Abstract
A tumor needs to initiate angiogenesis in order to develop its own blood supply, to grow, to invade, and to spread. Angiogenesis, under normal conditions, is a tightly regulated balance between endogenous pro- and antiangiogenic factors. In this study, we investigated, by microarray analysis, the effects of two known antiangiogenic agents (endostatin and fumagillin) on the gene expression profiles of human umbilical vein endothelial cells (HUVEC) in order to elucidate pathways common to the effects of these agents. We observed a majority of gene expression changes within 1 and 2 h of treatment. The genes demonstrating these early expression changes are involved in cell proliferation, gene transcription, and a number have unknown functions. We selected four genes (DOC1, KLF4, TC-1, ID1) from the microarray profile that showed a similar pattern of expression for both of the antiangiogenic agents we tested. We then used small interfering RNAs (siRNA) in an attempt to better understand the role of these selected genes in the inhibitory activity of these agents. Because the gene expression changes occurred within 1 and 2 h of treatment, these genes might be involved in the initial pathways of angiogenesis inhibition.
Carcinogenesis is a complex process, involving a number of different genetic events that lead a cell to acquire a malignant phenotype. However, these neoplastic properties may only be necessary, but not sufficient for the cancer cell to become invasive. A tumor also needs to initiate angiogenesis to develop its own blood supply and to invade and spread.
Angiogenesis is the formation of new blood vessels from pre-existing ones, and is essential for normal physiological processes such as reproduction, development, and repair. This process is a cascade of events involving endothelial cells, which, under normal conditions, is a tightly regulated balance between endogenous pro- and antiangiogenic factors (Jekunen and Kairemo 2003). The switch to an angiogenic phenotype involves a change in the equilibrium between these factors (Madhusudan and Harris 2002). The exposure of endothelial cells to proangiogenic factors leads to subsequent angiogenic processes characterized by invasion, migration, and proliferation of endothelial cells. This mechanism is reliant on the degradation of the extracellular matrix surrounding a tumor. The formation of new blood vessels is then determined by the sprouting from pre-existing microvessels, and the formation of tubes that can carry blood flow. The angiogenic switch is characterized by oncogene-driven tumor expression of angiogenic proteins such as VEGF, bFGF, IL8, PLGF, and TGF-β. Hypoxia can also stimulate the up-regulation of angiogenic factors by the down-regulation of angiogenesis suppressors, such as thrombospondin or endostatin (Kerbel and Folkman 2002). Tumors unable to induce angiogenesis remain dormant at a microscopic in situ size (Folkman 2000).
Endothelial cells proliferate much faster during angiogenesis than in normal tissues, and this characteristic makes them more sensitive to a wide variety of drugs. Inhibiting tumor angiogenesis is a promising strategy for treating cancer. Positive regulators are the best characterized, but many inhibitors have also been identified. The mechanism of action of these inhibitory agents has not yet been entirely elucidated. Various angiogenesis inhibitors have been developed to target endothelial cells and block tumor angiogenesis, and several of them have reached clinical trials. Hundreds of potential angiogenesis inhibitors have been identified and the pace of discovery is accelerating.
Although there are many potential targets for inhibiting angiogenesis, it is possible to classify antiagiogenic agents on the basis of their mechanisms of action. The two classes of angiogenesis inhibitors that have been described are indirect and direct inhibitors. Examples of indirect inhibitors are drugs able to target the matrix and the metalloproteinases (MMP). MMPs are crucial for tumor growth, migration, angiogenesis, invasion, and metastasis, because they degrade the extracellular matrix (Egeblad and Werb 2002). Thus, tissue inhibitors of MMPs (TIMPs) are used to inhibit this process, as are agents such as antisense oligonucleotide tyrosine kinase blockers, collagen peptidomimetics (Batismastata), heparanase inhibitors, or urokinase inhibitors (Madhusudan and Harris 2002). There are also drugs that target proangiogenic growth factors such as VEGF inhibitors, FGF2 inhibitors, oncogenic targets, and cytokine inhibitors (Madhusudan and Harris 2002). Direct inhibitors include compounds that target the vascular endothelium directly, such as angiostatin, endostatin, thrombospondin, tumstatin, and vitaxin. They are all involved in inhibiting endothelial cell migration, proliferation, and neo-vascularization (O'Reilly et al. 1994; Dixelius et al. 2002, 2003; Kerbel and Folkman 2002).
Many patients with cancer have received some form of experimental antiangiogenic therapy. Nevertheless, the impressive antitumor results obtained in murine models have not been entirely reproducible in patients (Madhusudan and Harris 2002). One reason for this may be that the molecular mechanisms for many of these antiangiogenic compounds are still not well understood.
In this study, we investigated the early effects of the antiangiogenic agents, endostatin and fumagillin, on the gene expression profiles in human umbilical vein endothelial cells (HUVEC) by microarray analysis.
Endostatin is a proteolityc fragment of a naturally occurring protein. It is the 20-kDa C-terminal fragment of collagen XVIII, and it was one of the first endogenous angiogenesis inhibitors to enter clinical trials (O'Reilly et al. 1997; Dixelius et al. 2003). Endostatin results in the inhibition of endothelial cell proliferation, migration, invasion, and tube formation. Several studies reported that recombinant endostatin was a potent inhibitor of endothelial cell proliferation in vitro, and in a large number of animal tumor studies, endostatin treatment resulted in inhibition of tumor growth, and in some cases, a decrease in the formation of metastasis was observed (Boehm et al. 1997; Sauter et al. 2000). Recombinant endostatin is currently being evaluated in the clinic (Herbst et al. 2001). Endostatin also affects vascular permeability (Hajitou et al. 2002). Additionally, it is involved in chronic inflammatory diseases and diabetic retinopathy, whereas, at the same time, not effecting normal wound healing (Abdollahi et al. 2004).
MacDonald et al. (2001) showed that recombinant endostatin binds tropomyosin in vitro in endothelial cells, resulting in the disruption of microfilament integrity (MacDonald et al. 2001). Recently, endostatin was shown to have an association with the integrin α5β1 and caveolin-1, inhibiting extracellular deposition of fibronectin, and consequently affecting migration (Wickstrom et al. 2002). The results of Calvo et al. (2002) are consistent with the notion that the major effect of endostatin is the prevention of cell recruitment, migration, and vessel formation. Different studies have shown that the inhibitory action of endostatin is also involved in the blockade of VEGF/VEGFR signaling (Kim et al. 2002), inhibition of metalloproteinases, and down-regulation of c-myc and cyclin D1. Recently, Abdollahi et al. (2004) identified an integrated endothelial response network consisting of well-known and not well-characterized pathways, which act in a orchestrated way by suppressing proangiogenic factors (ID1) together with up-regulating their antagonists (thrombospondin).
