of the downstream codon inhibits the production of a suitable HLA-E binding peptide (Lee et al. 1998a).
TCR Diversity and MHC Polymorphism
In all of the mammalian species studied thus far, the number of class I MHC genes per haplotype functioning in antigen presentation to TCR has been found to be limited to from one to three. The limit for the number of functional MHC loci is thought to be accounted for in relation to the T-cell repertoire restricted by MHC (Takahata 1995; Celada and Seiden 1996). Whereas diversity at MHC is advantageous in allowing for a diversity of pathogens that can be detected by TCR, major expansion of MHC diversity appears to act at the population level rather than the individual. Within an individual, two opposing forces of the MHC and TCR interaction consist of negative selection to eliminate potentially autoreactive T cells and positive selection to provide T cells capable of recognizing pathogens. Negative selection in humans and mice may eliminate as much as 95% of the TCR repertoire (Klein et al. 1990; Nossal 1994); thus, the available T-cell complexity is thought to be subject to the number of MHC loci interacting with TCR. In support of this, studies of tetraploid Xenopus demonstrated that the number of extra MHC loci that arose by duplicating the original diploid complement were rapidly suppressed or deleted (Flajnik 1996; Flajnik et al. 1999). In addition, models that calculate the survival probability versus the number of functional heterozygous loci (both class I and class II) suggest that the optimum number is about six, and when the number exceeds 12, the survival probability becomes negative (Takahata 1995). The simple explanation for this is that too many MHC loci remove too much of the TCR repertoire during negative selection, thus leaving a large hole in the TCR complement available to react with pathogen. This disadvantage is thought to strongly counterbalance the advantage of having many loci able to bind a larger diversity of foreign peptides.
It is clear that the rhesus has found a way around this limitation, especially in the case of the class I loci. The number of functional class I loci, as assessed by the existence of identical cDNA sequences, is at least nine per haplotype (two A loci and seven B loci), and may be as many as 16, judging by the number of loci that appear structurally intact (two A loci and 14 B loci). Whereas the functionality of some of these genes remains to be established, all appear intact and functional as judged by the prior demonstration of structurally similar B-like loci that bind peptide and present antigen to cytotoxic T cells (Dzuris et al. 2000; Evans et al. 2000). This supports an active role for a number of class I loci far in excess of that seen in other animals, and as noted above, predicted to be strongly deleterious. However, as is perhaps the only invariant rule in biology, all theoretical rules (except for this one) have exceptions, and theory must again yield to empirical evidence. New explanations need to be brought forward to explain this striking discrepancy, and further accountings of the expansion of class I loci as a consequence of selection unique to these nonhuman primates are now in order. It is possible that an increased MHC repertoire in an individual might be counterbalanced by a corresponding increase in the TCR repertoire, thus taking some advantage of an increased ability to present antigen without a corresponding loss in remaining TCR complexity.
Thus, the rhesus may provide a remarkable example of a unique reaction to pathogens along the divergent evolutionary road that separates these animals from humans. Perhaps this quantitative expansion in antigen-presenting capacity had consequences that enabled rhesus to modify its immune response by expanding the ability within individuals to detect foreign antigen. However, it is also possible that such an increase in gene number could alter qualitatively other aspects of the rhesus immune response, such as the interaction with the NK inhibitory and activatory network. The latter possibility would be important to understand in light of the extensive use of these animals as models for comprehending and evaluating human disease and vaccination.
Conclusions
The rhesus macaque is one of the most important nonhuman primates currently being used in clinical studies. Functional studies of the rhesus immune response can benefit from these data immediately by incorporating the new knowledge of mamu class I and class II gene content into their experimental design. Many of these studies are focused on CTL responses and are looking in detail at the MHC-peptide complexes that are associated with viral escape and disease progression (O'Connor et al. 2001). The consideration of all of the antigen-presenting genes that are available for participation in a specific immune response could reveal in more detail how that response is initiated, how it progresses, and under what circumstances it is ultimately evaded or completed. In this regard, the higher levels of polymorphism in immune-related genes in the MHC other than class I and class II could affect immune function in other ways that are qualitatively distinguishable from those in human. For example, the higher level of polymorphism in the TAP genes in the rat has functional consequences for peptide selection, a feature not found in the human (Powis et al. 1996; Daniel et al. 1997; Deverson et al. 1998). Whereas the rhesus TAP genes are more polymorphic than humans, whether that polymorphism has functional consequence is unknown.
