Abstract
All known vertebrate chromatin insulators interact with the highly conserved, multivalent 11-zinc finger nuclear factor CTCF to demarcate expression domains by blocking enhancer or silencer signals in a position-dependent manner. Recent observations document that the properties of CTCF include reading and propagating the epigenetic state of the differentially methylated H19 imprinting control region. To assess whether these findings may reflect a universal role for CTCF targets, we identified more than 200 new CTCF target sites by generating DNA microarrays of clones derived from chromatin-immunopurified (ChIP) DNA followed by ChIP-on-chip hybridization analysis. Target sites include not only known loci involved in multiple cellular functions, such as metabolism, neurogenesis, growth, apoptosis, and signalling, but potentially also heterochromatic sequences. Using a novel insulator trapping assay, we also show that the majority of these targets manifest insulator functions with a continuous distribution of stringency. As these targets are generally DNA methylation-free as determined by antibodies against 5-methylcytidine and a methyl-binding protein (MBD2), a CTCF-based network correlates with genome-wide epigenetic states.
The genome projects have revealed that most, if not all mammalian genes are organized in clusters. This organization presumably reflects the need to initiate and maintain proper expression domains that exploit common cis-regulatory elements in lineage-specific manners. Yet such domains must remain protected from unscheduled activation or silencing events emanating from within the cluster or from neighboring clusters or intergenic sequences, during a developmental window (Bell et al. 2001). This task is accomplished by chromatin insulator elements that demarcate expression domains by blocking enhancer or silencer signals only if physically positioned between the cis-regulatory element and pertinent gene promoters (Bell et al. 2001; Ohlsson et al. 2001).
The mechanisms which manifest this property remain unknown, although it has been noted that all known mammalian insulators interact with the highly conserved, multivalent 11-zinc finger nuclear factor CTCF (Bell et al. 2001; Ohlsson et al. 2001). Although chromatin insulators contribute to the organization of the human genome into different epigenetic landscapes, recent observations have revealed that not only is the interaction between CTCF and the chromatin insulator domain of the H19 imprinting control region (ICR) controlled by epigenetic marks in vitro (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000b) and in vivo (Holmgren et al. 2001; Kanduri et al. 2000b), but it also propagates the methylation-free epigenetic state of the maternally inherited H19 ICR (Pant et al. 2003; Schoenherr et al. 2003).
To assess whether these findings may reflect a universal role for CTCF, it was essential to map CTCF target sites genome-wide. This task was complicated, however, by the fact that the central portion of CTCF, which contains an 11-zinc finger DNA-binding domain, mediates binding to a wide range of target cis elements by varying contributions of individual zinc fingers (Ohlsson et al. 2001). To overcome this limitation, we created a CTCF target-site library derived from chromatin-immunopurified (ChIP) DNA, which was enriched in CTCF binding sites from mouse fetal liver. By exploiting a range of novel techniques, we examine here the link between occupancy of CTCF target sites and their epigenetic states.
RESULTS
Genome-Wide Occupancy of CTCF Target Sites in Mouse Fetal Liver
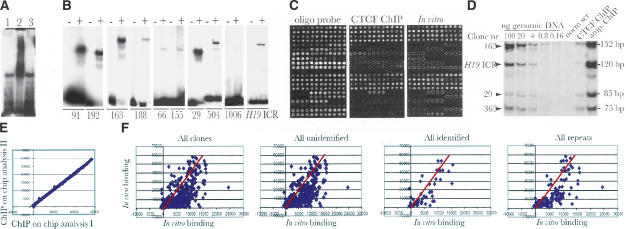
Following a 1000- to 2000-fold purification of crosslinked CTCF target sites from mouse fetal liver by using an antibody against the C-terminal domain of CTCF, and ligation of linkers and ChIP DNA into a pGEM vector, a plasmid library containing approximately 2200 clones was generated. The inserts of this library were size-selected (100–300 bp) to form a secondary library, in order to allow a more precise mapping of the CTCF binding sequences, reduce background from repetitive elements, and facilitate validation by EMSA analysis. A bandshift analysis revealed that a majority of the library sequences interacted with CTCF in vitro (Fig. 1A). This was verified by performing individual bandshift assays of nine randomly picked clones among the positive ones selected from in vivo hybridization, array-based binding assay, and PCR analysis (Fig. 1B). Following sequencing and elimination of duplicates, 266 unique clones could be identified and were spotted on glass slides.