Fumagillin (a natural product from Aspergillus fumigatus) and its synthetic analog TNP-470, were found to block angiogenesis both in vitro and in vivo by directly inhibiting endothelial cells (Ingber et al. 1990). Fumagillin inhibits HUVEC proliferation at low nM concentration, and it has partial or no effect on non-endothelial cells at concentrations up to 1 μM (J. Wang et al. 2000). It was proposed that fumagillin inhibits angiogenesis by covalently binding to the His-231 position of the enzyme MetAP2 (Griffith et al. 1997; Sin et al. 1997), and this finding provided a starting point for the rational design of fumagillin analogs (Rodeschini et al. 2004).
Endostatin and fumagillin were shown to target the endothelial cell cytoskeleton through altered regulation of heat-shock protein 27 and cofilin (Keezer et al. 2003). The actions of these two different angiogensis inhibitors seem to act via distinct pathways. However, the early responses (within 4 h) of endothelial cells to these agents have not been previously studied. To determine whether there were common early responses in endothelial cells to these agents, we treated endothelial cells and fibroblasts with endostatin and fumagillin and compared changes in gene expression using cDNA microarrays. To further identify genes specific to the activity of these compounds on endothelial cells, we also exposed cells to the cytotoxic agent 5-Fluorouracil (5-FU). Using this approach, we identified a group of genes whose expression patterns were commonly altered specifically in endothelial cells by endostatin and fumagillin. Small interfering RNAs (siRNAs) represents a very promising method for studying loss of function phenotypes by inducing efficient silencing of a target gene (McManus and Sharp 2002). In our study, we utilized this technique to obtain some insight into the interaction among some of the genes we identified.
RESULTS
HUVEC Proliferation Experiments
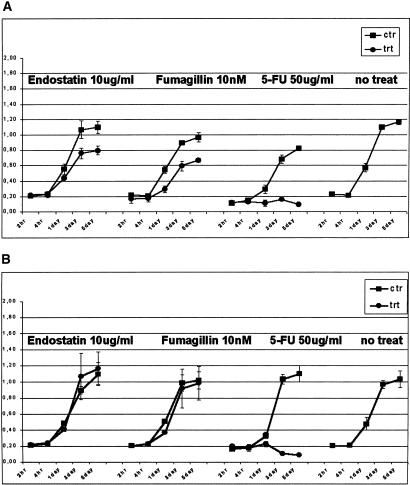
Endostatin (10 μg/mL), and fumagillin (10 nM) significantly inhibited the proliferation of HUVEC cells compared with human fibroblasts treated for 2 and 4 h, and 1, 2, and 5 d, whereas 5FU was cytotoxic for both (Fig. 1A,B). At 1 d, it is already possible to see a significant inhibition of proliferation for the cells treated with the two different antiangiogenic agents. 5FU was found to be cytotoxic at day 1.
Figure 1.
Effects of different antiangiogenic agents on cell proliferation using WST1 assay for HUVECs (A) and human fibroblasts (B). Between 2 and 4 × 103 P2-P3 cells were plated in a 96-well tray, and treated for 2–4 h, and then 1, 3, and 5 d with 10 μg/mL endostatin, 10 nM fumagillin, and 50 μg/mL 5-FU. In A, it is possible to see a significant difference in proliferation between the treated and nontreated cells for all antiangiogenic agents at day 1. The black line indicates the proliferation rate of cells treated with medium and carrier (DMSO for fumagillin and 5FU, a citrate-phosphate buffer for endostatin, see Methods). The gray lines represent the proliferation rate of cells treated with each different reagent. The line on the far right represents cells growing in medium alone. Each data point represents the average of three experiments. The values on the y-axis are the actual WST1 fluorescent intensity readings. Endostatin and fumagillin specifically inhibited the proliferation of endothelial cells, whereas 5FU was cytotoxic for both cell types.
Endostatin and Fumagillin Effect on HUVEC Gene Expression
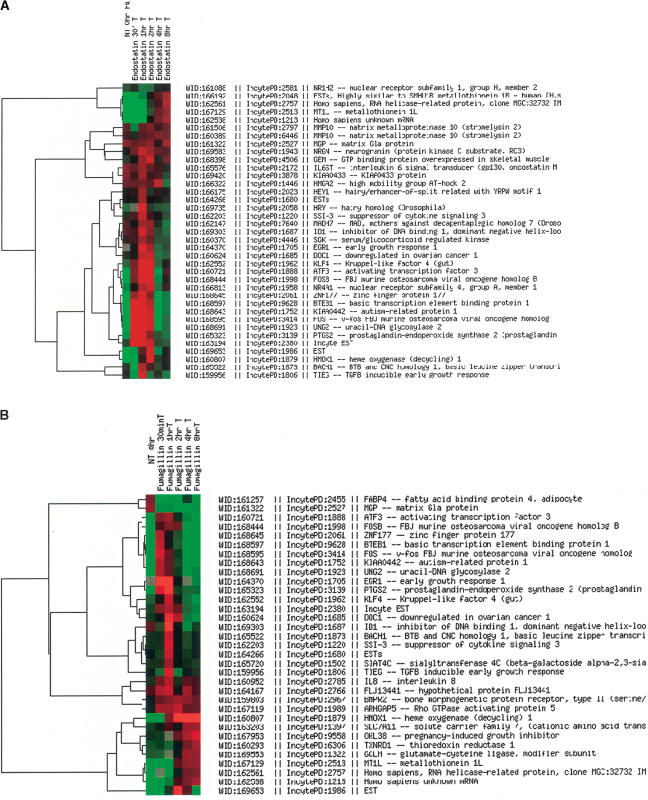
HUVEC were subsequently treated for a shorter period of time to investigate the early molecular changes after treatment. Cells were treated with endostatin (10 μg/mL) and fumagillin (10 nM) for 30 min, 1, 2, 4, and 8 h. At each time point, we extracted the total RNA, performed one round of amplification, and hybridized the products from both treated and nontreated cells on a human cDNA 10 k array (Hs-UniGem2, produced by the NCI/NIH ATC, Gaithersburg, MD). We performed basic filtering (Methods), and then a second filtering for only those genes that had a twofold up- or down-regulation (<2, <0.5) for at least one time point. We observed for both antiangiogenic agents a majority of gene expression changes within 1 and 2 h of treatment, whereas by 8 h, the levels came back to baseline as shown in Figure 2A,B. The genes demonstrating these early expression changes are involved in cell proliferation, gene transcription, and matrix organization; a number of the genes identified have unknown function. From the microarray analysis, we selected those genes for both agents (endostatin and fumagillin) that had a similar expression profile during the time course studied for further analysis. On the basis of this criteria, we selected DOC1 (XM_002964), KLF4 (BC029923), TC-1(BC021672), and ID1(S78825) (Fig. 3A,B).
