Abstract
The Giardia lamblia genome sequencing project affords us a unique opportunity to conduct comparative analyses of core cellular systems between early and late-diverging eukaryotes on a genome-wide scale. We report a survey to identify canonical transcription components in Giardia, focusing on RNA polymerase (RNAP) subunits and transcription-initiation factors. Our survey revealed that Giardia contains homologs to 21 of the 28 polypeptides comprising eukaryal RNAPI, RNAPII, and RNAPIII; six of the seven RNAP subunits without giardial homologs are polymerase specific. Components of only four of the 12 general transcription initiation factors have giardial homologs. Surprisingly, giardial TATA-binding protein (TBP) is highly divergent with respect to archaeal and higher eukaryotic TBPs, and a giardial homolog of transcription factor IIB was not identified. We conclude that Giardia represents a transition during the evolution of eukaryal transcription systems, exhibiting a relatively complete set of RNAP subunits and a rudimentary basal initiation apparatus for each transcription system. Most class-specific RNAP subunits and basal initiation factors appear to have evolved after the divergence of Giardia from the main eukaryotic line of descent. Consequently, Giardia is predicted to be unique in many aspects of transcription initiation with respect to paradigms derived from studies in crown eukaryotes.
Comparison of transcription in Archaea and Eucarya suggests that the most recent common ancestor to both lineages contained a core transcription apparatus composed of a single RNA polymerase and a recruitment mechanism corresponding to a TATA-binding protein (TBP)/Transcription Factor IIB (TFIIB)-type complex. The single, archaeal RNA polymerase shares a specific relationship with the three eukaryal polymerases, to the exclusion of bacterial RNA polymerase in phylogenetic analyses (Zillig et al. 1989; Iwabe et al. 1991), a relationship exemplified by shared subunit composition (Langer et al. 1995; Darcy et al. 1999) and a common architecture (Eloranta et al. 1998; Werner et al. 2000; Best and Olsen 2001). Basal initiation in archaeal transcription is dependent upon two factors, TBP and TFB (the archaeal homolog of TFIIB), recognizing TATA elements of gene promoters (Hausner et al. 1996). These common features reflect a shared evolutionary history. However, the eukaryal transcription apparatus has undergone extensive elaboration, unparalleled in the evolution of archaeal transcription.
Eukaryotes use three different RNA polymerases (RNAPI, RNAPII, and RNAPIII) to transcribe different gene classes, generally ribosomal RNAs, messenger RNAs, and transfer RNAs, respectively; each RNAP initiates transcription at specific gene promoters aided by different sets of transcription factors (Orphanides et al. 1996; Paule and White 2000). The textbook view of eukaryal transcription (e.g., Alberts et al. 1994) is derived principally from fungi, plants, and animals, representing a relatively small fraction of the diversity found in the eukaryal domain. Further, the elaborations seen in the eukaryal transcription apparatus are remarkable in scope and complexity, and the evolution of such a system is likely to have occurred in stages. These observations, coupled with our anecdotal observations of varying conservation among canonical eukaryotic transcription factors (Coulson and Ouzounis 2003), led us (1) to inquire about the broader applicability of the paradigm regarding transcription to Eucarya as a whole, and (2) to search for evidence of the hypothesized progression from a single polymerase-type system seen in Archaea to the complex system found in crown group eukaryotes.
A unique opportunity to examine the components of the transcription apparatus in a potentially early diverging eukaryote stems from the genome-sequencing project of the human parasite Giardia lamblia (McArthur et al. 2000). Phylogenetic analyses of small subunit rRNA (Sogin et al. 1989), translation elongation factor (EF) 1α (Hashimoto et al. 1994), EF-2 (Hashimoto et al. 1995), and eukaryotic release factor 3 (Inagaki and Doolittle 2000; Inagaki et al. 2003) support an early divergence of Giardia from the eukaryotic line of descent, although the basal position of Giardia in some phylogenetic reconstructions has been questioned (Hirt et al. 1999; also see Conclusions below). In addition, certain morphological features of Giardia cells hint at altered or simpler forms of characteristics found in recently diverging organisms (McCaffery and Gillin 1994; McCaffery et al. 1994). Regardless, it is most likely that divergent features of the transcription machinery observed in Giardia trace back to very early stages of eukaryal evolution (Gillin et al. 1996; Smith et al. 1998).
We have undertaken a survey of the nearly complete Giardia lamblia genome to distinguish more ancient from more recent innovations in basal transcription, focusing on RNA polymerase subunits and factors involved in initiation of transcription. First, we used evolutionary distance analyses of transcription components in crown eukaryotes to gain a better understanding of the relative conservation of the various proteins. Next, we queried the partial genome sequence of Giardia with canonical eukaryal and archaeal transcription components to learn which components were present at this stage of evolution. Finally, we used evolutionary distance and phylogenetic analyses of the crown eukaryal, giardial, and archaeal proteins to seek insights into the dramatic variations in tempo and mode of evolution of the transcription components. We predicted that if Giardia represents a transition state, it would possess components of transcription conserved between Archaea and Eucarya, whereas some factors involved in polymerase-specific initiation might not be present.
RESULTS AND DISCUSSION
What does the Giardia lamblia genome sequence tell us about the evolution of eukaryotic transcription? We have conducted a component-by-component analysis of archaeal and eukaryotic transcription proteins to address this question, identifying giardial homologs through similarity and profile searches, and characterizing identified components using evolutionary distance and phylogenetic analyses.
RNA Polymerase Subunits
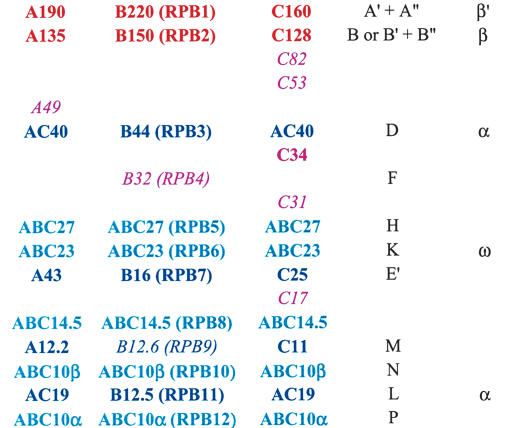
Table 1 lists subunits of eukaryal RNA polymerases (I, II, and III), archaeal RNA polymerase, and bacterial RNA polymerase. The table allows convenient cross-referencing of corresponding subunits among polymerases and provides a consistent nomenclature that we use throughout this work.
Table 1.
RNA Polymerase Subunit Correspondencesa
Subunits of the eukaryal RNA polymerases fall into three categories—shared, unique, and paralogous (Table 1). Shared (or common) subunits are present in all three polymerases. Unique subunits are found in only one of the three polymerases, and their genes do not share common ancestry with subunits from other RNAPs. Paralogous subunits are two or three distinct proteins in the different polymerases, but whose genes share common ancestry. These proteins generally perform related functions in their respective RNAP. The paralogous category subdivides into two groups—large and small. The large paralogous group includes the two largest subunits of each polymerase, which together form the active site of the enzyme. The small paralogous group contains the other sets of paralogs, which tend to have much lower molecular weights than the active site-forming subunits.
The three eukaryotic RNA polymerases share five subunits (Table 1), with RNAPI and RNAPIII sharing an additional two subunits (AC19 and AC40) (Geiduschek and Kassavetis 2001). The paralogs AC19 and AC40 are related to RNAPII subunits B12.5 and B44, respectively. Additional sets of paralogous subunits exist for each polymerase as follows: A12.2, B12.6, and C11 (Chedin et al. 1998); A43, B16, and C25 (Sadhale and Woychik 1994; Shpakovskii and Shematorova 1999); and the two largest subunits of each polymerase. The two largest subunits are conserved across the three domains of life (Table 1). Each polymerase also harbors unique subunits, the complement of which may vary among organisms (Huang and Maraia 2001; Shematorova and Shpakovski 2002). In Saccharomyces cerevisiae, three subunits are unique to RNAPI (Carles and Riva 1998; Ishihama et al. 1998), one is unique to RNAPII, and six are unique to RNAPIII (Geiduschek and Kassavetis 2001; Huang and Maraia 2001).
For each RNAP subunit, the average of interkingdom distance (plant–animal, plant–fungal, and animal–fungal) provides a measure of the relative divergence rate of the protein. Table 2 presents these distances, sorted in ascending order (most conserved to least). Several patterns are apparent in these data. The two largest subunits of RNAPII and RNAPIII are highly conserved, with distances ranging from 50 PAM units (estimated replacements per 100 amino acids [Dayhoff et al. 1978]) (B150) to 83 PAM units (C160). However, the two largest subunits of RNAPI are twice as diverged as the two largest subunits of RNAPII and RNAPIII. It was noted previously that RNAPI has evolved at a higher average rate than either RNAPII or RNAPIII among crown group organisms (Memet et al. 1988; Iwabe et al. 1991). The five common subunits show conservation comparable to the largest subunits. ABC10β exhibits the highest conservation of all RNAP subunits (37 PAM units), whereas ABC10α has a divergence of 122 PAM units, roughly three times that of ABC10β and comparable to that of A190. The divergence among the small paralogous subunits is generally equivalent to the more highly diverged representatives of the large and common subunits. Interestingly, the pattern seen among the two largest subunits—RNAPI subunits having a high rate of evolution relative to RNAPII and RNAPIII subunits—does not occur in comparisons of small paralogous subunits (with the exception of the A12.2/B12.6/C11 family). Corresponding subunits of each polymerase have evolved at a similar rate, suggesting little difference in constraints as a result of specialization of the three polymerases. Finally, polymerase-specific subunits (presumably the most recent elaborations) show the lowest level of conservation among crown eukaryote RNAP subunits, as though they serve less critical roles.