These data emphasize the need for genomic sequence data from other immune complexes within the rhesus macaque genome and from other nonhuman primates used in clinical research. At present, more than 30 species of nonhuman primates are targets for biomedical research (Austad 1997). It is likely that smaller, focused sequencing efforts will be able to rapidly provide essential genomic information about these animal MHCs and other immune response loci relevant to the clinical models for which they are being used (Eichler and DeJong 2002). The wealth of immunologic information that can be rapidly acquired through the generation of genomic sequence data can greatly benefit and advance the use of all of these animals in clinical disease studies.
METHODS
Rhesus Macaque Sequence-Ready BAC Contig
We had previously identified 580 human MHC-amplicons, designed as both unique sequence probes and unique PCR assays (Geraghty et al. 2002), and used this resource to test rhesus macaque DNA for cross reactivity. Of these, 384 were positive on rhesus genomic DNA in PCR-sequencing assays, showing from 80% to 99% sequence identity to the respective human sequence. These data were then used to outline a preliminary mapping strategy for the isolation of the rhesus macaque MHC. The CHORI-250 Rhesus macaque BAC library constructed at the Children's Hospital Oakland Research Institute, BACPAC Resources was purchased, and with high-density hybridization filters, were screened according to the manufacturer's protocol using combinations of MHC-amplicon probes spread over 500-kb segments. Positive BACs were organized as directed by the MHC PCR assays assuming colinearity of rhesus MHC with human. The rhesus blood donor used to create the library is of Indian origin and was born at the California Regional Primate Research Center (CRPRC). Blood and a cell line from this animal were generously provided to us by Marta L. Marthas (mlmarthas@ucdavis.edu) at the (CRPRC).
Covering about 70% of the MHC, PCR assays and finger-prints of overlapping BACs yielded confirmation of accurate representation of the rhesus genomic sequence. In regions where a low density of clones were isolated or where ambiguous PCR-sequencing results were obtained, additional probes from those regions were used in a second library screening. This approach succeeded in obtaining correct representation of the rhesus MHC with the exception of the class I A-like and class I B-like regions. At this point, shotgun sequencing of BACs was initiated and sequence data from BACs was used to design new probes for library screening and clone characterization. Considerable overlap between BACs in the class I-A and class I-B regions was needed to distinguish the haplotypes and to confirm the fidelity of clone coverage. This was due to the high similarity of duplicated segments in these regions (e.g., in the A-like region only eight nucleotide differences extending over two 80-kb segments). After two successive rounds of sequencing and probe design, the complete set of BACs spanning the rhesus MHC was obtained. At this point, it was also possible to assign unambiguously each BAC to one of the two haplotypes by PCR-sequence analysis. Five small gaps remained before contiguous sequence could be achieved, and these were filled by either PCR sequencing from BACs or by partial accumulation of whole BAC shotgun sequence. All of the haplotype assignments were confirmed by MHC–PCR analysis of the donor genomic DNA used to create the library.
DNA Sequencing
BAC DNAs were isolated according to established protocols. Shotgun libraries were produced from 15 μg of purified BAC DNA. Briefly, DNA was sonicated for four discreet timepoints and tested for appropriate levels of fragmentation on an agarose gel. Timepoints that collectively yielded a standard digestion pattern that provided near-random fragment generation (determined empirically) were combined and subjected to end repair using T4 DNA polymerase as described (Guillaudeux et al. 1998). Afterward, DNA was run on an agarose gel, and an appropriate size fraction was excised and purified using the Wizard DNA purification system (Promega) according to the manufacturer's instructions. The excised fragments were cloned into SmaI cut and dephosphorylated pUC18 and transformed into Escherichia coli strain XL1-Blue supercompetent cells (Stratagene). Colonies were picked by hand and plasmids were purified using a Qiabot 3000 robot (QIAGEN) according to the manufacturer's instructions. Purified plasmids were subjected to sequencing reactions with both forward and reverse primers using big dye terminator sequencing kits and provided protocols (Perkin Elmer). The sequences derived from this study were resolved on fluorescent-based capillary sequencers (Model 3700 and 3730, Applied Biosystems).