Figure 2.
Microarray cluster image of the gene expression profile of HUVEC treated with 10 μg/mL endostatin (A) and 10 nM fumagillin (B) over the time course of 0 h, 30 min, 1, 2, 4, and 8 h. The cluster is ordered by genes, and shows a list of genes that are up- or down-regulated by twofold for at least one time point. The majority of gene expression changes appeared to occur for both agents within 1 and 2 h of treatment. With Cy5, we labeled the mRNA from treated cells, and with Cy3, the mRNA from nontreated cells.
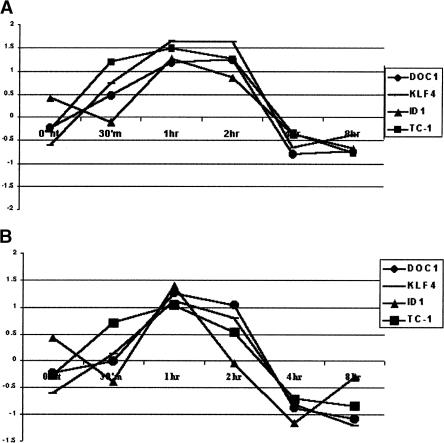
Figure 3.
Gene expression profile for DOC1, KLF4, ID1, and TC1 over the time course of treatment with 10 μg/mL endostatin (A) and 10 nM fumagillin (B). Of the list of genes with twofold up- or down-regulation in at least one time point, we chose these four genes because they have the same time course of expression in response to the two inhibitors we used. The y-axis represents the ratio between treated and nontreated cells (Cy5 intensity value for treated over Cy3 intensity value for untreated).
RT–PCR Validation Experiments
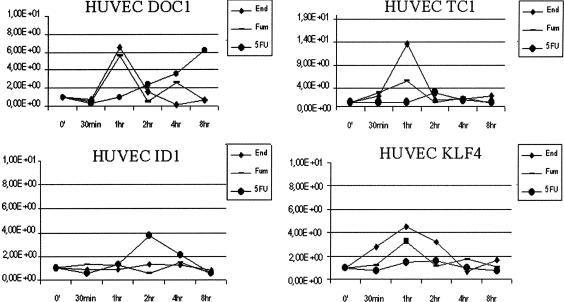
For each of these selected genes, we confirmed the changes in gene expression over time using real time quantitative RT–PCR (TaqMan assay, Applied Biosystems). In addition, to analyze the specificity of the antiangiogenic agents' effects on HUVEC cells, we treated HUVEC with 5FU for the same durations (30 min, 1, 2, 4, and 8 h) and analyzed the expression changes of the selected genes using TaqMan. TaqMan was performed in triplicate at each time point, for each gene and for each agent. We were able to confirm the same gene expression profile for DOC1, TC1, and KLF4, as was seen in the microarray data (Fig. 4). TaqMan for ID1 failed to confirm the same pattern as the microarray data, possibly due to cross-hybridization of the message for a similar gene with the ID1 spot on the cDNA array. 5FU treatment did not demonstrate considerable changes in gene expression profiles for KLF4, TC1, and ID1, whereas it caused a different pattern of expression of DOC1 compared with fumagillin and endostatin, with a progressive increase of gene expression over the time course, demonstrating the specific effects of the two angiogenic inhibitors on HUVEC cells (Fig. 4).
Figure 4.
TaqMan Real Time PCR verification performed for DOC1, KLF4, TC1, and ID1 on cDNA from HUVECs treated for 30 min, 1, 2, 4, and 8 h with both antiangiogenic agents (10 μg/mL endostatin and 10 nM fumagillin) and the cytotoxic agent (50 μg/mL 5FU). DOC1, KLF4, and TC1 showed the same gene expression changes as the microarray data. ID1 was not confirmed, most likely due to cross-hybridization with some similar gene on the cDNA array. DOC1 under 5FU treatment shows a progressive increase of gene expression over the time course. The y-axis represents a ratio between the number of ds-cDNA molecules of the gene of interest and the number of ds-cDNA molecules of the housekeeping gene GAPDH. Each dot is an average of three experiments.
Human Fibroblast Treatment
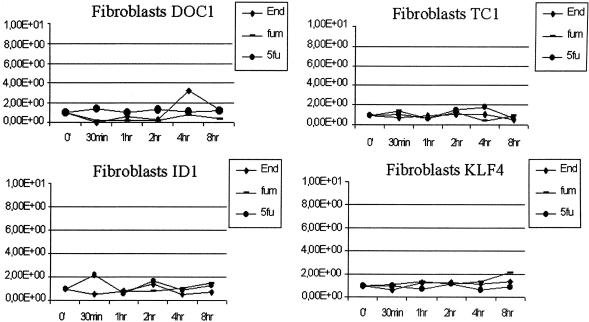
In a second experiment, we treated human fibroblasts with the same three agents in order to investigate whether the gene expression regulation of these four genes was specific for the antiangiogenic treatments and for the agents' effects on endothelial cells. As shown in Figure 5, all four genes had no significant change in expression over the course of treatment in fibroblasts compared with HUVEC, suggesting that their regulation is a unique feature of the response of endothelial cells to the angiogenesis inhibitors tested.
Figure 5.
TaqMan Real Time PCR verification performed for DOC1, KLF4, TC1, and ID1 on cDNA from human fibroblast cells treated for 30 min, 1, 2, 4, and 8 h with both antiangiogenic agents (10 μg/mL endostatin and 10 nM fumagillin) and the cytotoxic agent (50 μg/mL 5FU). No significant changes in gene expression for these four genes are detectable in fibroblasts after treatment with antiangiogenic and cytotoxic compounds. The y-axis represents a ratio between the number of ds-cDNA molecules of the gene of interest and the number of ds-cDNA molecules of the housekeeping gene GAPDH. Each dot is an average of three experiments.