Table 2.
Pairwise Distance Comparisons of RNAP Subunitsa
We surveyed the nearly completed genome sequence of Giardia lamblia using RNA polymerase subunits listed in Table 1 as queries in BLASTP, TBLASTN, and profile searches. Bold lettering in Table 1 indicates RNAP subunits that have homologs in Giardia. Table 2 lists the average pairwise distances between subunits within the crown groups, between the crown group and Giardia, between the crown group and Archaea, and between Giardia and the Archaea. Table 2 clearly demonstrates that highly conserved subunits within the crown also occur in Giardia, and evolutionary distances between homologous subunits are generally comparable to corresponding archaeal to crown group distances. In contrast, proteins that are highly diverged within the crown group are not identified in Giardia (see below for a discussion of interpreting gene absence).
Earlier analyses of giardial transcription identified components of RNAPII (Klenk et al. 1995) and RNAPIII (Lanzendorfer et al. 1992), but not of RNAPI, suggesting that Giardia might have diverged before the evolution of all three RNA polymerases in eukaryotic cells. This survey identified giardial ORFs corresponding to the two largest subunits of each RNA polymerase (Tables 1 and 2), unequivocally placing the divergence of Giardia after the duplication events giving rise to the catalytic cores of the three RNA polymerases. As was noted above for crown group organisms, the largest subunit of giardial RNAPI (i.e., A190) is roughly twice as divergent (289 PAM units) as the largest subunit of RNAPII (173 PAM units) or RNAPIII (115 PAM units; Table 2), suggesting an increased rate of evolution for the giardial RNAPI largest subunit. In contrast, the second largest subunits of each polymerase in Giardia have similar levels of divergence (Table 2).
To more clearly see the levels of divergence among large subunits within and between the crown group, Giardia, and Archaea, we plotted pairwise distances of each RNAP subunit versus small subunit rRNA pairwise distances (Fig. 1A–F), that is, transcription component divergence (vertical axis) as a function of organismal divergence (horizontal axis). The most striking observation is that some interdomain (archaeal to crown group) distances are smaller than the intradomain distances from giardial to crown group proteins (Fig. 1A,B). On the basis of rRNA distances, one would expect an approximately equal amount of divergence of giardial components from both archaeal and higher eukaryotic homologs (as in Fig. 1C,D). These data then suggest an increased rate of divergence for some giardial proteins, a theme that recurs throughout our analyses. The patterns of conservation seen in Figure 1, A–F are representative of patterns exhibited by many of the other giardial transcription components (data not shown).
Figure 1.
Comparisons of transcription component divergence with organismal divergence. Graphical representation of component distance (amino acid replacements per position) plotted on the y-axis vs. organismal small subunit rRNA distance (nucleotide substitutions per position) plotted on the x-axis. Data points are coded as indicated in the key. Error bars, one standard deviation uncertanties derived from 10 bootstrap replicates of protein distance calculations, are not visible at this scale. The identity of the RNAP subunit or transcription factor is indicated in A–G.
The complement of small subunits surrounding the large subunit core has undergone a high degree of elaboration during the evolution of the three eukaryal RNA polymerases; however, a set of five subunits remains common to each polymerase. In keeping with this conservation, giardial homologs to each of these subunits were identified (Tables 1 and 2). Intriguingly, two giardial ORFs with similarity to ABC27 were identified during BLAST searches (Table 2; Seshadri et al. 2003). Phylogenetic analysis indicates that the duplication event giving rise to these ORFs occurred subsequent to the divergence of the giardial lineage from the main eukaryal line of descent (data not shown). Each ABC27 gene is transcribed (Seshadri et al. 2003), but whether the proteins are components of specific polymerases (I, II, or III) has not been determined. If the giardial ABC27 paralogs are members of distinct RNA polymerases, this could be a lineage-specific elaboration of the shared archaeal/eukaryal RNA polymerase core (Table 1, common subunits), perhaps analogous to duplications leading to the paralogous subunit families of B44/AC40 and B12.5/AC19 found in all (known) extant eukaryotes.
In addition to the largest subunits of each polymerase, we identified nine of the 10 proteins comprising four sets of paralogous subunits (Table 1). Specifically, homologs of A43, B16, and C25; A12.2 and C11; AC19 and B12.5; and AC40 and B44 were identified in the giardial genome. The distance of giardial A43 to crown group representatives is very large (332 PAM units), but this is consistent with the distances observed within the crown group itself (306 PAM units; Table 2). The giardial A43 homolog was retrieved with an expectation value of 1 × 10–4 in a BLAST analysis, but the expectation value improved to 1 × 10–36 with a single iteration of PSI-BLAST, consistent with the assertion that the giardial ORF is truly related to eukaryal A43 homologs. Phylogenetic analysis using the PSI-BLAST alignments also supports this relationship (Supplemental Fig. S1).
A giardial ortholog of B12.6, a member of the A12.2/B12.6/C11 protein family, was not unambiguously identified. This protein family is involved in transcription elongation and/or transcript cleavage in each of the three RNA polymerase systems (Awrey et al. 1997; Chedin et al. 1998; Hemming and Edwards 2000; Hemming et al. 2000; Imazawa et al. 2001; Van Mullem et al. 2002a,b), and members are related to the transcription elongation factor TFIIS through a C-terminal Zn-ribbon motif (Qian et al. 1993; Kaine et al. 1994). The genome of Giardia contains three ORFs with this Zn-ribbon domain, which we denote as Zrd1, Zrd2, and Zrd3. Zrd1 and Zrd2 exhibit domain structures and sequence lengths characteristic of eukaryotic RNAP subunits. Consistent with these observations, phylogenetic analyses identified Zrd1 and Zrd2 as A12.2 and C11 homologs, respectively (Supplemental Fig. S2). In contrast, the length of Zrd3 is comparable to TFIIS, although sequence similarity outside of the C-terminal Zn-ribbon domain is minimal. Based solely upon the shared Zn-ribbon domain, Zrd3 exhibited a weakly supported relationship to the B12.6 clade (Supplemental Fig. S2). Interestingly, the phylogeny also suggested a relationship between TFIIS and B12.6 sequences. We caution that the small number of amino acid positions available for analysis limits the statistical reliability of these inferences. Nonetheless, these observations lead to the hypothesis that the Zn-ribbon motif of eukaryotic TFIIS proteins could be derived from an ancestral, RNAPII-specific member of the A12.2/B12.6/C11 family, in which case, Zrd3 could represent a relative of this hypothetical B12.6/TFIIS ancestor.
Of the seven subunits specific to just one of the three eukaryal RNA polymerases (Table 1), only one was found in the genome of Giardia, consistent with unique subunits being more recent additions. A giardial ORF with weak similarity (395 PAM units) to subunit C34 of RNAPIII was identified (Table 2). In yeast, subunit C34 interacts with subunits C82 and C31 to form a subcomplex with a role in RNAPIII initiation (Werner et al. 1993). Mutants in C34 or C31 can initiate transcription nonspecifically, but are deficient in specific initiation (Thuillier et al. 1995; Brun et al. 1997). The presence of a critical component of this subcomplex in Giardia suggests it represents a rudimentary form of the RNAPIII initiation system that occurs in canonical systems. Of special note is the apparent absence of a giardial homolog of B32. Biochemical methods (Darcy et al. 1999; Werner et al. 2000) recently identified an archaeal homolog of B32 (subunit F), and subsequent sequence analyses revealed very weak conservation. This finding suggests the existence of a B32/subunit F ancestor prior to the divergence of Archaea and Eucarya; it is possible that Giardia has lost this protein, or, given the low similarity of archaeal and eukaryal B32 sequences (311 PAM units; Table 2), a giardial homolog may exist, but be unrecognizable due to divergence.
The inability to detect particular RNAP subunits (or transcription factors) in the genome of Giardia could be explained by incomplete genome sequence information, true absence (due to loss or ancestral state), or high levels of sequence divergence. The current coverage of the Giardia genome is >98%, assembling into contigs with a total length of 11.4 megabases. Unless the missing RNAP subunit genes reside within contiguous regions of the genome not represented in the shotgun clone library, it is unlikely that greater genome coverage will lead to their identification. Two primary lines of evidence suggest that certain RNAP subunits are truly absent, as opposed to being undetectable due to low levels of sequence similarity. (1) The identification of highly diverged components of RNAPI transcription (A43 and see below) suggests that detection of other components with a similarly large divergence in crown eukaryotes is possible, and (2) a pattern exists with regard to unidentified components. That is, the group of unidentified components is comprised of six of the seven polymerase-specific subunits (Table 1) rather than a random sampling of RNA polymerase subunits.
Thus, the genome survey reveals that Giardia harbors the catalytic core of each eukaryal RNA polymerase, subunits common to the three polymerases and archaeal RNAP, and four sets of noncatalytic, paralogous subunits. However, the lack of clear homologs to many of the unique subunits suggests that Giardia may have diverged before most of the class-specific polymerase subunits arose. If the giardial lineage has evolved analogous proteins to perform one or more of the corresponding functions, the use of a purely bioinformatics (similarity-based) approach could not identify them.