Figure 1.
Characterization of the CTCF target-site library. (A) The bulk of the CTCF target-site library of mouse fetal liver interacts with CTCF in vitro as determined by bandshift analysis. Lane 1 depicts inserts from the library cut with NotI as probe and no protein; lane 2 shows band-shift with recombinant CTCF. The specificity of the band shift was ascertained by including a 100-fold molar excess of cold H19 ICR as competitor (lane 3). (B) Band shift assays with nine randomly picked individual clones. The first eight clones are positive, whereas clone 1006 is negative for band-shift. The – and + symbols indicate absence and presence of recombinant CTCF, respectively. (C) Microarray analysis of the CTCF target-site library. From left to right, the images show total DNA estimation using an oligo hybridization probe, pattern of in vivo (using ChIP DNA from mouse fetal liver) and in vitro (using recombinant CTCF) interactions. Each clone is represented by four adjacent spots that were printed in duplicates on each microarray. (D) The random amplification of ChIP DNA is unbiased for five randomly selected sequences. The first five lanes show the genomic DNA concentration to ascertain that the PCR amplifications were performed under semiquantitative conditions. (E) Comparison of two independent ChIP-on-chip hybridization experiments. (F) Quantitative scatterplots of in vivo/in vitro binding patterns distributed over different classes of sequences as indicated in the images. The red lines represent an estimate of in vivo/in vitro CTCF binding efficiency based on the 10 highest values.
To determine which of the inserts displayed inherent CTCF binding activity, we immunopurified in vitro-formed complexes between library sequences and recombinant CTCF. The immunopurified DNA was amplified and labeled with Cy3, whereas input library DNA was labeled with Cy5. Following simultaneous hybridization to microarrays of the plasmid library and normalization to DNA concentration in the spots, as determined by oligo DNA hybridization, we identified numerous sequences that were specifically enriched as a result of the in vitro interaction with CTCF (Fig.1C).
In order to examine the in vivo pattern of CTCF target-site occupancy, we amplified and Cy5-labeled CTCF ChIP DNA derived from mouse fetal liver using an affinity-purified antibody against the N-terminal portion of CTCF. By hybridizing the ChIP probe to the microarray in the presence of a vast excess of herring sperm and Cot1 DNA to suppress hybridization from contaminating repetitive sequences, we were able to visualize the pattern of CTCF target-site occupancy in mouse fetal liver (Fig. 1C). To ascertain that the amplification protocol did not introduce any significant bias in sequence representation, we performed multiplex PCR analysis of the original and amplified ChIP DNA. Figure 1D shows that the amplification of randomly picked sequences from CTCF ChIP DNA did not cause any significant bias. Moreover, the ChIP-on-chip analyses were highly reproducible, as an essentially identical pattern of occupancy could be demonstrated by comparing independent experiments, as exemplified in Figure 1E.