SiRNA Experiments
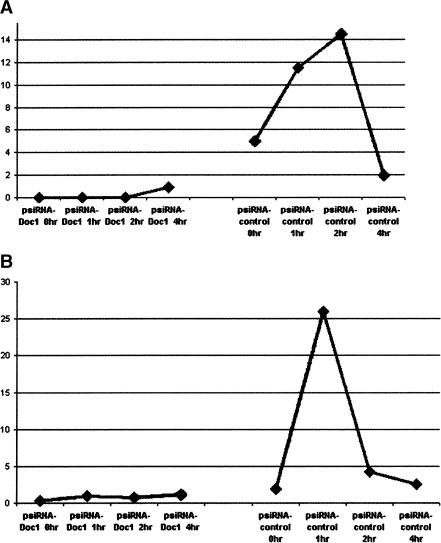
For a better understanding of the possible relationship between these genes, we performed siRNA knock-down experiments. We separately transfected HUVECs with a vector containing siRNA targeting the DOC1 gene (psiRNA-Neo-Doc1) and a vector containing a null sequence (psiRNA-Neo-Control), which, after BLAST search, resulted in complementary binding only to the intronic sequence of the human Tyrosine Hydroxylase gene. Figure 6 demonstrates how the expression of the DOC1 gene in psiRNA-Neo-Doc1-transfected cells is silenced after endostatin (A) and fumagillin (B) treatment compared with its change of expression at 1 h in psiRNA-Neo-Control-transfected cells. On the basis of these results, we decided to investigate the expression of KLF4 and TC1 in the psiRNA-Neo-Control-transfected cells (Fig. 7A) after treatment with endostatin. In addition, we also evaluated the expression of KLF4 and TC1 in HUVECs transfected with psiRNA-DOC1 after treatment with endostatin and fumagillin (Fig. 7B,C) in order to determine the relationship between DOC1 gene expression and the expression of KLF4 and TC1.
Figure 6.
The expression of the DOC1 gene in psiRNA-Neo-Doc1-transfected cells is silenced after endostatin (A) and fumagillin (B) treatment compared with its change of expression at 1 h in psiRNA-Neo-Control-transfected cells. The lack of change in gene expression after 1 h of treatment with both agents is a reliable sign that the DOC1 gene is being silenced by the appropriate siRNA. Each dot represents a ratio between the number of ds-cDNA molecules of the gene of interest and the number of ds-cDNA molecules of the housekeeping gene GAPDH. Each dot is an average of three experiments.
Figure 7.
(A) KLF4 and TC1 gene expression changes in HUVECs transfected with psiRNA-Neo-Control treated with endostatin. As expected, the two genes were up-regulated after 1 h of treatment. (B) shows how there is no longer up-regulation of KLF4 and TC1 gene expression after 1 h of endostatin treatment when the DOC1 gene is silenced by the appropriate siRNA. (C), in contrast, shows how the up-regulation in gene expression of KLF4 and TC1 in response to fumagillin is unaffected by DOC1.
Our results showed that TC1 and KLF4 failed to demonstrate an up-regulation in expression in response to endostatin treatment when DOC1 was silenced (Fig. 7A). However, after fumagillin treatment, despite DOC1 silencing, TC1 and KLF4 were up-regulated by the treatment at a 1-h time point in a similar fashion to the response of control transfected cells (Fig. 7B).
DISCUSSION
Angiogenesis is the formation of new blood vessels from pre-existing ones, and it plays a critical role in the regulation of embryonic development as well as postnatal pathophysiological processes, including wound healing and tumor growth (Lode et al. 2002). Tumors need to initiate angiogenesis in order to create a blood supply and grow beyond a size of 0.2–3.0 mm. A key feature of this process is the proliferation of endothelial cells. During tumor growth, endothelial cells proliferate more rapidly than in normal tissues, and their unique phenotype provides a distinct target for a wide range of agents.
Under normal conditions, angiogenesis is a tightly regulated equilibrium between pro- and antiangiogenic molecules. Many positive regulators have been identified and characterized (VEGF, FGF, IL8, PLGF). Endogenous inhibitors have likewise been identified, such as endostatin, angiostatin, and EMAP-II (Kayton and Libutti 2001). Exogenous inhibitors have been discovered as biological products produced by other organisms such as fumagillin from a fungus (Madhusudan and Harris 2002). Although several aspects of how these inhibitors work is known (Feldman and Libutti 2000; Kerbel and Folkman 2002; Madhusudan and Harris 2002; Jekunen and Kairemo 2003), much of their molecular mechanisms of action are still not clear. This is especially true with respect to common pathways involved in the activity of a variety of angiogenesis inhibitors. The elucidation of such pathways might enable us to develop more effective targeted therapies and to better understand the process of tumor vessel formation.
In our study, we investigated the early molecular changes occurring in endothelial cells after treatment with endostatin, as an endogenous compound, and fumagillin as an exogenous compound. The mechanism of action of endostatin in inhibiting angiogenesis involves the induction of endothelial cell apoptosis. The most consistent effect on endothelial cells in vitro is inhibition of migration, which may be due to disruption of cell matrix interactions and also involvement in cytoskeleton organization. The structure of a primary receptor is not known (MacDonald et al. 2001; Wickstrom et al. 2002; Dixelius et al. 2003; Keezer et al. 2003).
Fumagillin is a fungal metabolite and its analog, TNP-470, has been evaluated in clinical trials (Kerbel and Folkman 2002). The molecular target of fumagillin and its analog is methionine aminopeptidase-2 (MetAP-2). Fumagillin binds MetAP-2 on His-231, inactivating the enzyme. MetAP-2 removes the N-terminal methionine from most proteins involved in cell cycle regulation as a part of the translocation process, so its inhibition results in cell cycle arrest and apoptosis (Fardis et al. 2003).
We first confirmed (Fig. 1) that the antiangiogenic compounds we tested in this study are specific for the inhibition of endothelial cell proliferation. Whereas 5FU had the same inhibitory effect on both endothelial cells and fibroblasts, fumagillin and endostatin's inhibitory effect was only seen on the endothelial cells.