Basal Transcription Initiation Factors
Eukaryotic transcription initiation is mediated by multiple factors specific to each polymerase and represents a complex evolutionary progression to enhance regulation of transcript levels. Despite many differences in detail (for reviews, see Orphanides et al. 1996; Paule and White 2000; Geiduschek and Kassavetis 2001), common or paralogous components are utilized in all three eukaryal systems. The shared components extend across domain boundaries to archaeal transcription initiation (for reviews, see Bell et al. 2001; Soppa 2001). The single factor common to each of the four transcription systems is TBP. In three of the transcription systems (archaeal, RNAPII, and RNAPIII), TBP recognizes specific sequences in promoter regions and facilitates recruitment of RNA polymerase through a second transcription factor (TFB, TFIIB, or BRF, each sharing a common ancestry). Although TBP is required in RNAPI transcription, its role is not understood, and a counterpart to TFB/TFIIB/BRF has not been identified. In Archaea, TBP and TFB alone mediate basal transcription initiation, whereas RNAPI, RNAPII, and RNAPIII each requires additional, system-specific transcription initiation factors.
RNAPII basal transcription factors are universally distributed among the five crown group organisms; however, this is not the case for RNAPI or RNAPIII transcription-initiation factors. RNAPI transcription initiation requires two multisubunit transcription factors serving common functions in both fungi and metazoa, but the polypeptides comprising the factors do not share sequence similarity (Paule and White 2000). These factors are apparently analogous, although homology obscured by high divergence cannot be dismissed. Beyond TBP, the single homologous factor shared among RNAPI transcription systems in crown eukaryotes is the recruitment factor Rrn3 (S. cerevisiae nomenclature). RNAPIII transcription initiation also requires two main factors, TFIIIB and TFIIIC (a third factor, TFIIIA, is required at 5S rRNA promoters) (Geiduschek and Kassavetis 2001). Components of TFIIIB (TBP and BRF) are conserved among crown group organisms, but only two of the six polypeptides comprising TFIIIC share clear homology between fungal and metazoan systems (τ 95 and τ 131 from S. cerevisiae with hTFIIIC63 and hTFIIIC102 from Homo sapiens, respectively). These observations highlight the diversity and complexity of eukaryal transcription, even among crown group organisms (Reichmann et al. 2000; Coulson and Ouzounis 2003).
Distance analyses conducted on homologous transcription factors within the crown group underscore this diversity (Table 3). In general, RNAPII transcription factors are more conserved than either RNAPI or RNAPIII transcription factors. However, RNAPII factors show a wide range of conservation (from 73 PAM units to 349 PAM units), demonstrating the large degree of variation within the universally distributed components of the transcription initiation system (among the crown group). Comparison of distance values between transcription factors (Table 3) and RNAP subunits (Table 2) reveals that transcription factors are, on average, less conserved than RNAP subunits. An exception is TBP, the sole factor common to each of the transcription systems. The conservation of TBP within the crown group (41 PAM units, Table 3) is comparable to conservation of the catalytic subunits of the RNA polymerases (50–80 PAM units, Table 2). This high level of sequence conservation reflects the pivotal role that this protein plays in transcription initiation.
Table 3.
Pairwise Distance Comparisons of Transcription Factorsa
| Transcription Factor | Component of RNAP I, II or III | Crown Distanceb | Crown/Giardia Distance | Crown/Archaea Distancec | Archaea/Giardia Distance | ||
|---|---|---|---|---|---|---|---|
| TBP | I | II | III | 41 | 246 | 123 | 217 |
| TFIIH P90 | II | 73 | 158 | ||||
| TFIIH P80 | II | 74 | 181 | ||||
| TFIIH Cdk7 | II | 92 | 110 | ||||
| TFIIA γ | II | 99 | |||||
| TFIIB | II | 113 | 151 | ||||
| TFIIH P44 | II | 120 | 256 | ||||
| TFIIH P52 | II | 126 | |||||
| TFIIH MAT1 | II | 159 | |||||
| TFIIH P34 | II | 176 | 289 | ||||
| BRF | III | 191 | 259 | 204 | 247 | ||
| TFIIH Cyclin H | II | 199 | |||||
| TFIIF β | II | 210 | |||||
| TFIIA αβ | II | 222 | |||||
| TFIIE β | II | 232 | |||||
| TFIIE α | II | 244 | 298 | ||||
| TFIIH P62 | II | 265 | |||||
| TFIIIC 102 | III | 270 | |||||
| RRN3 | I | 287 | 481 | ||||
| TFIIIC 63 | III | 303 | |||||
| TFIIIA | III | 339 | |||||
| TFIIF α | II | 349 | |||||
| TFIIIB B90 | III | 385 | |||||
Distance is the estimated number of replacements per 100 amino acids. Proteins highlighted in bold have a giardial homolog. Factors are sorted from least to greatest Crown Distance.
Inter-kingdom distances only; the animal—animal and fungus—fungus values are excluded
Archaea are represented by Methanococcus jannaschii and Sulfolobus solfataricus
We used the transcription factors listed in Table 3 and nonhomologous factors known to play a role in transcription systems within the crown group as queries in BLAST and profile analyses against the giardial genome. Components of only four of twelve basal initiation factors (comprised of >40 separate peptides) functioning in RNAPI, RNAPII, and RNAPIII transcription in crown eukaryotes were identified in the genome survey of Giardia (Table 3). This is in contrast to the relatively complete complement of RNAP subunits identified, suggesting that Giardia uses modes of initiation very different from those seen in crown group eukaryotes. This suggestion is buttressed by closer observation of the identified components and by noting conspicuous absences.
At minimum, we expected to identify transcription factors common to Archaea and Eucarya. Of these factors (TBP and the TFB/TFIIB/BRF family), only TBP and BRF were identified. Interestingly, giardial TBP is highly divergent with respect to both archaeal and other eukaryal TBPs (Table 3). Comparative distance analyses indicate that giardial TBP is twice as divergent from crown group TBPs (246 PAM units) as are archaeal TBPs (123 PAM units; Table 3). This divergence is dramatically illustrated in a plot of TBP pairwise distances versus small subunit rRNA distances (Fig. 1G). Moreover, inspection of a multiple alignment of TBP sequences reveals that giardial TBP contains substitutions of key residues important for TATA-element binding (Supplemental Fig. S3). For instance, four intercalating phenylalanines, responsible for inducing a severe DNA bend (Kim et al. 1993a,b), are absolutely conserved from Archaea to crown eukaryotes (with the exception of a replacement of the first phenylalanine by an isoleucine in Plasmodium falciparum and a tyrosine in Cenarchaeum symbiosum). Giardial TBP contains replacements for three of the four phenylalanines; one that is a nonconservative change to serine, removing the intercalating phenolic ring. These data suggest a dramatic relaxation of functional constraints on giardial TBP.
Possibly related to the unusual giardial TBP, we did not detect an ortholog to TFIIB. Instead, queries using TFIIB sequences returned a giardial ORF more similar to BRF, the TFIIB homolog specific for RNAPIII transcription. Phylogenetic analysis of an aligned set of eukaryal TFIIB, BRF, and archaeal TFB proteins, masked to exclude the BRF-specific C-terminal domain, confirms that this giardial ORF is specifically related to eukaryotic BRF sequences (Fig. 2). The strongly supported position of giardial BRF argues for the loss of TFIIB in the giardial lineage. This is the first example of a eukaryotic organism lacking a TFIIB homolog, and given its crucial role in eukaryotic transcription, the result must be viewed with some caution. Although it is possible that the gene for a giardial TFIIB may remain unsequenced, we believe this is unlikely, as both assembled sequence and single-pass sequencing reads have been queried with TFIIB, BRF, and TFB proteins. If Giardia has truly lost TFIIB, one intriguing possibility is that the single giardial BRF plays a dual role in RNAPII and RNAPIII transcription. The demonstrations that (1) TFIIB and the N-terminal, TFIIB-like domain of BRF both interact with the C-terminal stirrup of TPB (Schröder et al. 2003), (2) the N-terminal domain of BRF is sufficient to assemble a transcriptionally active TFIIIB/DNA complex, whereas the absence of this domain in TFIIIB/DNA complexes inhibits transcription (Kassavetis et al. 1998), and (3) mutations in the common Zn-ribbon domains of TFIIB, BRF, and archaeal TFB lead to similar phenotypes (see Shröder et al. 2003, and references therein) suggest a common, critical role in transcription initiation for TFIIB and BRF proteins (Schröder et al. 2003). These data lend support to the hypothesis of a dually functioning giardial BRF, one that would be very interesting to pursue experimentally.
Figure 2.
Phylogeny of archaeal TFB, eukaryal TFIIB, and eukaryal BRF transcription factors. Archaeal TFB sequences are used as the outgroup. Support values for the class V consensus tree (see Methods) are indicated next to each branch present in >50% of the significant topologies. Branch lengths of the consensus tree were estimated using PAML. The scale bar represents 10 amino acid replacements per 100 positions. Abbreviated sequence identifiers are Drosophila melanogaster, Giardia lamblia, Homo sapiens, Methanococcus jannaschii, Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Sulfolobus solfataricus.
The remaining transcription initiation factors are specific to the eukaryal domain, and our survey revealed giardial homologs to only two additional factors—Rrn3 for RNAPI transcription and a subset of TFIIH subunits for RNAPII transcription. Rrn3 recruits RNAPI to promoters by acting as a bridge between subunit A43 of the polymerase and Rrn6 of core factor in S. cerevisiae (Bodem et al. 2000; Moorefield et al. 2000; Peyroche et al. 2000). Beyond the two largest subunits, subunit A43 and factor Rrn3 are the only RNAPI-specific components identified in Giardia. Even though there is very low overall similarity between the giardial ORF and Rrn3, signature motifs identified in higher organisms are present (Moorefield et al. 2000). Thus, it would appear that this mode of recruitment arose early in the evolution of RNAPI transcription, followed by invention of additional RNAPI transcription factors in separate lineages.