The visualization of the in vivo (i.e., chromatin immunoprecipitation from living cells) and in vitro (i.e., array-based binding assay and EMSA) CTCF binding data in scatter plots (Fig. 1F) allows us to draw several conclusions: First, there seems to be only moderate agreement between in vivo and in vitro binding patterns for some of the weaker binding sequences, a deduction further visualized by the introduction of a red line that crosses a point averaging the 10 strongest in vivo/in vitro values (Fig. 1F). Second, ChIP-on-chip and PCR analyses (data not shown) also revealed that some sequences bound CTCF very weakly if at all in vitro but strongly in vivo, such as one site located in Gsk-3β (clone 1006, Fig. 1B), suggesting that protein–protein interactions can localize CTCF to pivotal, non-CTCF target sites in the genome. Third, a continuous distribution of binding affinity between CTCF and the target sites is seen, suggesting that most sequences in the library interact to some degree with CTCF. Fourth, CTCF target sites could be documented in various types of repeat sequences, such as long terminal repeats (LTRs) and CpG islands (data not shown).
Identification of In Vivo Utilized CTCF Target Sites
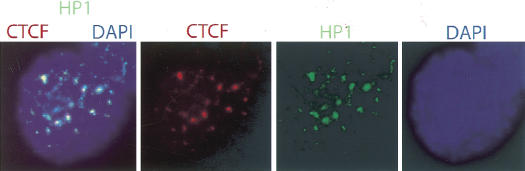
Sequences interacting with CTCF in vitro and/or in vivo in mouse fetal liver have been submitted to GenBank (accession nos. AY457177–AY457567). A surprising outcome of our analysis is that more than two-thirds of the library of CTCF target sites could not be identified in mouse genome databanks (Celera, NCBI), although Southern blot and quantitative PCR analyses ruled out contamination of DNA from other sources and confirmed the presence of these sequences in the mouse genome (data not shown). As the genome sequencing projects have little or no coverage of heterochromatin (The Mouse Genome Sequencing Consortium 2002), we propose that many of these unidentified CTCF target sites map to heterochromatic regions of the genome. To examine this possibility in some detail, we analyzed the nuclear patterns of distribution of CTCF versus a marker for heterochromatin, HP1 (James and Elgin 1986). Figure 2 shows that that CTCF does indeed colocalize with HP1β in an almost identical manner supporting the notion of a link between CTCF and heterochromatin. The scatterplots therefore include a separation of data with respect to all identified and unidentified loci, highlighting the possibility that CTCF target sites display the same range of characteristics in both euchromatic and heterochromatic domains.
Figure 2.
Immunofluorescent analysis of CTCF and HP1β in murine lung fibroblast cells. Colocalization of CTCF (Cy-3) and HP1β (FITC) clusters within the nuclei (DAPI) was seen after double immunostaining using rabbit anti-CTCF and goat anti-HP1β antibodies. Magnification ×1000. From left to right, images show merged CTCF/HP1/DAPI, CTCF (red), HP1 (green), DAPI (blue).
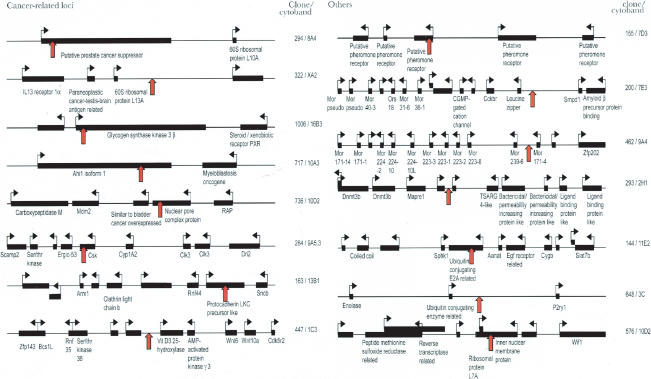
A selection of the identified CTCF target sites is displayed in Figure 3 and Table 1. In light of the known regulation of genes more than 90 kb distant from CTCF target sites (Pant et al. 2003; Schoenherr et al. 2003), we have displayed a 300-kb window of the chromosomal context surrounding several clones. Of 55 sequences which could be unambiguously identified, 21 mapped to introns, two to exons, one to an exon-intron boundary, and the remaining to intergenic regions, including two in known imprinted domains. Using the Gene Ontology database (see Methods; The Gene Ontology Consortium 2000), we determined that nine clones mapped to potential loci implicated in cancer, four to loci involved in the ubiquitin pathway, five to the G protein signaling pathway, three to the Wnt signaling pathway, three to apoptotic pathways, six to neurogenesis loci, and four within or adjacent to clusters of olfactory or pheromone receptor genes, to mention just a few. A complete list of identified genes at or adjacent to CTCF target sites, with corresponding Celera and GenBank accession numbers, can be found in the Supplemental material.