We used cDNA microarray analysis to identify early response changes in gene expression in reaction to the exposure of endothelial cells to endostatin and fumagillin. This technology allowed us to study thousands of genes simultaneously. We selected genes for further study that had at least a twofold up- or down-regulation in at least one time point as shown in Figure 2A. It appeared that the majority of gene expression changes were occurring for both agents at the 1- or 2-h time point. We examined the filtered genes identified, and found a group of four genes that had the same gene expression profile over the time course studied for both agents tested (Fig. 3A,B). The genes are DOC1, KLF4, TC1, and ID1. KLF4 and ID1 are transcription factors that have been well studied in mice and are involved in development, neurogenesis, and angiogenesis and are specific to the endothelium (Yet et al. 1998; Lyden et al. 1999). The function of DOC1 and TC1 is unknown, and only limited information on their gene structures and their tissue expression is available (Mok et al. 1994; Chua et al. 2000). The DOC1 gene has some similarities to the heavy myosin chain of the mouse gene, suggesting that it might be involved in the organization of the cytoskeleton. We were able to confirm the expression changes in DOC1, KLF4, and TC1 by TaqMan, whereas ID1 did not show any significant changes. Because the TaqMan assay is more sensitive and specific than the microarray technique, it is possible that our microarray result was due to another member of the ID family cross-hybridizing and giving us a false-positive signal.
To determine whether the gene changes we observed were specific to the antiangiogenic agents we tested, 5FU, a well known cytotoxic agent, was used to contrast its effects with the specific effects of the antiangiogenic compounds on these four genes. In Figure 4, it is clear that these genes have a significant change in gene expression at 1 h only with the two angiogenesis inhibitors and not with 5-FU, suggesting that the significant changes in gene profiles was a unique feature of the endothelial cell response to the angiogenesis inhibitors tested. These genes might be early response genes involved in some of the initial pathways of angiogenesis inhibition. Interestingly, 5FU treatment caused a progressive increase of DOC1 gene expression over the time course, suggesting that this gene might be involved in cell death as well. Furthermore, we treated human fibroblasts with the same agents and there are no significant changes for any of the genes over the course of treatment (Fig. 5), supporting the notion that DOC1, KLF4, and TC1 are specific for the endothelial cells response to endostatin and fumagillin.
To elucidate any possible interactions between these potential early response genes we focused our attention on the DOC1 gene. The rationale for the decision is based on the fact that this gene consistently demonstrated altered expression in several different experiments, performed in our lab with other antiangiogenic compounds (data not shown). Moreover, DOC1 up-regulation after 1 h of treatment in the different repeat experiments was consistent and higher than the other genes tested. Mok et al. (1994) discovered that this gene was present in normal ovarian surface epithelial cells, but consistently absent in all of the ovarian cancer cell lines tested in their study. A microarray data set search reveals the presence of the DOC1 gene hidden in the long list of genes that are informative in class prediction algorithm used to define human tumor type and aggressiveness, being down-regulated or absent in malignant tumors (Ross et al. 2000; Su et al. 2001). This also suggests DOC1 has a possible role as a tumor-suppressor gene. Using InvivoGen technology, we cloned sequences into an RNA polymerase III on the basis of expression vector (psiRNA-Neo) that enabled the endogenous production of small dsRNAs targeting the human DOC1 gene. Endothelial cells were transfected with either the vector containing the DOC1 targeting siRNA or a vector containing a null sequence as a control. The presence in the vector of a resistance gene to Neomycin allowed us to select the HUVECs that were transfected. We were able to silence the expression of DOC1 as shown in Figure 6 using this vector. In response to both endostatin and fumagillin treatment, there was reversal of the up-regulation of DOC1 expression at a 1-h time point after treatment, compared with the control HUVECs (Fig. 6, right curve). In addition, the up-regulation in expression of KLF4 and TC1 is also shutdown when DOC1 is silenced, but only after endostatin treatment (Fig. 7B). As expected, the control HUVECs continue to demonstrate up-regulation of both genes at the1-h time point after treatment (Fig. 7A). The abrogation of KLF4 and TC1 response by DOC1 silencing is not seen following fumagillin treatment (Fig. 7C).
These results suggest that DOC1 may be upstream from KLF4 and TC1 in the cascade of events following endostatin treatment, whereas for fumagillin, they may be involved in the mechanism of angiogenesis inhibition, albeit via an alternative signaling pathway. DOC1, considering its early expression, could be considered an early response gene that acts as a transcription factor that causes the up-regulation of KLF4 and TC1 in the endostatin cascade. Alternatively, it could also very likely represent a membrane protein that interacts with endostatin, and after some specific intracellular signaling, causes the up-regulation of KLF4 and TC1, whereas with fumagillin treatment, it appears there is some other player with a specific role between DOC1 and KLF4 and TC1. Moreover, the presence of DOC gene sequence homology to myosin heavy-chain sequences (as noted in the GenBank sequence file: XM_002964) could suggest an additional role of the gene in the organization of the cytoskeleton, therefore effecting migration and proliferation (Dixelius et al. 2002). Recent studies present KLF4 as a possible tumor-suppressor gene in colon cancer, suggesting that the lack of the gene favors tumor growth (Zhao et al. 2004). KLF4 was frequently down-regulated in bladder cancer cell lines and cancer tissues. When the KLF4 gene was transduced into the bladder cancer cell lines using adenoviral, it suppressed cell growth and induced apoptosis (Ohnishi et al. 2003).
No information is available regarding sequence homology or protein function of the TC1 gene, except that it seems to be differentially expressed in thyroid tumors (Chua et al. 2000).
Further studies are necessary to clarify these early mechanisms and to better understand the function of these genes. DOC1, KLF4, TC1, and their related pathway may provide new targets for the inhibition of endothelial cell proliferation in cancer gene therapy, as well as a marker for the characterization and development of new antiangiogenic compounds.
METHODS
Cell Proliferation Assay
To measure cell proliferation, we used the reagent WST1. This is a colorimetric assay for the quantification of cell proliferation and cell viability, based on the cleavage of the tetrazolium salt WST-1 by mitochondrial dehydrogenases in viable cells (Roche Molecular Biochemicals). HUVEC and human fibroblasts were exposed to human endostatin (10 μg/mL; EntreMed Inc.), fumagillin (10 nM; Sigma), 5-FU (50 μg/mL; Sigma), or control medium (EBM, Cambrex) for varying time durations as follows: 2 and 4 h, 1–3–5 d. A total of 2000/3000 cells were plated in a 96-well culture plate with 100 μL of medium with and without treatment. At the end of each time point, we added 10 μL of WST1 reagent and took the readings after 4 h of incubation. Endostatin was resuspended in 66 mM sodium phosphate, 17 mM citric acid, and 59 mM NaCl buffer (pH 6.2). Fumagillin and 5FU were resuspended in DMSO100%.
Cell Culture Treatment for Microarray Analysis
HUVECs were maintained <70% confluent with EGM and DMEM medium in T150 mL flasks (Cambrex) and exposed to endostatin (10 μg/mL), fumagillin (10 nM), and control medium for varying time durations as follows: 30 min, 1, 2, 4, and 8 h.