Sequencing of all BACs proceeded through three phases of analysis as follows: (1) shotgun sequencing, where ∼2500 forward and reverse reads were produced and assembled using phredphrap-consed (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998); (2) identification of low-quality regions, single-stranded regions, and gaps in the sequence, and subjecting those having two or more plasmid sequences spanning the gaps to primer-directed sequencing of appropriate plasmid DNAs. In cases where one or no plasmid spanned such regions, appropriate primers were designed and PCR sequencing of the respective BAC DNA was performed. Plasmids spanning weak regions were automatically identified using a custom Java script produced in our lab (D.E. Geraghty, unpubl.). High-quality discrepancies were examined to identify possible misassembly of the shotgun data. In most cases, user input could resolve these discrepancies, although when highly duplicated regions were contained within a BAC, additional shotgun data from overlapping BACs was needed to resolve them completely; (3) in a subset of cases, primer-directed plasmid sequencing or BAC PCR sequencing did not resolve the weak sequence. In the former case, it was often sufficient to repurify the plasmid DNA and repeat the sequencing reaction with larger quantities to yield adequate sequence data. For the remaining refractory regions, either spanning plasmid DNAs or PCR fragments were subjected to sonication to yield mini-libraries of 200–600 bp inserts, and sequencing was carried out as above. Of the 54 BACs this project included, this was necessary on only three occasions. The submitted sequence data is estimated by phred values to have an accuracy of well over 99.99%. All of the BACs were subjected to physical fingerprint analysis, which showed complete agreement with electronic digestion patterns.
Sequence Data Analysis
Sequence data acquisition was managed using a data tracking, management, and storage system custom built in our laboratory, updated from that previously described (Geraghty et al. 2000). This system allows for tracking lab workflow and data quality, and its regular use led to significant cost savings over our previous experience. Shotgun sequence assembly and editing was done with the Phred-Phrap-Consed package developed by Phil Green and coworkers (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998). Cross_match was used extensively in annotation of the sequence data as was RepeatMasker with the current human repeat matrix developed by A. Smit (Smit 1999). Primer selection was done using Primer 3 (Rozen and Skaletsky 2000) embedded within custom in-house scripts to enable convenient primer design and ordering.
Annotation of the MHC started with the previous annotation to the human MHC (MHC Consortium 1999) using updated cDNA sequences from public databases. Human EST databases were screened for homology against the rhesus sequence masked both for repeats and previously identified cDNAs using combinations of cross_match and the BLASTN and BLASTX-BEAUTY and BLAT database search tools (Worley et al. 1998; Hernandez et al. 2000; Kent 2002). Analysis of interspersed repeats and G+C content was done using RepeatMasker version 2002/05/15 with the human repeat library (Smit 1999) and the GESTALT Work-bench (Glusman and Lancet 2000). The MAP (multiple alignment program), written by Xiaoqiu Huang, was used for aligning the two monkey and two human sequences using max match 10, min mismatch -1, gap-open penalty 70, gap-extension penalty 1 (http://deepc2.zool.iastate.edu/aat/map/mapdoc.html). Potential rhesus genes were verified by constructing virtual rhesus cDNAs from genomic DNA using the human cDNA as a guide. This allowed for the classification of rhesus genes as probable pseudogenes due to significant alterations in the coding sequences or as modified genes due to altered start or stop codons. For rhesus MHC class I sequences, we relied on the database of class I sequences accumulated in the Watkins laboratory at the University of Wisconsin, Wisconsin Regional Primate Research Center. For MHC rhesus class II cDNA sequences, we relied on GenBank and the Bontrop laboratory at the Biomedical Primate Research Centre in the Netherlands.
Acknowledgments
Ruihan Wang provided outstanding informatics support for all phases of the project. Simon Fortelny provided important analysis support and sequence data curation. Quyen Vu, Luke Williams, Bethany Richards, Jana Stonehocker, and Barrett Nelson at the FHCRC and Brian Birditt, Scott Bloom, and Ericka Johnson at the ISB provided valuable help with sequencing at various stages of the project. The generous help of David O'Connor and David Watkins who provided unpublished mamu cDNA sequences for analysis, and Ronald Bontrop who provided genomic DNA from unrelated animals was essential for analysis. We thank Lee Hood for reading the manuscript. This work was supported by a grant from the NIH National Center for Research Resources (R24 RR17186) to D.E.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2134504.
Footnotes
[Supplemental material is available online at www.genome.org and www.fhcrc.org/labs/geraghty. The sequence data from this study have been submitted to GenBank under accession nos. AC148659–AC148717. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: D. Watkins, D. O'Connor, R. Bontrop, and M.L. Marthas.]