Figure 3.
Depiction of positions of CTCF target sites showing significant in vivo binding. A 300-kb window surrounding each target site (red arrow) is displayed for a selection of loci, including those with known functions in oncogenesis. Additional loci are described in Table 1 and in the Supplemental material.
Table 1.
Selected Genes At or Adjacent To Newly Identified CTCF Target Sites
| Clone | Accession # | Gene | Biological function | |||
|---|---|---|---|---|---|---|
| Intronic CTCF target sites | ||||||
| 140 | AY457222 | DOCK-1 | Apoptosis, phagocytosis, integrin receptor pathway | |||
| 144 | AY457225 | Ubiquitin conjugating enzyme E2A related | Ubiquitin-dependent protein degradation | |||
| 163 | AY457233 | Protocadherin LKC precursor like | Regulation of cell proliferation | |||
| 294 | AY457286 | Putative prostate cancer suppressor | Electron transport | |||
| 411 | AY457336 | Coagulation factor II | Apoptosis, JAK-STAT cascade, caspase activation | |||
| 717 | AY457431 | Ahi1 isoform 1 | Mannosyl-oligosaccharide glucosidase | |||
| 1006 | AY457543 | Glycogen synthase kinase3 beta | Anti-apoptosis, morphogenesis | |||
| Exonic CTCF target sites | ||||||
| 284 | AY457278 | C-src tyrosine kinase | Mitotic S-specific transcription, zygotic axis determination | |||
| 906 | AY457503 | Translation initiation factor 3 subunit | Protein biosynthesis | |||
| Genes adjacent to CTCF target site | ||||||
| 6 | AY457178 | Cbp/p300-interacting transactivator | Transcription regulation | |||
| 94 | AY457205 | Fgd1 related F-actin binding protein | Transcription factor, morphogenesis, & organogenesis | |||
| 200 | Sphingomyelin phosphodiesterase | Neurogenesis | ||||
| 398 | AY457331 | Grb10 | Neuropeptide, insulin & EGF receptor, cell-cell signalling | |||
| 398 | AY457331 | Cordon-bleu | Neural tube formation | |||
| 447 | AY457350 | Vitamin D3 25-hydroxylase | Lipid metabolism, Ca2+ homeostasis, electron transport | |||
| 648 | AY457400 | Ubiquitin conjugating enzyme E2-related | Ubiquitin-dependent protein degradation, cell cycle control | |||
| 648 | AY457400 | Purinergic receptor P2Y | Cytosolic Ca2+ concentration elevator | |||
| 797 | AY457461 | Tolloid-like | Skeletal development | |||
| Neighboring genes | ||||||
| 144 | AY457225 | Sphingosine kinase 1 | Sphingolipid metabolism, cell communication | |||
| 163 | AY457233 | Synuclein beta | Anti-apoptosis, neurogenesis | |||
| 200 | Amyloid beta A4 precursor binding B1 | Intracellular signalling cascade | ||||
| 200 | Cholecystokinin B receptor | G protein signalling linked to IP3 2nd messenger | ||||
| 265 | AY457268 | FoxC1 | Segment polarity determination, morphogenesis | |||
| 284 | AY457278 | Cytochrome P450 1a2 | Cell growth & maintenance, electron transport | |||
| 293 | AY457285 | Mapre1 | Cell cycle control, cell proliferation | |||
| 322 | AY457302 | Paraneoplastic C-T-B related | Neurogenesis, tumor antigen | |||
| 447 | AY457350 | AMP-activated protein kinase gamma 3 | Spermatogenesis, stresss response | |||
| 717 | AY457431 | Myeloblastosis oncogene | Anti-apoptosis | |||
| 736 | AY457440 | Transformed mouse 3T3 cell double minute 2 | Negative control of cell proliferation, ubiquitin protein ligase | |||
| 1006 | AY457543 | Pregnane X receptor | Steroid metabolismolism, skeletal development | |||
See Supplemental information for a complete list, including Celera and GenBank ID, Gene Ontology information and a summary of functional data for each clone.