Microarray Analysis
Total RNA was extracted using RNeasy kits (QIAGEN). Total RNA was then subjected to one round of amplification following the modified Eberwine method (Eberwine 1996; E. Wang et al. 2000) resulting in ∼15 μg of messenger RNA (mRNA). The quality of the extracted RNA was tested by spectrophotometer and by evaluations on minichips (BioAnalyzer, Agilent Tecnologies).
Microarray analysis was conducted on the RNA extracted from HUVEC treated with endostatin, fumagillin, and carrier control. Three micrograms of amplified mRNA was used for the hybridization on 10 k human cDNA microarrays, (Hs-UniGem2, produced by the NCI/NIH ATC). Comparisons were made between the amplified mRNA of treated HUVECs at each time point to the same control, which consisted of a pool of amplified mRNA extracted at each time point from the untreated cells. Fluorescent marker dyes (Cy5 and Cy3) were used to label the treated and untreated samples, respectively. The respective dyes and samples were also switched to test for any labeling bias. The mixture of the two populations of RNA species was then hybridized to the same microarray and incubated for 16 h at 42°C. cDNA microarrays were then washed and scanned using the GenePix 4000B (Axon Instruments Inc.) and images were analyzed with GenePix software version 3.0. For each sample, a file containing the image of the array and an Excel file containing the expression ratio values for each gene was uploaded onto the MadbArray Web site (National Center for Biotechnology Information/NIH; http://nciarray.nci.nih.gov) for further analysis. To accurately compare measurements from different experiments, the data was normalized and the ratio (Signal Cy5/Signal Cy3) was calculated so that the median (Ratio) was 1.0.
A basic filtering was performed on the arrays, consisting of selecting only the genes in which the signal to background was >2, spot size for both channels was >30%, and intensity values for both signals >500. The second filtering that we applied to the data consisted of analyzing only the genes that had at least a twofold up- or down-regulation (ratio Cy5/CY3 = >2, <0.5). Each of these analyses was performed using the MadbArray Web site (National Center for Biotechnology Information/NIH) that offers this service to all the NCI users.
cDNA Synthesis and Real-Time PCR Assay
HUVEC and fibroblast cultures were maintained <70% confluent with EGM and DMEM medium in T150-mL flasks (Cambrex) and exposed to 5-FU (50 μg/mL) or control medium for varying time durations as follows: 30 min, 1, 2, 4, and 8 h.
TaqMan assay (Applied Biosystems) Real Time PCR analysis was conducted for DOC1, KLF4, TC1, and ID1 from total RNA extracted from all of the treatments (endostatin, fumagillin, and 5FU) at all of the time points (30 min, 1, 2, 4, and 8 h). CDNA was synthesized from Total RNA using Superscript Kit/Gibco (InvitroGen Corp.). Real Time PCR was performed by TaqMan assay as follows: four primers and probes were designed on intron–exon junctions in the 3′ end of the gene using Primer Express software 2.0 and following standard experimental protocols from Applied Biosystems. Real time PCR reactions were run and analyzed on a GeneAmp 5700 Sequence Detection System (Applied Biosystems). All PCRs were normalized against human GAPDH, a housekeeping gene provided as a control by the same company. Standard curves for each gene were made of a series of dilutions with known concentrations of the PCR product amplified by the same two primers used for the detection. Dilution ranged from 1010 number of ds-cDNA molecules to 103. Sequences of primers and probes are as follows.
DOC1 (XM_002964)
DOC1F-Forward Primer: AGTCCATACTGATATTTTTGCAAG GAA; anneals between residues 2774 and 2800, with a Tm of 59°. DOC1R-Reverse Primer: CCCAAAGTACGAGTTCAGTCAGTCT; anneals between residues 2861 and 2837, with a Tm of 59°. DOC1P-TaqMan Probe: ATCCTTTTTTAATCATCCCTCCATATC CCCC; anneals between residues 2805 and 2835, with a Tm of 68°.
KLF4 (BC029923)
KLF4F-Forward Primer: ACCAGGCACTACCGTAAACACA; anneals between residues 1806 and 1827, with a Tm of 58°. KLF4R-Reverse Primer: GGTCCGACCTGGAAAATGCT; anneals between residues 1884 and 1865, with a Tm of 60°. KLF4P-TaqMan Probe: CCGTTCCAGTGCCAAAAATGCGA; anneals between residues 1839 and 1861, with a Tm of 69°.
TC-1 (BC021672)
TC-1F-Forward Primer: AGCCAAGATCATTTTTGCCATAGA; anneals between residues 258 and 281, with a Tm of 60°. TC-1R-Reverse Primer: GAAACTGGAAAAGCTTGTCTTTTGT; anneals between residues 355 and 331, with a Tm of 58°. TC-1P-TaqMan Probe: AAAACGCGTGCCCTGATGGCCT; anneals between residues 298 and 319, with a Tm of 69°.
ID1 (S78825)
ID1F-Forward Primer: AGAACCGCAAGGTGAGCAA; anneals between residues 254 and 272, with a T of 60°.ID1R-Reverse Primer: CCAACTGAAGGTCCCTGATGTA; anneals between residues 322 and 301, with a T of 59°. ID1P-TaqMan Probe: TGGAGATTCTC CAGCACGTCATCGAC; anneals between residues 275 and 300, with a T of 68°.
Small Interfering RNA Assay and Real-Time PCR
InvivoGen has developed the psiRNA system. Four 19-nt dsDNA fragments complementary to a portion of the mRNA molecule of DOC1 were cloned by InvivoGen into a RNA polymerase III on the basis of expression vector (psiRNA-Neo) that enabled the endogenous production of small dsRNAs targeting the gene of interest. This vector has a self-replicating cassette, allowing the vector to propagate during cell division. InvivoGen provided us with three 19-nt sequences for the DOC1 gene, and for the cloning, we chose the one that gave the least number of hits after a BLAST search. A vector containing a null sequence (intron1 repeat polymorphism of the human tyrosine hydroxylase gene) was created as a control. The 19-nt sequences cloned into the 2.5-kb expression vectors were as follows: PsiRNA-Neo-DOC1: 5′-AGCG TAACCAAGGAGAGAGAT-3′ (accession no. XM_002964, position 1172–1192); PsiRNA-Neo-Control: 5′-ATTCATTCATT CATTCACCAT-3′ (accession no. D00269, position 1192–1212).
