Abstract
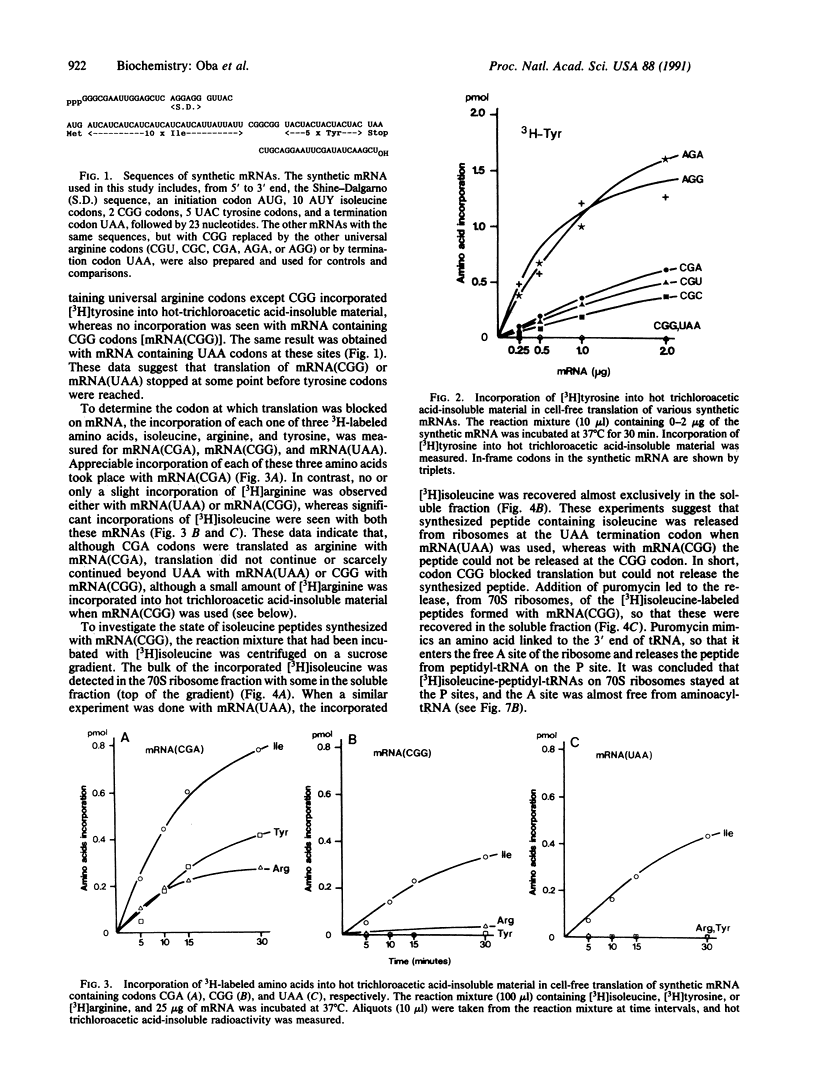
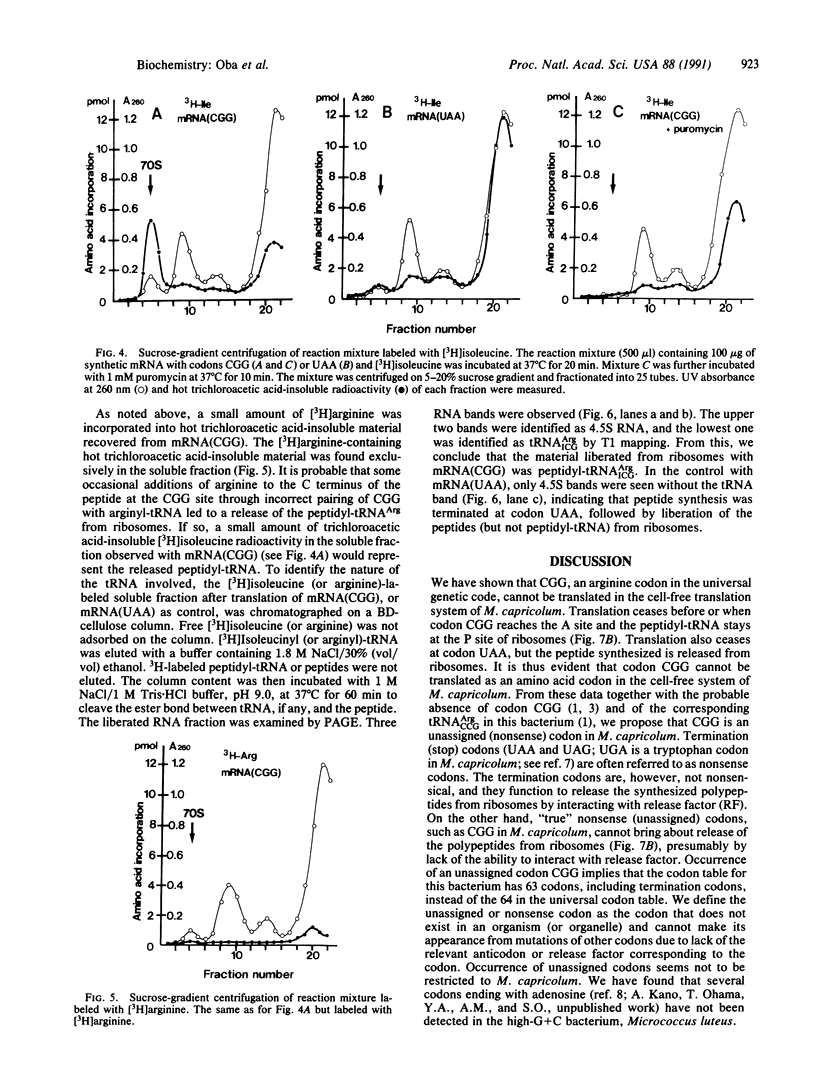
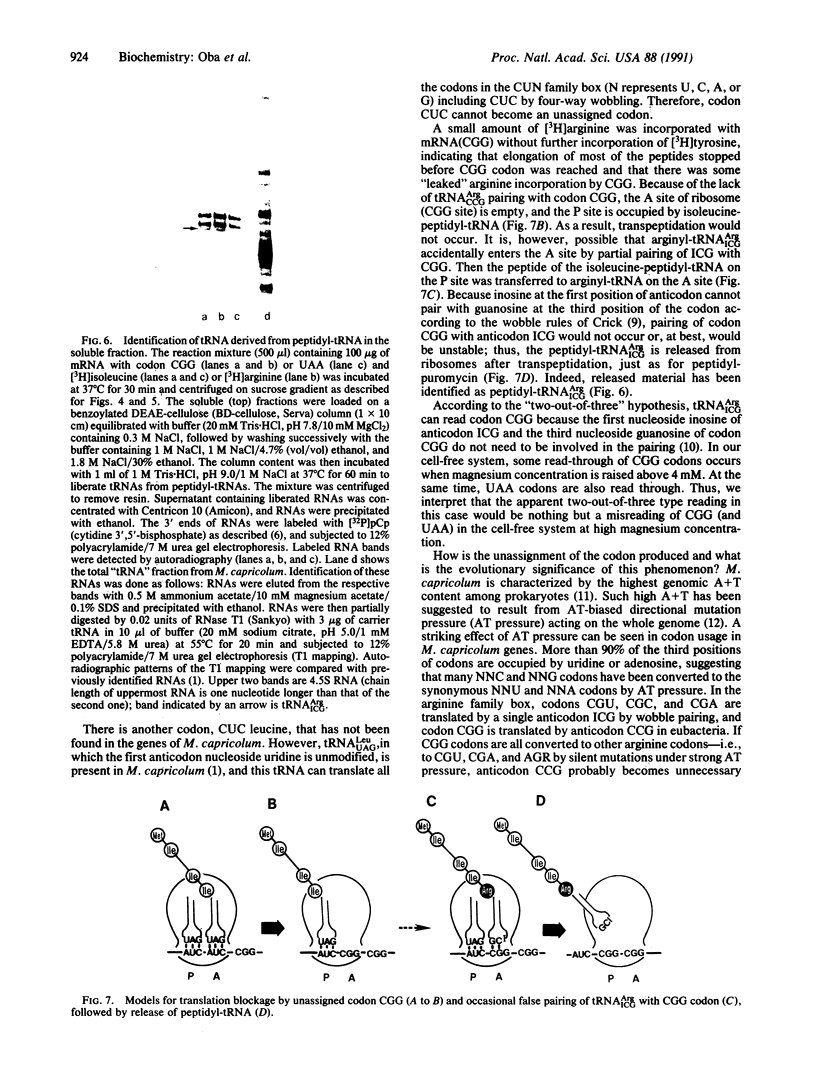
CGG is an arginine codon in the universal genetic code. We previously reported that in Mycoplasma capricolum, a relative of Gram-positive eubacteria, codon CGG did not appear in coding frames, including termination sites, and tRNA(ArgCCG) pairing with codon CGG, was not detected. These facts suggest that CGG is a nonsense (unassigned and untranslatable) codon--i.e., not assigned to arginine or to any other amino acid. We have investigated whether CGG is really an unassigned codon by using a cell-free translation system prepared from M. capricolum. Translation of synthetic mRNA containing in-frame CGG codons does not result in "read-through" to codons beyond the CGG codons--i.e., translation ceases just before CGG. Sucrose-gradient centrifugation profiles of the reaction mixture have shown that the bulk of peptide that has been synthesized is attached to 70S ribosomes and is released upon further incubation with puromycin. The result suggests that the peptide is in the P site of ribosome in the form of peptidyl-tRNA, leaving the A site empty. When in-frame CGG codons are replaced by UAA codons in mRNA, no read-through occurs beyond UAA, just as in the case of CGG. However, the synthesized peptide is released from 70S ribosomes, presumably by release factor 1. These data suggest strongly that CGG is an unassigned codon and differs from UAA in that CGG is not used for termination.