TFIIH plays roles in both RNAPII transcription initiation and nucleotide excision repair (NER) in higher eukaryotes (for review, see Egly 2001). The factor is composed of nine subunits, divided into a five-subunit core and a three-subunit CDK-activating kinase (CAK). The remaining subunit is a helicase that bridges CAK with the core TFIIH. The largest core subunit, p90, is the helicase responsible for the ATP-dependent unwinding of promoter DNA. The roles of other core subunits are less clear, but mutations in p44 inhibit promoter escape of RNAPII (Egly 2001). In Giardia, three of the core subunits are present (p90, p44, and p34). Although the core is not entirely conserved, apparently vital components are conserved in subunits p90 and p44. The bridging factor and helicase, p80, is also identified in Giardia, raising the possibility that it recruits an activating kinase.
However, the identity of an activating kinase in Giardia is questionable. The CAK serves to increase the efficiency of transcription through phosphorylation of the C-terminal domain (CTD) of RNAPII. The kinase responsible for phosphorylating the CTD is Cdk7 (Kin28 in yeast), which is regulated by both Cyclin H and Mat1 (Egly 2001). There are over 20 CDK-like ORFs in the giardial genome; however, none of them shows a specific relationship to the Cdk7 clade in phylogenetic analyses (data not shown). In addition, the lack of homologs in Giardia to either cyclin H or Mat1 raises questions regarding further regulation of a kinase component of TFIIH. Nonetheless, one of the CDK-like proteins may be a member of a giardial TFIIH-like complex.
The roles of TFIIH in RNAPII transcription are varied, involving interactions not only with RNA polymerase and other general transcription factors, but also with numerous regulators of transcription (Zurita and Merino 2003). Further underscoring the critical nature of TFIIH, data indicate that TFIIH is essential for RNAPI transcription (Iben et al. 2002), and that TFIIH complexes are dynamically shared among RNAPI, RNAPII, and nucleotide excision repair processes in vivo (Hoogstraten et al. 2002). Consistent with the multiplicity and importance of TFIIH functions is the conservation of all subunits of TFIIH between yeast and human (Takagi et al 2003). In contrast to an earlier report (Coulson and Ouzounis 2003), we also find potential homologs for each TFIIH subunit in plants (Supplemental Table S1). These observations support the hypothesis that TFIIH is an ancient transcription factor, and suggest that the components observed in Giardia could be a minimal structure of TFIIH.
Transcriptional Protein Evolution Rates in Archaea and Eucarya
Each set of homologous proteins is subject to different constraints, and hence, display intrinsic patterns of conservation. To normalize for this effect, we compared the Crown/Giardia and Crown/Archaea divergences with the sequence divergence within the crown group. That is, we normalized the Crown/Giardia and Crown/Archaea distances by the respective intracrown distances (Crown Distance in Tables 2 and 3). For all of the universal proteins except TBP, the geometric average of Crown/Giardia divergences is 1.66 times the intracrown group divergences, reflecting both the greater phylogenetic depth and the accelerated clock of Giardia. The most substantial departure from this ratio is TBP, for which Giardia is 6.0-fold more diverged from the crown group as the latter sequences are from one another (discussed above in terms of the highly modified sequence observed in Giardia). The 10 proteins shared among Eucarya, but not found in Archaea, are on average less conserved than the universal proteins, having 1.56 times the divergence within the crown group. As would be expected for a stable level of functional constraint, their Crown/Giardia divergence is 1.62 times their intracrown group divergence (c.f., 1.66 times for universal proteins). This suggests that although these proteins were new to Eucarya, and they have been changing more than the universal (ancestral) proteins, there is no evidence for a fundamentally different character to their evolution.
The geometric average of Crown/Archaea divergences of universal proteins (less TBP) is 1.74 times that of the intracrown group divergences, not much greater than the ratio for the giardial divergence. The only protein with a higher divergence than TBP (3.00 times the crown distance) is ABC10β (at 3.37 times the intracrown group divergence), but the latter value is almost entirely due to a highly diverged Methanococcus jannaschii sequence, not representative of most Archaea.
If we reverse our reference point and consider divergences of universal proteins (less TBP) from the Archaea, the geometric average of the Giardia protein divergences is 1.19 times that of the Archaea to crown group divergences. Thus, as has been noted for ribosomal RNA (Sogin et al. 1989) and a variety of proteins (Stiller et al. 1998; Wu et al. 2000; Morrison et al. 2001), there has been a systematic tendency for giardial proteins to diverge at a higher rate than those of crown group Eucarya. In this comparison, it is A190, the largest subunit of RNAPI, that is the most extreme outlier (with Giardia 1.90 times more diverged from the Archaea as is the crown group). As discussed previously, this is due to the highly diverged Giardia sequence. The high divergence of the giardial TBP results in its being 1.76 times as diverged from the Archaea as is the crown group. The remaining divergence ratios are relatively tightly clustered, with a range of from 0.92 to 1.46.
In summary, prior to the divergence of Giardia from other eukaryal groups, the transcription apparatus diversified from a single system present in the eukaryal-archaeal common ancestor to yield three distinct systems. In spite of the functional changes, the relative levels of conservation for most of the proteins are constant (give or take a factor of two) throughout this process. The most dramatic exceptions are TBP, A190, and ABC10β, all of which display greater volatility in some portions of their history than in others (Memet et al. 1988; Iwabe et al. 1991).
Conclusions
Model for Evolution of Transcription in Eucarya
Given the evolutionary strategy of elaboration used in Eucarya (that is, the propensity to add complexity to existing core systems), it seems probable that evidence of transition states in various cellular systems might exist in early diverging lineages. Therefore, one of the questions that intrigued us was whether the genome of Giardia would contain evidence of the hypothesized progression from a simpler, single-polymerase transcription system to the highly complex system seen in crown eukaryotes. Although components of the three RNA polymerase systems were identified, we believe that the pattern of presence or absence of specific higher eukaryotic components in Giardia represents evidence of this progression in the evolution of eukaryotic transcription.
The evolution of the eukaryal transcription apparatus could be divided into two major segments. The first is the evolution of the three polymerases and a minimal mode of specific initiation—the evolution of a core cellular process. The second is the evolution of the transcription factors and associated regulatory proteins—the evolution of a peripheral set of functions surrounding the core machinery. We propose that the core of each eukaryal transcription system had evolved before the divergence of Giardia from the main eukaryal line of descent, whereas peripheral functions represent more recent evolutionary innovations.
Two main lines of evidence suggest that the core machinery had evolved before the divergence of Giardia. First, sets of paralogous subunits found in each polymerase are present (A190, B220, and C160; A135, B150, and C128; AC19 and B12.5; AC40 and B44; A43, B16, and C25; A12 and C11), indicating that duplications leading to specific polymerases had occurred. Second, rudimentary machinery for basal initiation appears to have evolved for each of the polymerases. Factor Rrn3 and subunit A43 of RNAPI initiation and BRF and subunit C34 of RNAPIII initiation are present in the genome of Giardia. The conservation of TBP/TFIIB between Archaea and Eucarya leads to the conclusion that this mode of initiation was established before the divergence of these domains.
In contrast, the development of peripheral functions has likely occurred throughout eukaryal evolution. The multiple origins of analogous transcription factors of RNAPI initiation are a clear example of this. Likewise, the high level of divergence seen among many transcription factor subunits of RNAPII initiation within crown group organisms (Table 3) could be indicative of recent evolution, as high rates of change would be expected for proteins whose functions have yet to be fully optimized. Factors of RNAPIII initiation also display a high level of divergence, and the existence of a second TFIIIC in metazoa, which is absent in other crown groups (Geiduschek and Kassavetis 2001), indicates continued emergence of complexity, as do the numerous lineage-specific transcription factors found in plants, animals, and fungi (Reichmann et al. 2000; Coulson and Ouzounis 2003). The apparent absence of giardial homologs to most canonical transcription factors and RNA polymerase-specific subunits is fully consistent with the evolution of these functions subsequent to the divergence of Giardia.
Truly Ancestral or Simply Reductive Evolution?
The data could be interpreted in terms of an alternative hypothesis—the observed transcription system in Giardia is a result of reductive evolution, perhaps caused by a parasitic lifestyle. In an effort to distinguish between these two hypotheses, we compare the giardial case with that of the Microsporidia, focusing on (1) the preponderance of the evidence concerning the evolutionary position of Giardia relative to the evidence for a late emergence of Microsporidia, and (2) the set of identified transcription components in the complete genome of the microsporidian, Encephalitozoon cuniculi.
The deep placement of Diplomonads, Microsporidia, and other protist lineages in early molecular phylogenies has undergone intense scrutiny over the past decade. It has been argued that their deep position is an artifact due to systematic errors in phylogenetic reconstruction methods, specifically, compositional biases of molecular markers and/or long-branch attraction (e.g., Hirt et al. 1999). Although both microsporidial and giardial/diplomonad molecular markers exhibit characteristics that theoretically make them susceptible to long-branch attraction, the evidence leads to very different conclusions in the two cases regarding the effects of long-branch attraction. The “Microsporidia early” observations are almost certainly due to long-branch attraction, whereas the strongest possible statement regarding the “Giardia/diplomonads early” observations is that they are possibly due to long-branch attraction.