HUVECs were transfected with 1 μg of each psiRNA-Neo expression vector using a lipofectamine technique (Gibco/Invitrogen). After 48 h of transfection, cells were grown on medium containing Neomycin (Gibco/Invitrogen). The clones were selected and grown further to use them in the RT–PCR assays. The transfection protocol is as follows: Lipofectamine (4 μL) was diluted in 100 μL of Opti MEMI medium (Gibco/InvitroGen Corp) and incubated for 15 min. A total of 1 μg of psiRNA-Neo was diluted in 100 μL of Opti MEMI. The two solutions were then mixed together and incubated for 15 min. HUVECs (70%–80% confluent) in 6-well cell culture plates were washed with PBS and replaced with 800 μL of Opti MEMI. A total of 200 μL of Opti MEMI containing lipofectamine and psiRNA, previously mixed, were added to each well, mixed, and incubated for 1 h at 37°C. After 1 h, 3 mL of complete medium (EBM-2) was added to each well and incubated for 1 d. After 1 d, the medium was changed with fresh medium, and after a second day, it was added to each well with the selective antibiotic Zeocin. When cells reached the appropriate number, they were plated (2000/4000) in a 96-well culture plate and treated with either endostatin, fumagillin, or control medium for different durations as follows: 0, 1, 2, and 4 h. Total RNA was then extracted at each time point, and mRNA levels analyzed for the expression of DOC1, TC1, and KLF4 by TaqMan Real Time PCR.
Acknowledgments
We would like to acknowledge the editorial assistance of Romi Sawhney.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2552804.
References
- Abdollahi, A., Hahnfeldt, P., Maercker, C., Grone, H.J., Debus, J., Ansorge, W., Folkman, J., Hlatky, L., and Huber, P.E. 2004. Endostatin's antiangiogenic signaling network. Mol. Cell 13: 649–663. [DOI] [PubMed] [Google Scholar]
- Boehm, T., Folkman, J., Browder, T., and O'Reilly, M.S. 1997. Antiangiogenic therapy of experimental cancer does not induce acquired drug resistance. Nature 390: 404–407. [DOI] [PubMed] [Google Scholar]
- Calvo, A., Feldman, A.L., Libutti, S.K., and Green, J.E. 2002. Adenovirus-mediated endostatin delivery results in inhibition of mammary gland tumor growth in C3(1)/SV40 T-antigen transgenic mice. Cancer Res. 62: 3934–3938. [PubMed] [Google Scholar]
- Chua, E.L., Young, L., Wu, J.R., Turtle, W.M., and Dong, Q. 2000. Cloning of TC-1 (C8orf4), a novel gene found to be overexpressed in thyroid cancer. Genomics 69: 342–347. [DOI] [PubMed] [Google Scholar]
- Dixelius, J., Cross, M., Matsumoto, T., Sasaki, T., Timpl, R., and Claesson-Welsh, L. 2002. Endostatin regulates endothelial cell adhesion and cytoskeletal organization. Cancer Res. 62: 1944–1947. [PubMed] [Google Scholar]
- Dixelius, J., Cross, M.J., Matsumoto, T., and Claesson-Welsh, L. 2003. Endostatin action and intracellular signaling: β-catenin as a potential target? Cancer Lett. 196: 1–12. [DOI] [PubMed] [Google Scholar]
- Eberwine, J. 1996. Amplification of mRNA populations using aRNA generated from immobilized oligo(dT)-T7 primed cDNA. Biotechniques 20: 584–591. [DOI] [PubMed] [Google Scholar]
- Egeblad, M. and Werb, Z. 2002. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2: 161–174. [DOI] [PubMed] [Google Scholar]
- Fardis, M., Pyun, H.J., Tario, J., Jin, H., Kim, C.U., Ruckman, J., Lin, Y., Green, L., and Hicke, B. 2003. Design, synthesis and evaluation of a series of novel fumagillin analogues. Bioorg. Med. Chem. 11: 5051–5058. [DOI] [PubMed] [Google Scholar]
- Feldman, A.L. and Libutti, S.K. 2000. Progress in antiangiogenic gene therapy of cancer. Cancer 89: 1181–1194. [PubMed] [Google Scholar]
- Folkman, J. 2000. Tumor angiogenesis. In Cancer medicine (eds. R. Bast et al.). BC Decker Inc., Hamilton, Ontario, Canada.
- Griffith, E.C., Su, Z., Turk, B.E., Chen, S., Chang, Y.H., Wu, Z., Biemann, K., and Liu, J.O. 1997. Methionine aminopeptidase (type 2) is the common target for angiogenesis inhibitors AGM-1470 and ovalicin. Chem. Biol. 4: 461–471. [DOI] [PubMed] [Google Scholar]
- Hajitou, A., Grignet, C., Devy, L., Berndt, S., Blacher, S., Deroanne, C.F., Bajou, K., Fong, T., Chiang, Y., Foidart, J.M., et al. 2002. The antitumoral effect of endostatin and angiostatin is associated with a down-regulation of vascular endothelial growth factor expression in tumor cells. FASEB J. 16: 1802–1804. [DOI] [PubMed] [Google Scholar]
- Herbst, R.S., Lee, A.T., Tran, H.T., and Abbruzzese, J.L. 2001. Clinical studies of angiogenesis inhibitors: The University of Texas MD Anderson Center Trial of Human Endostatin. Curr. Oncol. Rep. 3: 131–140. [DOI] [PubMed] [Google Scholar]
- Ingber, D., Fujita, T., Kishimoto, S., Sudo, K., Kanamaru, T., Brem, H., and Folkman, J. 1990. Synthetic analogues of fumagillin that inhibit angiogenesis and suppress tumour growth. Nature 348: 555–557. [DOI] [PubMed] [Google Scholar]
- Jekunen, A. and Kairemo, K. 2003. Inhibition of angiogenesis at endothelial cell level. Microsc. Res. Tech. 60: 85–97. [DOI] [PubMed] [Google Scholar]
- Kayton, M.L. and Libutti, S.K. 2001. Endothelial monocyte activating polypeptide II (EMAP II) enhances the effect of TNF on tumor-associated vasculature. Curr. Opin. Investig. Drugs 2: 136–138. [PubMed] [Google Scholar]
- Keezer, S.M., Ivie, S.E., Krutzsch, H.C., Tandle, A., Libutti, S.K., and Roberts, D.D. 2003. Angiogenesis inhibitors target the endothelial cell cytoskeleton through altered regulation of heat shock protein 27 and cofilin. Cancer Res. 63: 6405–6412. [PubMed] [Google Scholar]