References
- Accolla, R.S., Adorini, L., Sartoris, S., Sinigaglia, F., and Guardiola, J. 1995. MHC: Orchestrating the immune response. Immunol. Today 16: 8–11. [DOI] [PubMed] [Google Scholar]
- Adams, E.J. and Parham, P. 2001. Species-specific evolution of MHC class I genes in the higher primates. Immunol. Rev. 183: 41–64. [DOI] [PubMed] [Google Scholar]
- Anzai, T., Shiina, T., Kimura, N., Yanagiya, K., Kohara, S., Shigenari, A., Yamagata, T., Kulski, J.K., Naruse, T.K., Fujimori, Y., et al. 2003. Comparative sequencing of human and chimpanzee MHC class I regions unveils insertions/deletions as the major path to genomic divergence. Proc. Natl. Acad. Sci. 100: 7708–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austad, S.N. 1997. Small nonhuman primates as potential models of human aging. Ilar J. 38: 142–147. [DOI] [PubMed] [Google Scholar]
- Bontrop, R.E. 2001. Non-human primates: Essential partners in biomedical research. Immunol. Rev. 183: 5–9. [DOI] [PubMed] [Google Scholar]
- Boyson, J.E., McAdam, S.N., Gallimore, A., Golos, T.G., Liu, X., Gotch, F.M., Hughes, A.L., and Watkins, D.I. 1995. The MHC E locus in macaques is polymorphic and is conserved between macaques and humans. Immunogenetics 41: 59–68. [DOI] [PubMed] [Google Scholar]
- Boyson, J.E., Shufflebotham, C., Cadavid, L.F., Urvater, J.A., Knapp, L.A., Hughes, A.L., and Watkins, D.I. 1996. The MHC class I genes of the rhesus monkey. Different evolutionary histories of MHC class I and II genes in primates. J. Immunol. 156: 4656–4665. [PubMed] [Google Scholar]
- Boyson, J.E., Iwanaga, K.K., Golos, T.G., and Watkins, D.I. 1997. Identification of a novel MHC class I gene, Mamu-AG, expressed in the placenta of a primate with an inactivated G locus. J. Immunol. 159: 3311–3321. [PubMed] [Google Scholar]
- Celada, F. and Seiden, P.E. 1996. Affinity maturation and hypermutation in a simulation of the humoral immune response. Eur. J. Immunol. 26: 1350–1358. [DOI] [PubMed] [Google Scholar]
- Colonna, M. 1996. Natural killer cell receptors specific for MHC class I molecules. Curr. Opin. Immunol. 8: 101–107. [DOI] [PubMed] [Google Scholar]
- Daniel, S., Caillat-Zucman, S., Hammer, J., Bach, J.F., and van Endert, P.M. 1997. Absence of functional relevance of human transporter associated with antigen processing polymorphism for peptide selection. J. Immunol. 159: 2350–2357. [PubMed] [Google Scholar]
- Deverson, E.V., Leong, L., Seelig, A., Coadwell, W.J., Tredgett, E.M., Butcher, G.W., and Howard, J.C. 1998. Functional analysis by site-directed mutagenesis of the complex polymorphism in rat transporter associated with antigen processing. J. Immunol. 160: 2767–2779. [PubMed] [Google Scholar]
- Dzuris, J.L., Sidney, J., Appella, E., Chesnut, R.W., Watkins, D.I., and Sette, A. 2000. Conserved MHC class I peptide binding motif between humans and rhesus macaques. J. Immunol. 164: 283–291. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E. and DeJong, P.J. 2002. Biomedical applications and studies of molecular evolution: A proposal for a primate genomic library resource. Genome Res. 12: 673–678. [DOI] [PubMed] [Google Scholar]
- Evans, D.T., Jing, P., Allen, T.M., O'Connor, D.H., Horton, H., Venham, J.E., Piekarczyk, M., Dzuris, J., Dykhuzen, M., Mitchen, J., et al. 2000. Definition of five new simian immunodeficiency virus cytotoxic T-lymphocyte epitopes and their restricting major histocompatibility complex class I molecules: Evidence for an influence on disease progression. J. Virol. 74: 7400–7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Flajnik, M.F. 1996. The immune system of ectothermic vertebrates. Vet. Immunol. Immunopathol. 54: 145–150. [DOI] [PubMed] [Google Scholar]