The case for Microsporidia can be summarized by three points (for review, see Keeling and Fast 2002). First, there are discrepancies in phylogenetic position (deep divergence vs. late/crown divergence) between early rRNA (Vossbrinck et al. 1987) and elongation factor protein (Kamaishi et al. 1996a,b) analyses and most protein phylogenies (Edlind et al. 1996; Keeling and Doolittle 1996; Germot et al. 1997; Hirt et al. 1997, 1999; Peyretaillade et al. 1998; Fast et al. 1999). Second, Microsporidia are consistently placed specifically with or near the fungal clade in protein phylogenies (see Table 1 of Keeling and Fast 2002). Third, when methods are used to minimize the effects of long-branch attraction (e.g., accounting for among-site rate variation) a consistent, generally well-supported, nonbasal position for Microsporidia is recovered. Further, reanalysis of molecular markers initially supporting a basal position of Microsporidia using the improved phylogenetic methods results in a nonbasal position (generally associated with fungi; Hirt et al. 1999; Van de Peer et al. 2000; Tanabe et al. 2002). These data provide a sound argument for the nonbasal position of Microsporidia, and strongly suggest that initial results were due to long-branch attraction artifacts.
The giardial/diplomonad case can be summarized in a similar manner. First, Giardia is consistently placed among early-branching eukaryotes in both rRNA and protein phylogenies (for review, see Adam 2001). Second, Diplomonads have not consistently associated with any specific, late-emerging phylogenetic groups (see Table 3 in Adam 2001). Third, accounting for among-site rate variation does not result in a consistent, well-supported, nonbasal position of Giardia or relatives (Hirt et al. 1999; Inagaki and Doolittle 2000; Bapteste et al. 2002; Dacks et al. 2002; Inagaki et al. 2003). Nor does reanalysis using improved methods result in altered phylogenetic positions (with respect to early vs. late emergence) for giardial sequences, although bootstrap support is reduced in some cases (Hirt et al. 1999). In fact, the basal position of Giardia/diplomonads is well supported by several protein phylogenies, using the best available phylogenetic methods (Inagaki and Doolittle 2000; Bapteste et al. 2002; Dacks et al. 2002; Inagaki et al. 2003). Rather than asserting that a position basal to most eukaryotes must be long-branch attraction that cannot be overcome by current phylogenetic reconstruction methods (Bapteste et al. 2002), perhaps we should consider the possibility that the data are yielding an accurate recounting of evolutionary history. Thus, whereas the Microsporidia are a good example of the early-divergence as artifact argument, the case for Diplomonads is circumstantial at best. Even so, were it determined that Diplomonads are misplaced in current phylogenies, the inability to identify a relationship with any particular, late-emerging group suggests that they are an independent lineage in an unresolved radiation. Therefore, Giardia would still offer glimpses into the state of cellular systems just prior to the plant/animal/fungal radiation events.
It is also important to divorce the question of the phylogenetic position of Diplomonads from the issues surrounding whether they once harbored mitochondria or not. When it was assumed that they never had mitochondria, it was appealing to note their early divergence (Cavalier-Smith 1983). With questions arising regarding their amitochondrial history (e.g., Roger et al. 1998), this was taken to diminish support for early divergence. One must recognize that even if Diplomonads once had mitochondria, this only diminishes the appeal for an early divergence, not the evidence per se.
The highly reduced genome of Encephalitozoon cuniculi (a mere 1997 genes; much less than the complement of Giardia, and less than many bacteria; Katinka et al. 2001), coupled with the phylogenetic position of Microsporidia, suggests that this is a potentially good example of a late emerging, reduced eukaryotic transcription system. Significantly, the genome sequencing project of Encephalitozoon cuniculi identified a full set of RNA polymerase II general transcription factors (GTFs), including subunits of TFIID (TBP associated factors; Katinka et al. 2001). These results indicate that the fully evolved, highly complex RNAPII transcription system requires each of the GTFs for initiation, even under the immense reductive pressure encountered by Encephalitozoon cuniculi. This is in stark contrast to the absence of RNAPII GTFs in the genome of Giardia, suggesting that the RNAPII transcription system had not attained the complexity of the canonical version found in crown eukaryotes.
Other analyses of transcription in Giardia are consistent with our conclusions. Several studies have recently addressed promoter requirements of protein-encoding giardial transcripts (for review, see Adam 2001; Elmendorf et al. 2001b; Ong et al. 2002). With the exception of the highly expressed variant-specific surface protein (VSP) class of genes, the consensus that is emerging revolves around a generally A-T rich region surrounding the start site of transcription (Holberton and Marshall 1995; Elmendorf et al. 2001b). This element, functionally akin to Inr in higher eukaryotes, is sufficient and required to direct transcription of some genes (Elmendorf et al. 2001b). The regulatory information at these promoters is contained within a very short region upstream of the start site, in many cases <50 bp. Through comparisons of upstream sequences from known giardial promoters, consensus sequences corresponding to TATA-elements found in higher eukaryotes are not observed (Holberton and Marshall 1995; Elmendorf et al. 2001b). However, there appear to be loosely conserved regulatory elements located upstream of the start site, and mutation or deletion of these regions influences levels of transcription (Sun and Tai 1999; Yee et al. 2000; Elmendorf et al. 2001b; Ong et al. 2002). In addition, up to 20% of total cellular polyadenylated RNA isolated from Giardia is comprised of antisense, nonproductive RNA transcripts (Elmendorf et al. 2001a), a finding explained in part by evidence for bidirectional transcription from many giardial promoters, despite the absence of a divergently transcribed upstream gene (H. Elmendorf, pers. comm.). These data suggest relaxed constraints on transcription initiation relative to other Eucarya. Our findings provide a context for this relaxation—adoption of an altered strategy for transcription initiation at protein promoters derived from a primitive RNAPII transcription system. Coupled with our findings that Giardia has a highly degenerate TBP and lacks most of the canonical RNAPII transcription factors (including TFIIB), the data support an altered strategy for transcription initiation at protein promoters in Giardia.
Despite idiosyncrasies such as those just described, the core of each eukaryal transcription system is present in Giardia, whereas peripheral components are absent, arguing for a transitory position of the giardial system in the evolution of eukaryal transcription. The possibility remains that the parasitic lifestyle of Giardia lamblia has led to a reduction of transcription system components, resulting in absence or high divergence. We believe this scenario is unlikely, but it would be of great interest to sequence the genome of a closely related, free-living protist, providing opportunities for comparative analyses to address issues such as these.
METHODS
Sequences of RNA polymerase subunits and transcription factors used in this study were obtained through the Entrez system at the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/Entrez/) (Benson et al. 2002). Ribosomal RNA sequences were retrieved from the Ribosomal Database Project (http://rdp.cme.msu.edu/html/; Maidak et al. 2001). Genome assemblies of Giardia lamblia were from the Giardia genome project (McArthur et al. 2000). ORFs were called using the program CRITICA (Badger and Olsen 1999) and TBLASTN (Altschul et al. 1997). Protein sequences (Supplemental Table S1) from crown group eukaryotes (Arabidopsis thaliana, Drosophila melanogaster, Homo sapiens, Saccharomyces cerevisiae, and Schizosaccharomyces pombe) were used to query Giardia partial genome assemblies using BLAST and profile searches [PSI-BLAST (Altschul et al. 1997) and HMMER (S. Eddy, http://hmmer.wustl.edu/) algorithms] to identify giardial homologs. These organisms were chosen because each has a completed genome sequence and is used routinely in transcriptional studies. Identified transcription components in Giardia were aligned with eukaryal and archaeal (Methanococcus jannaschii and Sulfolobus solfataricus) homologs using ClustalW (version 1.7, http://www.ebi.ac.uk/clustalw; Thompson et al. 1994). Alignments were either used directly in distance analyses or manually refined in the AE editor (T. Macke, unpubl.; Larsen et al. 1993) for subsequent phylogenetic analyses.
Protein pairwise distances were calculated with PROTDIST (J. Felsenstein, PHYLIP version 3.5c, http://evolution.genetics.washington.edu/phylip.html), and the uncertainty due to sampling was estimated from 10 bootstrap samples. Ribosomal RNA pairwise distances were calculated with PAUP* (version 4.0b7, D. Swofford, Sinauer Associates, Inc., http://paup.csit.fsu.edu) under the distance optimality criterion using a Gamma shape parameter estimated from maximum likelihood analysis. Plots were produced with Cricket Graph III (v. 1.01, Computer Associates International, Inc.).
Phylogenetic analyses were performed on selected transcription components using a combination of parsimony and maximum likelihood methods (Marsh et al. 1994). Briefly, the 500 most parsimonious tree topologies were generated in PAUP* using a cost matrix based on BLOSUM45 or BLOSUM62 (Henikoff and Henikoff 1992). These trees were evaluated using maximum likelihood criteria accounting for among-site rate variation (four discreet rate categories and α parameter estimation) with codeml in the PAML package (version 3.13d, http://abacus.gene.ucl.ac.uk/software/paml.html; Yang 1997). Tree topologies were rejected as significantly worse (P > 0.05) than the maximum likelihood topology according to the AU test (Shimodaira 2002) using the program CONSEL (version 0.1f, http://www.is.titech.ac.jp/∼shimo/prog/consel; Shimodaira and Hasegawa 2001). A weighted consensus tree (class V; Jermiin et al. 1997) was constructed from the set of remaining topologies using the programs TreeCons (version 1.0, http://jcsmr.anu.edu.au/dmm/humgen/lars/treeconssub.htm; Jermiin et al. 1997) and Consense (J. Felsenstein, PHYLIP version 3.6a3). Figures were prepared with TreeView (v. 1.6.6, http://taxonomy.zoology.gla.ac.uk/rod/treeview.html; Page 1996).