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andachi Y., Yamao F., Muto A., Osawa S. Codon recognition patterns as deduced from sequences of the complete set of transfer RNA species in Mycoplasma capricolum. Resemblance to mitochondria. J Mol Biol. 1989 Sep 5;209(1):37–54. doi: 10.1016/0022-2836(89)90168-x. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966 Aug;19(2):548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Jukes T. H. A change in the genetic code in Mycoplasma capricolum. J Mol Evol. 1985;22(4):361–362. doi: 10.1007/BF02115692. [DOI] [PubMed] [Google Scholar]
- Lagerkvist U. "Two out of three": an alternative method for codon reading. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1759–1762. doi: 10.1073/pnas.75.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Andachi Y., Yuzawa H., Yamao F., Osawa S. The organization and evolution of transfer RNA genes in Mycoplasma capricolum. Nucleic Acids Res. 1990 Sep 11;18(17):5037–5043. doi: 10.1093/nar/18.17.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Osawa S. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc Natl Acad Sci U S A. 1987 Jan;84(1):166–169. doi: 10.1073/pnas.84.1.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto A., Yamao F., Osawa S. The genome of Mycoplasma capricolum. Prog Nucleic Acid Res Mol Biol. 1987;34:29–58. doi: 10.1016/s0079-6603(08)60492-4. [DOI] [PubMed] [Google Scholar]