- Kerbel, R. and Folkman, J. 2002. Clinical translation of angiogenesis inhibitors. Nat. Rev. Cancer 2: 727–739. [DOI] [PubMed] [Google Scholar]
- Kim, Y.M., Hwang, S., Pyun, B.J., Kim, T.Y., Lee, S.T., Gho, Y.S., and Kwon, Y.G. 2002. Endostatin blocks vascular endothelial growth factor-mediated signaling via direct interaction with KDR/Flk-1. J. Biol. Chem. 277: 27872–27879. [DOI] [PubMed] [Google Scholar]
- Lode, H.N., Wenkel, J., and Gaedicke, G. 2002. Angiogenesis, views and news. The 93rd Annual Meeting of the American Association for Cancer Research. Expert. Opin. Biol. Ther. 2: 671–676. [DOI] [PubMed] [Google Scholar]
- Lyden, D., Young, A.Z., Zagzag, D., Yan, W., Gerald, W., O'Reilly, R., Bader, B.L., Hynes, R.O., Zhuang, Y., Manova, K., et al. 1999. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature 401: 670–677. [DOI] [PubMed] [Google Scholar]
- MacDonald, N.J., Shivers, W.Y., Narum, D.L., Plum, S.M., Wingard, J.N., Fuhrmann, S.R., Liang, H., Holland-Linn, J., Chen, D.H., and Sim, B.K. 2001. Endostatin binds tropomyosin. A potential modulator of the antitumor activity of endostatin. J. Biol. Chem. 276: 25190–25196. [DOI] [PubMed] [Google Scholar]
- Madhusudan, S. and Harris, A.L. 2002. Drug inhibition of angiogenesis. Curr. Opin. Pharmacol. 2: 403–414. [DOI] [PubMed] [Google Scholar]
- McManus, M.T. and Sharp, P.A. 2002. Gene silencing in mammals by small interfering RNAs. Nat. Rev. Genet. 3: 737–747. [DOI] [PubMed] [Google Scholar]
- Mok, S.C., Wong, K.K., Chan, R.K., Lau, C.C., Tsao, S.W., Knapp, R.C., and Berkowitz, R.S. 1994. Molecular cloning of differentially expressed genes in human epithelial ovarian cancer. Gynecol. Oncol. 52: 247–252. [DOI] [PubMed] [Google Scholar]
- Ohnishi, S., Ohnami, S., Laub, F., Aoki, K., Suzuki, K., Kanai, Y., Haga, K., Asaka, M., Ramirez, F., and Yoshida, T. 2003. Downregulation and growth inhibitory effect of epithelial-type Kruppel-like transcription factor KLF4, but not KLF5, in bladder cancer. Biochem. Biophys. Res. Commun. 308: 251–256. [DOI] [PubMed] [Google Scholar]
- O'Reilly, M.S., Holmgren, L., Shing, Y., Chen, C., Rosenthal, R.A., Moses, M., Lane, W.S., Cao, Y., Sage, E.H., and Folkman, J. 1994. Angiostatin: A novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell 79: 315–328. [DOI] [PubMed] [Google Scholar]
- O'Reilly, M.S., Boehm, T., Shing, Y., Fukai, N., Vasios, G., Lane, W.S., Flynn, E., Birkhead, J.R., Olsen, B.R., and Folkman, J. 1997. Endostatin: An endogenous inhibitor of angiogenesis and tumor growth. Cell 88: 277–285. [DOI] [PubMed] [Google Scholar]
- Rodeschini, V., Boiteau, J.G., Van de Weghe, P., Tarnus, C., and Eustache, J. 2004. MetAP-2 inhibitors based on the fumagillin structure. Side-chain modification and ring-substituted analogues. J. Org. Chem. 69: 357–373. [DOI] [PubMed] [Google Scholar]
- Ross, D.T., Scherf, U., Eisen, M.B., Perou, C.M., Rees, C., Spellman, P., Iyer, V., Jeffrey, S.S., Van de Rijn, M., Waltham, M., et al. 2000. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 24: 227–235. [DOI] [PubMed] [Google Scholar]
- Sauter, B.V., Martinet, O., Zhang, W.J., Mandeli, J., and Woo, S.L. 2000. Adenovirus-mediated gene transfer of endostatin in vivo results in high level of transgene expression and inhibition of tumor growth and metastases. Proc. Natl. Acad. Sci. 97: 4802–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin, N., Meng, L., Wang, M.Q., Wen, J.J., Bornmann, W.G., and Crews, C.M. 1997. The antiangiogenic agent fumagillin covalently binds and inhibits the methionine aminopeptidase, MetAP-2. Proc. Natl. Acad. Sci. 94: 6099–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, A.I., Welsh, J.B., Sapinoso, L.M., Kern, S.G., Dimitrov, P., Lapp, H., Schultz, P.G., Powell, S.M., Moskaluk, C.A., Frierson Jr., H.F., et al. 2001. Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res. 61: 7388–7393. [PubMed] [Google Scholar]
- Wang, E., Miller, L.D., Ohnmacht, G.A., Liu, E.T., and Marincola, F.M. 2000. High-fidelity mRNA amplification for gene profiling. Nat. Biotechnol. 18: 457–459. [DOI] [PubMed] [Google Scholar]
- Wang, J., Lou, P., and Henkin, J. 2000. Selective inhibition of endothelial cell proliferation by fumagillin is not due to differential expression of methionine aminopeptidases. J. Cell. Biochem. 77: 465–473. [PubMed] [Google Scholar]
- Wickstrom, S.A., Alitalo, K., and Keski-Oja, J. 2002. Endostatin associates with integrin α5β1 and caveolin-1, and activates Src via a tyrosyl phosphatase-dependent pathway in human endothelial cells. Cancer Res. 62: 5580–5589. [PubMed] [Google Scholar]
- Yet, S.F., McA'Nulty, M.M., Folta, S.C., Yen, H.W., Yoshizumi, M., Hsieh, C.M., Layne, M.D., Chin, M.T., Wang, H., Perrella, M.A., et al. 1998. Human EZF, a Kruppel-like zinc finger protein, is expressed in vascular endothelial cells and contains transcriptional activation and repression domains. J. Biol. Chem. 273: 1026–1031. [DOI] [PubMed] [Google Scholar]
- Zhao, W., Hisamuddin, I.M., Nandan, M.O., Babbin, B.A., Lamb, N.E., and Yang, V.W. 2004. Identification of Kruppel-like factor 4 as a potential tumor suppressor gene in colorectal cancer. Oncogene 23: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://nciarray.nci.nih.gov; MadebArray NCI/NIH