The A43/B16/C25 protein family phylogeny (Supplemental Fig. S1) was constructed using a combination of distance and maximum likelihood methods as described below. A set of 500 bootstrap replicates were created using Seqboot (J. Felsenstein, PHYLIP version 3.6a3) on a multiple alignment of 62 taxa derived from PSI-BLAST analyses of the putative giardial A43 homolog. Phylogenetic trees were constructed using the neighbor-joining algorithm as implemented in Neighbor (J. Felsenstein, PHYLIP version 3.6a3). The distance matrices used in these reconstructions were created using TREE-PUZZLE (version 5.1, http://www.tree-puzzle.de/; Strimmer and von Haeseler 1996) and PUZZLEBOOT (M. Holder and A. Roger, version 1.03, http://hades.biochem.dal.ca/Rogerlab/Software/), accounting for among-site rate variation. A consensus tree was constructed using Consense (J. Felsenstein, PHYLIP version 3.6a3); branch lengths for the consensus tree were estimated using codeml from the PAML package. Accession numbers of giardial sequences identified as transcription components in the study are as follows: EAA37424.1, EAA38645.1, EAA38649.1, EAA39106.1, EAA39710.l, EAA39886.1, EAA40003.1, EAA40027.1, EAA40465.1, EAA40851.1, EAA41218.1, EAA41267.1, EAA41416.1, EAA41681.1, EAA42291.1, EAA42523.1, EAA42548.1, EAA42665.1. See also Supplemental Table S1.
Acknowledgments
We thank Claudia Reich for insightful discussions and critical reading of the manuscript and Rodney Adam and Heidi Elmendorf for sharing data prior to publication. This work was supported in part by NIH grant AI43273 to M.L.S., by NIH grant AI51089 to A.G.M, and DOE grant DE-FG02-01ER63201 to G.J.O. Additional support was provided by the G. Unger Vetlesen Foundation and LI-COR Biotechnology. Sequences that are part of the genome project can be accessed at the Web site: http://www.mbl.edu/Giardia.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2256604.
Footnotes
[Supplemental material is available online at www.genome.org. The sequences of Giardia lamblia transcription components identified in this study have been submitted to GenBank under accession numbers listed in the Methods section and in Supplemental Table S1. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: H. Elmendorf and R. Adam.]
References
- Adam, R.D. 2001. Biology of Giardia lamblia. Clin. Microbiol. Rev. 14: 447–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts, B., Bray, D., Lewis, J., Raff, M., Roberts, K., and Watson, J.D. 1994. Molecular Biology of the Cell, Third Edition. Garland Publishing, Inc. New York.
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awrey, D.E., Weilbaecher, R.G., Hemming, S.A., Orlicky, S.M., Kane, C.M., and Edwards, A.M. 1997. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem. 272: 14747–14754. [DOI] [PubMed] [Google Scholar]
- Badger, J.H. and Olsen, G.J. 1999. CRITICA: Coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16: 512–524. [DOI] [PubMed] [Google Scholar]
- Bapteste, E., Brinkmann, H., Lee, J.A., Moore, D.V., Sensen, C.W., Gordon, P., Durufle, L., Gaasterland, T., Lopez, P., Muller, M., et al. 2002. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc. Natl. Acad. Sci. 99: 1414–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S.D., Magill, C.P., and Jackson, S.P. 2001. Basal and regulated transcription in Archaea. Biochem. Soc. Trans. 29: 392–395. [DOI] [PubMed] [Google Scholar]
- Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., Rapp, B.A., and Wheeler, D.L. 2002. GenBank. Nucleic Acids Res. 30: 17–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best, A.A. and Olsen, G.J. 2001. Similar subunit architecture of archaeal and eukaryal RNA polymerases. FEMS Microbiol. Lett. 195: 85–90. [DOI] [PubMed] [Google Scholar]
- Bodem, J., Dobreva, G., Hoffmann-Rohrer, U., Iben, S., Zentgraf, H., Delius, H., Vingron, M., and Grummt, I. 2000. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 1: 171–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun, I., Sentenac, A., and Werner, M. 1997. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 16: 5730–5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles, C. and Riva, M. 1998. Yeast RNA polymerase I subunits and genes. In Transcription of ribosomal RNA genes by eukaryotic RNA polymerase I (ed. M.R. Paule), pp. 9–38. Springer-Verlag, New York.
- Cavalier-Smith, T. 1983. A 6-kingdom classification and a unified phylogeny. In Endocytobiology II (eds. H.E.A. Schenk and W. Schwemmler), pp. 1027–1034. De Gruyter, Berlin, Germany.
- Chedin, S., Ferri, M.L., Peyroche, G., Andrau, J.C., Jourdain, S., Lefebvre, O., Werner, M., Carles, C., and Sentenac, A. 1998. The yeast RNA polymerase III transcription machinery: A paradigm for eukaryotic gene activation. Cold Spring Harb. Symp. Quant. Biol. 63: 381–389. [DOI] [PubMed] [Google Scholar]
- Coulson, R.M.R. and Ouzounis, C.A. 2003. The phylogenetic diversity of eukaryotic transcription. Nucleic Acids Res. 31: 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks, J.B., Marinets, A., Ford Doolittle, W., Cavalier-Smith, T., and Logsdon Jr., J.M. 2002. Analyses of RNA polymerase II genes from free-living protists: Phylogeny, long branch attraction, and the eukaryotic big bang. Mol. Biol. Evol. 19: 830–840. [DOI] [PubMed] [Google Scholar]
- Darcy, T.J., Hausner, W., Awery, D.E., Edwards, A.M., Thomm, M., and Reeve, J.N. 1999. Methanobacterium thermoautotrophicum RNA polymerase and transcription in vitro. J. Bacteriol. 181: 4424–4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff, M.O., Schwart, R.M., and Orcutt, B.C. 1978. A model of evolutionary change in proteins. In: Atlas of protein sequence and structure 5 (ed. M.O. Dayhoff), pp. 345–352. National Biomedical Research Foundation, Washington, DC.
- Edlind, T.D., Li, J., Visvesvara, G.S., Vodkin, M.H., McLaughlin, G.L., and Katiyar, S.K. 1996. Phylogenetic analysis of b-tubulin sequences from amitochondrial protozoa. Mol. Phylogen. Evol. 5: 359–367. [DOI] [PubMed] [Google Scholar]
- Egly, J.M. 2001. The 14th Datta lecture. TFIIH: From transcription to clinic. FEBS Lett. 498: 124–128. [DOI] [PubMed] [Google Scholar]
- Elmendorf, H.G., Singer, S.M., and Nash, T.E. 2001a. The abundance of sterile transcripts in Giardia lamblia. Nucleic Acids Res. 29: 4674–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmendorf, H.G., Singer, S.M., Pierce, J., Cowan, J., and Nash, T.E. 2001b. Initiator and upstream elements in the a2-tubulin promoter of Giardia lamblia. Mol. Biochem. Parasitol. 113: 157–169. [DOI] [PubMed] [Google Scholar]
- Eloranta, J.J., Kato, A., Teng, M.S., and Weinzierl, R.O. 1998. In vitro assembly of an archaeal D-L-N RNA polymerase subunit complex reveals a eukaryote-like structural arrangement. Nucleic Acids Res. 26: 5562–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast, N., Logsdon Jr., J., and Doolittle, W. 1999. Phylogenetic analysis of the TATA box binding protein (TBP) gene from Nosema locustae: Evidence for a microsporidia-fungi relationship and spliceosomal intron loss. Mol. Biol. Evol. 16: 1415–1419. [DOI] [PubMed] [Google Scholar]
- Geiduschek, E.P. and Kassavetis, G.A. 2001. The RNA polymerase III transcription apparatus. J. Mol. Biol. 310: 1–26. [DOI] [PubMed] [Google Scholar]
- Germot, A., Philippe, H., and Le Guyader, H. 1997. Evidence for loss of mitochondria in Microsporidia from a mitochondrial-type HSP70 in Nosema locustae. Mol. Biochem. Parasitol. 87: 159–168. [DOI] [PubMed] [Google Scholar]
- Gillin, F.D., Reiner, D.S., and McCaffery, J.M. 1996. Cell biology of the primitive eukaryote Giardia lamblia. Annu. Rev. Microbiol. 50: 679–705. [DOI] [PubMed] [Google Scholar]
- Hashimoto, T., Nakamura, Y., Nakamura, F., Shirakura, T., Adachi, J., Goto, N., Okamoto, K., and Hasegawa, M. 1994. Protein phylogeny gives a robust estimation for early divergences of eukaryotes: Phylogenetic place of a mitochondria-lacking protozoan, Giardia lamblia. Mol. Biol. Evol. 11: 65–71. [DOI] [PubMed] [Google Scholar]
- Hashimoto, T., Nakamura, Y., Kamaishi, T., Nakamura, F., Adachi, J., Okamoto, K., and Hasegawa, M. 1995. Phylogenetic place of mitochondrion-lacking protozoan, Giardia lamblia, inferred from amino acid sequences of elongation factor 2. Mol. Biol. Evol. 12: 782–793. [DOI] [PubMed] [Google Scholar]
- Hausner, W., Wettach, J., Hethke, C., and Thomm, M. 1996. Two transcription factors related with the eucaryal transcription factors TATA-binding protein and transcription factor IIB direct promoter recognition by an archaeal RNA polymerase. J. Biol. Chem. 271: 30144–30148. [DOI] [PubMed] [Google Scholar]