- Ohama T., Muto A., Osawa S. Role of GC-biased mutation pressure on synonymous codon choice in Micrococcus luteus, a bacterium with a high genomic GC-content. Nucleic Acids Res. 1990 Mar 25;18(6):1565–1569. doi: 10.1093/nar/18.6.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkubo S., Muto A., Kawauchi Y., Yamao F., Osawa S. The ribosomal protein gene cluster of Mycoplasma capricolum. Mol Gen Genet. 1987 Dec;210(2):314–322. doi: 10.1007/BF00325700. [DOI] [PubMed] [Google Scholar]
- Osawa S., Jukes T. H. Codon reassignment (codon capture) in evolution. J Mol Evol. 1989 Apr;28(4):271–278. doi: 10.1007/BF02103422. [DOI] [PubMed] [Google Scholar]
- Osawa S., Muto A., Jukes T. H., Ohama T. Evolutionary changes in the genetic code. Proc Biol Sci. 1990 Jul 23;241(1300):19–28. doi: 10.1098/rspb.1990.0060. [DOI] [PubMed] [Google Scholar]
- Sawada M., Osawa S., Kobayashi H., Hori H., Muto A. The number of ribosomal RNA genes in Mycoplasma capricolum. Mol Gen Genet. 1981;182(3):502–504. doi: 10.1007/BF00293942. [DOI] [PubMed] [Google Scholar]
- Yamao F., Muto A., Kawauchi Y., Iwami M., Iwagami S., Azumi Y., Osawa S. UGA is read as tryptophan in Mycoplasma capricolum. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2306–2309. doi: 10.1073/pnas.82.8.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]