- Flajnik, M.F., Ohta, Y., Namikawa-Yamada, C., and Nonaka, M. 1999. Insight into the primordial MHC from studies in ectothermic vertebrates. Immunol. Rev. 167: 59–67. [DOI] [PubMed] [Google Scholar]
- Geraghty, D.E. 1993. Structure of the HLA class I region and expression of its resident genes. Curr. Opin. Immunol. 5: 3–7. [DOI] [PubMed] [Google Scholar]
- Geraghty, D.E., Koller, B.H., Hansen, J.A., and Orr, H.T. 1992a. The HLA class I gene family includes at least six genes and twelve pseudogenes and gene fragments. J. Immunol. 149: 1934–1946. [PubMed] [Google Scholar]
- Geraghty, D.E., Koller, B.H., Pei, J., and Hansen, J.A. 1992b. Examination of four HLA class I pseudogenes. Common events in the evolution of HLA genes and pseudogenes. J. Immunol. 149: 1947–1956. [PubMed] [Google Scholar]
- Geraghty, D.E., Fortelny, S., Guthrie, B., Irving, M., Pham, H., Wang, R., Daza, R., Nelson, B., Stonehocker, J., Williams, L., et al. 2000. Data acquisition, data storage, and data presentation in a modern genetics laboratory. Rev. Immunogenet. 2: 532–540. [PubMed] [Google Scholar]
- Geraghty, D.E., Daza, R., Williams, L.M., Vu, Q., and Ishitani, A. 2002. Genetics of the immune response: Identifying immune variation within the MHC and throughout the genome. Immunol. Rev. 190: 69–85. [DOI] [PubMed] [Google Scholar]
- Glusman, G. and Lancet, D. 2000. GESTALT: A workbench for automatic integration and visualization of large-scale genomic sequence analyses. Bioinformatics 16: 482–483. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Guillaudeux, T., Janer, M., Wong, G.K., Spies, T., and Geraghty, D.E. 1998. The complete genomic sequence of 424,015 bp at the centromeric end of the HLA class I region: Gene content and polymorphism. Proc. Natl. Acad. Sci. 95: 9494–9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, P., Martin, A., and Dorado, G. 2000. The BLAST algorithms: Practical application in molecular cloning, marker-assisted selection (MAS) and introgression of wheat. DNA Seq. 11: 339–347. [DOI] [PubMed] [Google Scholar]
- Hershberger, K.L., Shyam, R., Miura, A., and Letvin, N.L. 2001. Diversity of the killer cell Ig-like receptors of rhesus monkeys. J. Immunol. 166: 4380–4390. [DOI] [PubMed] [Google Scholar]
- Horton, R., Niblett, D., Milne, S., Palmer, S., Tubby, B., Trowsdale, J., and Beck, S. 1998. Large-scale sequence comparisons reveal unusually high levels of variation in the HLA-DQB1 locus in the class II region of the human MHC. J. Mol. Biol. 282: 71–97. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, J., Kasahara, M., Gutknecht, J., and Figueroa, F. 1990. Origin and function of Mhc polymorphism. Chem. Immunol. 49: 35–50. [PubMed] [Google Scholar]
- Knapp, L.A., Cadavid, L.F., and Watkins, D.I. 1998. The MHC-E locus is the most well conserved of all known primate class I histocompatibility genes. J. Immunol. 160: 189–196. [PubMed] [Google Scholar]
- LaBonte, M.L., Hershberger, K.L., Korber, B., and Letvin, N.L. 2001. The KIR and CD94/NKG2 families of molecules in the rhesus monkey. Immunol. Rev. 183: 25–40. [DOI] [PubMed] [Google Scholar]
- Lee, N., Goodlett, D.R., Ishitani, A., Marquardt, H., and Geraghty, D.E. 1998a. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J. Immunol. 160: 4951–4960. [PubMed] [Google Scholar]
- Lee, N., Llano, M., Carretero, M., Ishitani, A., Navarro, F., Lopez-Botet, M., and Geraghty, D.E. 1998b. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. 95: 5199–5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MHC Consortium. 1999. Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature 401: 921–923. [DOI] [PubMed] [Google Scholar]
- Miller, M.D., Yamamoto, H., Hughes, A.L., Watkins, D.I., and Letvin, N.L. 1991. Definition of an epitope and MHC class I molecule recognized by gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys. J. Immunol. 147: 320–329. [PubMed] [Google Scholar]