- Hemming, S.A. and Edwards, A.M. 2000. Yeast RNA polymerase II subunit RPB9: Mapping of domains required for transcription elongation. J. Biol. Chem. 275: 2288–2294. [DOI] [PubMed] [Google Scholar]
- Hemming, S.A., Jansma, D.B., Macgregor, P.F., Goryachev, A., Friesen, J.D., and Edwards, A.M. 2000. RNA polymerase II subunit Rpb9 regulates transcription elongation in vivo. J. Biol. Chem. 275: 35506–35511. [DOI] [PubMed] [Google Scholar]
- Henikoff, S. and Henikoff, J.G. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. 89: 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt, R.P., Healy, B., Vossbrinck, C.R., Canning, E.U., and Embley, T.M. 1997. A mitochondrial Hsp70 orthologue in Vairimorpha necatrix: Molecular evidence that microsporidia once contained mitochondria. Curr. Biol. 7: 995–998. [DOI] [PubMed] [Google Scholar]
- Hirt, R.P., Logsdon Jr., J.M., Healy, B., Dorey, M.W., Doolittle, W.F., and Embley, T.M. 1999. Microsporidia are related to Fungi: Evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl. Acad. Sci. 96: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holberton, D.V. and Marshall, J. 1995. Analysis of consensus sequence patterns in Giardia cytoskeleton gene promoters. Nucleic Acids Res. 23: 2945–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogstraten, D., Nigg, A.L., Heath, H., Mullenders, L.H., van Driel, R., Hoeijmakers, J.H., Vermeulen, W., and Houtsmuller, A.B. 2002. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell 10: 1163–1174. [DOI] [PubMed] [Google Scholar]
- Huang, Y. and Maraia, R.J. 2001. Comparison of the RNA polymerase III transcription machinery in Schizosaccharomyces pombe, Saccharomyces cerevisiae and human. Nucleic Acids Res. 29: 2675–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iben, S., Tschochner, H., Bier, M., Hoogstraten, D., Hozak, P., Egly, J.M., and Grummt, I. 2002. TFIIH plays an essential role in RNA polymerase I transcription. Cell 109: 297–306. [DOI] [PubMed] [Google Scholar]
- Imazawa, Y., Imai, K., Yao, Y., Yamamoto, K., Hisatake, K., Muramatsu, M., and Nogi, Y. 2001. Isolation and characterization of the fission yeast gene Sprpa12+ reveals that the conserved C-terminal zinc-finger region is dispensable for the function of its product. Mol. Gen. Genet. 264: 852–859. [DOI] [PubMed] [Google Scholar]
- Inagaki, Y. and Doolittle, W.F. 2000. Evolution of the eukaryotic translation termination system: Origins of release factors. Mol. Biol. Evol. 17: 882–889. [DOI] [PubMed] [Google Scholar]
- Inagaki, Y., Blouin, C., Susko, E., and Roger, A.J. 2003. Assessing functional divergence in EF-1a and its paralogs in eukaryotes and archaebacteria. Nucleic Acids Res. 31: 4227–4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama, A., Kimura, M., and Mitsuzawa, H. 1998. Subunits of yeast RNA polymerases: Structure and function. Curr. Opin. Microbiol. 1: 190–196. [DOI] [PubMed] [Google Scholar]
- Iwabe, N., Kuma, K., Kishino, H., Hasegawa, M., and Miyata, T. 1991. Evolution of RNA polymerases and branching patterns of the three major groups of Archaebacteria. J. Mol. Evol. 32: 70–78. [DOI] [PubMed] [Google Scholar]
- Jermiin, L., Olsen, G., Mengerson, K., and Easteal, S. 1997. Majority-rule consensus of phylogenetic trees obtained by maximum-likelihood analysis. Mol. Biol. Evol. 14: 1296–1302. [Google Scholar]
- Kaine, B.P., Mehr, I.J., and Woese, C.R. 1994. The sequence, and its evolutionary implications, of a Thermococcus celer protein associated with transcription. Proc. Natl. Acad. Sci. 91: 3854–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaishi, T., Hashimoto, T., Nakamura, F., Murata, S., Okada, N., Okamoto, K., Shimizu, M., and Hasegawa, M. 1996a. Protein phylogeny of translation elongation factor EF-1a suggests microsporidians are extremely ancient eukaryotes. J. Mol. Evol. 42: 257–263. [DOI] [PubMed] [Google Scholar]
- Kamaishi, T., Hashimoto, T., Nakamura, Y., Masuda, Y., Nakamura, F., Okamoto, K., Shimizu, M., and Hasegawa, M. 1996b. Complete nucleotide sequences of the genes encoding translation elongation factors 1a and 2 from a microsporidian parasite, Glugea plecoglossi: Implications for the deepest branching of eukaryotes. J. Biochem. (Tokyo) 120: 1095–1103. [DOI] [PubMed] [Google Scholar]
- Kassavetis, G.A., Kumar, A., Ramirez, E., and Geidusche, E.P. 1998. The functional and structural organization of Brf, the TFIIB-related component of the RNA polymerase III transcription initiation complex. Mol. Cell. Biol. 18: 5587–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka, M.D., Duprat, S., Cornillot, E., Metenier, G., Thomarat, F., Prensier, G., Barbe, V., Peyretaillade, E., Brottier, P., Wincker, P., et al. 2001. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414: 450–453. [DOI] [PubMed] [Google Scholar]
- Keeling, P. and Doolittle, W. 1996. α-tubulin from early-diverging eukaryotic lineages and the evolution of the tubulin family. Mol. Biol. Evol. 13: 1297–1305. [DOI] [PubMed] [Google Scholar]
- Keeling, P.J. and Fast, N.M. 2002. MICROSPORIDIA: Biology and evolution of highly reduced intracellular parasites. Annu. Rev. Microbiol. 56: 93–116. [DOI] [PubMed] [Google Scholar]
- Kim J.L., Nikolov, D.B., and Burley, S.K. 1993a. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 365: 520–527. [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger, J.H., Hahn, S., and Sigler, P.B. 1993b. Crystal structure of a yeast TBP/TATA-box complex. Nature 365: 512–520. [DOI] [PubMed] [Google Scholar]
- Klenk, H.P., Zillig, W., Lanzendorfer, M., Grampp, B., and Palm, P. 1995. Location of protist lineages in a phylogenetic tree inferred from sequences of DNA-dependent RNA polymerases. Archiv. Protisten. Kunde 145: 221–230. [Google Scholar]
- Langer, D., Hain, J., Thuriaux, P., and Zillig, W. 1995. Transcription in Archaea: Similarity to that in Eucarya. Proc. Natl. Acad. Sci. 92: 5768–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzendorfer, M., Palm, P., Grampp, B., Peattie, D.A., and Zillig, W. 1992. Nucleotide sequence of the gene encoding the largest subunit of the DNA-dependent RNA polymerase III of Giardia lamblia. Nucleic Acids Res. 20: 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen, N., Olsen, G.J., Maidak, B.L., McCaughey, M.J., Overbeek, R., Macke, T.J., Marsh, T.L., and Woese, C.R. 1993. The ribosomal database project. Nucleic Acids Res. 21: 3021–3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maidak, B.L., Cole, J.R., Lilburn, T.G., Parker Jr., C.T., Saxman, P.R., Farris, R.J., Garrity, G.M., Olsen, G.J., Schmidt, T.M., and Tiedje, J.M. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29: 173–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, T.L., Reich, C.I., Whitelock, R.B., and Olsen, G.J. 1994. Transcription factor IID in the Archaea: Sequences in the Thermococcus celer genome would encode a product closely related to the TATA-binding protein of eukaryotes. Proc. Natl. Acad. Sci. 91: 4180–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur, A.G., Morrison, H.G., Nixon, J.E., Passamaneck, N.Q., Kim, U., Hinkle, G., Crocker, M.K., Holder, M.E., Farr, R., Reich, C.I., et al. 2000. The Giardia genome project database. FEMS Microbiol. Lett. 189: 271–273. [DOI] [PubMed] [Google Scholar]
- McCaffery, J.M. and Gillin, F.D. 1994. Giardia lamblia: Ultrastructural basis of protein transport during growth and encystation. Exp. Parasitol. 79: 220–235. [DOI] [PubMed] [Google Scholar]
- McCaffery, J.M., Faubert, G.M., and Gillin, F.D. 1994. Giardia lamblia: Traffic of a trophozoite variant surface protein and a major cyst wall epitope during growth, encystation, and antigenic switching. Exp. Parasitol. 79: 236–249. [DOI] [PubMed] [Google Scholar]
- Memet, S., Saurin, W., and Sentenac, A. 1988. RNA polymerases B and C are more closely related to each other than to RNA polymerase A. J. Biol. Chem. 263: 10048–10051. [PubMed] [Google Scholar]
- Moorefield, B., Greene, E.A., and Reeder, R.H. 2000. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl. Acad Sci. 97: 4724–4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, H.G., Roger, A.J., Nystul, T.G., Gillin, F.D., and Sogin, M.L. 2001. Giardia lamblia expresses a proteobacterial-like DnaK homolog. Mol. Biol. Evol. 18: 530–541. [DOI] [PubMed] [Google Scholar]
- Ong, S.J., Huang, L.C., Liu, H.W., Chang, S.C., Yang, Y.C., Bessarab, I., and Tai, J.H. 2002. Characterization of a bi-directional promoter for divergent transcription of a PHD-zinc finger protein gene and a ran gene in the protozoan pathogen Giardia lamblia. Mol. Microbiol. 43: 665–676. [DOI] [PubMed] [Google Scholar]
- Orphanides, G., Lagrange, T., and Reinberg, D. 1996. The general transcription factors of RNA polymerase II. Genes & Dev. 10: 2657–2683. [DOI] [PubMed] [Google Scholar]
- Page, R.D. 1996. TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12: 357–358. [DOI] [PubMed] [Google Scholar]
- Paule, M.R. and White, R.J. 2000. Survey and summary: Transcription by RNA polymerases I and III. Nucleic Acids Res. 28: 1283–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyretaillade, E., Biderre, C., Peyret, P., Duffieux, F., Metenier, G., Gouy, M., Michot, B., and Vivares, C. 1998. Microsporidian Encephalitozoon cuniculi, a unicellular eukaryote with an unusual chromosomal dispersion of ribosomal genes and a LSU rRNA reduced to the universal core. Nucleic Acids Res. 26: 3513–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche, G., Milkereit, P., Bischler, N., Tschochner, H., Schultz, P., Sentenac, A., Carles, C., and Riva, M. 2000. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 19: 5473–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, X., Jeon, H., Agarwal, K., and Weiss, M.A. 1993. The Zn ribbon: Structure of the nucleic-acid binding domain of elongation factor TFIIS. Nature 365: 277–279. [DOI] [PubMed] [Google Scholar]
- Reichmann, J.L., Heard, J., Martin, G., Reuber, L., Jiang, C.-Z., Keddie, J., Adam, L., Pineda, O., Ratcliffe, O.J., Samaha, R.R., et al. 2000. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290: 2105–2110. [DOI] [PubMed] [Google Scholar]
- Roger, A.J., Svard, S.G., Tovar, J., Clark, C.G., Smith, M.W., Gillin, F.D., and Sogin, M.L. 1998. A mitochondrial-like chaperonin 60 gene in Giardia lamblia: Evidence that diplomonads once harbored an endosymbiont related to the progenitor of mitochondria. Proc. Natl. Acad. Sci. 95: 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhale, P.P. and Woychik, N.A. 1994. C25, an essential RNA polymerase III subunit related to the RNA polymerase II subunit RPB7. Mol. Cell. Biol. 14: 6164–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder, O., Bryant, G.O., Geiduschek, E.P., Berk, A.J., and Kassevetis, G.A. 2003. A common site on TBP for transcription by RNA polymerases II and III. EMBO J. 22: 5115–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri, V., McArthur, A.G., Sogin, M.L., and Adam, R.D. 2003. Giardia lamblia RNA polymerase II: Amanitin-resistant transcription. J. Biol. Chem. 278: 27804–27810. [DOI] [PubMed] [Google Scholar]
- Shematorova, E.K. and Shpakovski, G.V. 2002. Structure and functions of eukaryotic nuclear DNA-dependent RNA polymerase I. Mol. Biol. 36: 3–26. [PubMed] [Google Scholar]
- Shimodaira, H. 2002. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51: 492–508. [DOI] [PubMed] [Google Scholar]
- Shimodaira, H. and Hasegawa, M. 2001. CONSEL: For assessing the confidence of phylogenetic tree selection. Bioinformatics 17: 1246–1247. [DOI] [PubMed] [Google Scholar]
- Shpakovskii, G.V. and Shematorova, E.K. 1999. Characteristics of the cDNA of the Schizosaccharomyces pombe rpa43+ gene: Structural similarity of the Rpa43 subunit of RNA-polymerase I with the Rpc25 subunit of RNA-polymerase III. Bioorg. Khim. 25: 791–796. [PubMed] [Google Scholar]
- Smith, M.W., Aley, S.B., Sogin, M., Gillin, F.D., and Evans, G.A. 1998. Sequence survey of the Giardia lamblia genome. Mol. Biochem. Parasitol. 95: 267–280. [DOI] [PubMed] [Google Scholar]
- Sogin, M.L., Gunderson, J.H., Elwood, H.J., Alonso, R.A., and Peattie, D.A. 1989. Phylogenetic meaning of the kingdom concept: An unusual ribosomal RNA from Giardia lamblia. Science 243: 75–77. [DOI] [PubMed] [Google Scholar]
- Soppa, J. 2001. Basal and regulated transcription in Archaea. Adv. Appl. Microbiol. 50: 171–217. [DOI] [PubMed] [Google Scholar]
- Stiller, J.W., Duffield, E.C., and Hall, B.D. 1998. Amitochondriate amoebae and the evolution of DNA-dependent RNA polymerase II. Proc. Natl. Acad. Sci. 95: 11769–11774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer, K. and von Haeseler, A. 1996. Quartet puzzling: A quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13: 964–969. [Google Scholar]
- Sun, C.H. and Tai, J.H. 1999. Identification and characterization of a ran gene promoter in the protozoan pathogen Giardia lamblia. J. Biol. Chem. 274: 19699–19706. [DOI] [PubMed] [Google Scholar]
- Takagi, Y., Komori, H., Chang, W.-H., Hudmon, A., Erdjument-Bromage, H., Tempst, P., and Kornberg, R.D. 2003. Revised subunit structure of yeast transcription factor IIH (TFIIH) and reconciliation with human TFIIH. J. Biol. Chem. 278: 43897–43900. [DOI] [PubMed] [Google Scholar]
- Tanabe, Y., Watanabe, M.M., and Sugiyama, J. 2002. Are Microsporidia really related to Fungi?: A reappraisal based on additional gene sequences from basal fungi. Mycol. Res. 106: 1380–1391. [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuillier, V., Stettler, S., Sentenac, A., Thuriaux, P., and Werner, M. 1995. A mutation in the C31 subunit of Saccharomyces cerevisiae RNA polymerase III affects transcription initiation. EMBO J. 14: 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer, Y., Ben Ali, A., and Meyer, A. 2000. Microsporidia: Accumulating molecular evidence that a group of amitochondriate and suspectedly primitive eukaryotes are just curious fungi. Gene 246: 1–8. [DOI] [PubMed] [Google Scholar]
- Van Mullem, V., Landrieux, E., Vandenhaute, J., and Thuriaux, P. 2002a. Rpa12p, a conserved RNA polymerase I subunit with two functional domains. Mol. Microbiol. 43: 1105–1113. [DOI] [PubMed] [Google Scholar]
- Van Mullem, V., Wery, M., Werner, M., Vandenhaute, J., and Thuriaux, P. 2002b. The Rpb9 subunit of RNA polymerase II binds transcription factor TFIIE and interferes with the SAGA and Elongator histone acetyltransferases. J. Biol. Chem. 277: 10220–10225. [DOI] [PubMed] [Google Scholar]
- Vossbrinck, C.R., Maddox, J.V., Friedman, S., Debrunner-Vossbrinck, B.A., and Woese, C.R. 1987. Ribosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature 326: 411–414. [DOI] [PubMed] [Google Scholar]
- Werner, M., Chaussivert, N., Willis, I.M., and Sentenac, A. 1993. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J. Biol. Chem. 268: 20721–20724. [PubMed] [Google Scholar]
- Werner, F., Eloranta, J.J., and Weinzierl, R.O. 2000. Archaeal RNA polymerase subunits F and P are bona fide homologs of eukaryotic RPB4 and RPB12. Nucleic Acids Res. 28: 4299–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, G., McArthur, A.G., Fiser, A., Sali, A., Sogin, M.L., and Mllerm, M. 2000. Core histones of the amitochondriate protist, Giardia lamblia. Mol. Biol. Evol. 17: 1156–1163. [DOI] [PubMed] [Google Scholar]
- Yang, Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yee, J., Mowatt, M.R., Dennis, P.P., and Nash, T.E. 2000. Transcriptional analysis of the glutamate dehydrogenase gene in the primitive eukaryote, Giardia lamblia: Identification of a primordial gene promoter. J. Biol. Chem. 275: 11432–11439. [DOI] [PubMed] [Google Scholar]
- Zillig, W., Klenk, H.P., Palm, P., Puhler, G., Gropp, F., Garrett, R.A., and Leffers, H. 1989. The phylogenetic relations of DNA-dependent RNA polymerases of archaebacteria, eukaryotes, and eubacteria. Can. J. Microbiol. 35: 73–80. [DOI] [PubMed] [Google Scholar]
- Zurita, M. and Merino, C. 2003. The transcriptional complexity of the TFIIH complex. Trends Genet. 19: 578–584. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/Entrez/; Entrez Query Page, National Center for Biotechnology Information.
- http://rdp.cme.msu.edu/html/; Ribosomal Database Project homepage.
- http://hmmer.wustl.edu/; HMMER Homepage.
- http://www.ebi.ac.uk/clustalw; European Bioinformatics Institute ClustalW page.
- http://evolution.genetics.washington.edu/phylip.html; PHYLIP homepage.
- http://paup.csit.fsu.edu; PAUP* homepage.
- http://abacus.gene.ucl.ac.uk/software/paml.html; PAML homepage.
- http://www.is.titech.ac.jp/∼shimo/prog/consel; CONSEL homepage.
- http://jcsmr.anu.edu.au/dmm/humgen/lars/treeconssub.htm; TreeCons homepage.
- http://taxonomy.zoology.gla.ac.uk/rod/treeview.html; Treeview homepage.
- http://www.tree-puzzle.de/; TREE-PUZZLE homepage.
- http://hades.biochem.dal.ca/Rogerlab/Software/; PUZZLE-BOOT distribution page.
- http://www.mbl.edu/Giardia; Giardia lamblia genome sequencing project homepage.