- Momburg, F., Armandola, E.A., Post, M., and Hammerling, G.J. 1996. Residues in TAP2 peptide transporters controlling substrate specificity. J. Immunol. 156: 1756–1763. [PubMed] [Google Scholar]
- Mothe, B.R., Sidney, J., Dzuris, J.L., Liebl, M.E., Fuenger, S., Watkins, D.I., and Sette, A. 2002. Characterization of the peptide-binding specificity of Mamu-B*17 and identification of Mamu-B*17-restricted epitopes derived from simian immunodeficiency virus proteins. J. Immunol. 169: 210–219. [DOI] [PubMed] [Google Scholar]
- Nossal, G.J. 1994. Negative selection of lymphocytes. Cell 76: 229–239. [DOI] [PubMed] [Google Scholar]
- O'Connor, D., Friedrich, T., Hughes, A., Allen, T.M., and Watkins, D. 2001. Understanding cytotoxic T-lymphocyte escape during simian immunodeficiency virus infection. Immunol. Rev. 183: 115–126. [DOI] [PubMed] [Google Scholar]
- Otting, N. and Bontrop, R.E. 1993. Characterization of the rhesus macaque (Macaca mulatta) equivalent of HLA-F. Immunogenetics 38: 141–145. [DOI] [PubMed] [Google Scholar]
- Parham, P. 1999. Virtual reality in the MHC. Immunol. Rev. 167: 5–15. [DOI] [PubMed] [Google Scholar]
- Pearson, W.R., Wood, T., Zhang, Z., and Miller, W. 1997. Comparison of DNA sequences with protein sequences. Genomics 46: 24–36. [DOI] [PubMed] [Google Scholar]
- Powis, S.J., Young, L.L., Joly, E., Barker, P.J., Richardson, L., Brandt, R.P., Melief, C.J., Howard, J.C., and Butcher, G.W. 1996. The rat cim effect: TAP allele-dependent changes in a class I MHC anchor motif and evidence against C-terminal trimming of peptides in the ER. Immunity 4: 159–165. [DOI] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386. [DOI] [PubMed] [Google Scholar]
- Ryan, A.F., Grendell, R.L., Geraghty, D.E., and Golos, T.G. 2002. A soluble isoform of the rhesus monkey nonclassical MHC class I molecule Mamu-AG is expressed in the placenta and the testis. J. Immunol. 169: 673–683. [DOI] [PubMed] [Google Scholar]
- Saitou, N. and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Slierendregt, B.L., Otting, N., van Besouw, N., Jonker, M., and Bontrop, R.E. 1994. Expansion and contraction of rhesus macaque DRB regions by duplication and deletion. J. Immunol. 152: 2298–2307. [PubMed] [Google Scholar]
- Smit, A.F. 1999. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9: 657–663. [DOI] [PubMed] [Google Scholar]
- Takahata, N. 1995. MHC diversity and selection. Immunol. Rev. 143: 225–247. [DOI] [PubMed] [Google Scholar]
- Townsend, A. and Bodmer, H. 1989. Antigen recognition by class I-restricted T lymphocytes. Annu. Rev. Immunol. 7: 601–624. [DOI] [PubMed] [Google Scholar]
- Urvater, J.A., Otting, N., Loehrke, J.H., Rudersdorf, R., Slukvin, I.I., Piekarczyk, M.S., Golos, T.G., Hughes, A.L., Bontrop, R.E., and Watkins, D.I. 2000. Mamu-I: A novel primate MHC class I B-related locus with unusually low variability. J. Immunol. 164: 1386–1398. [DOI] [PubMed] [Google Scholar]
- Watkins, D.I. 1994. MHC of nonhuman primates. Curr. Top. Microbiol. Immunol. 188: 145–159. [DOI] [PubMed] [Google Scholar]
- Watkins, D.I., Kannagi, M., Stone, M.E., and Letvin, N.L. 1988. Major histocompatibility complex class I molecules of nonhuman primates. Eur. J. Immunol. 18: 1425–1432. [DOI] [PubMed] [Google Scholar]
- Worley, K.C., Culpepper, P., Wiese, B.A., and Smith, R.F. 1998. BEAUTY-X: Enhanced BLAST searches for DNA queries. Bioinformatics 14: 890–891. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.sanger.ac.uk/HGP/Chr6/MHC/; The Sanger Institute/The MHC haplotype project.
- http://deepc2.zool.iastate.edu/aat/map/mapdoc.html; Information resource on the Multiple Sequence Alignment.
- www.fhcrc.org/labs/geraghty; Geraghty lab Web data resource.
- www.chori.org/bacpac/; BACPAC Resources center homepage.