A Genome-Wide Screen of Insulator Function by a Novel Assay
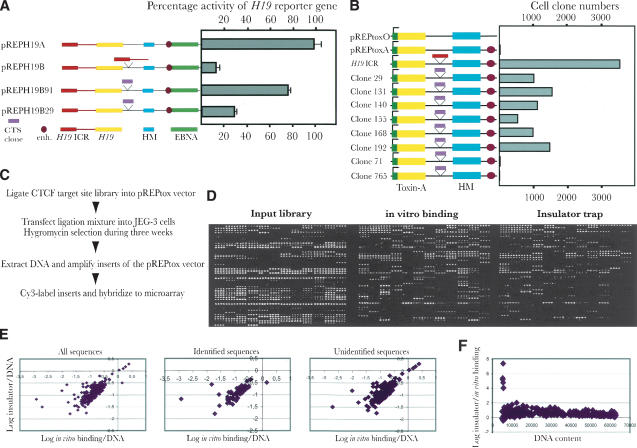
Given the strong link between CTCF binding and chromatin insulator function (Pant et al. 2003; Schoenherr et al. 2003), we examined some randomly selected clones for their ability to block enhancer-promoter communications, using the episomal insulator assay (Kanduri et al. 2000a,b). Figure 4A shows that the two clone sequences were indeed able to block activation of the H19 reporter gene to various degrees. To more efficiently examine the presence or absence of insulator function of sequences in the entire CTCF library, we developed a novel approach based on the ability of intervening sequences, inserted into a multiple cloning site, to interfere with SV40 enhancement of toxin-A reporter gene activation. The episomal vector also included a hygromycin resistance gene placed outside the toxin-A-insulator axis and hence monitored for silencing activity (Fig. 4B). To assess the validity of this technique, we ligated the H19 ICR into the multiple cloning site of the pREPtoxA vector (see Fig. 4B,C), followed by transfection into JEG-3 cells and hygromycin selection for 3 wks. Figure 4B shows that the number of cell clones increased dramatically (more than 400-fold from a background of 5–10 clones) when the toxin-A gene was insulated from the effects of the SV40 enhancer by the H19 ICR. Although it could be argued that this assay does not formally distinguish between insulator and silencer function, silencers are, in contrast to insulators, expected to repress the adjacent hygromycin gene and hence be selected against under the hygromycin pressure. Although this insulator assay cannot establish insulator activity at the endogenous locus, it does give a good indication of potential insulator function.
Figure 4.
The insulator trap assay. (A) Schematic maps of the various constructs used in the classical insulator study. Symbols explained at the bottom of the panel. Each construct is linked to its performance in the enhancer-blocking assays, which were normalized to RNA input and episome copy number. The SV40 enhancer-driven expression of the pREPH19A construct was assigned a value of 100 whereas all other samples were normalized relative to this value. The mean deviation of three different experiments is indicated for each vector construct. (B) Schematic maps of the different pREPtox vectors. Cerise circle: the position of the SV40 enhancer. Green square: the H19 promoter. Pink and red blocks: the different inserts from clones (indicated by its original number) and H19 ICR, respectively. The numbers of the surviving clones were estimated from a colony count assay. (C) Outline of the strategy of the toxin-A assay and its application in microarray analysis of the CTCF target-site library. (D) An example of hybridization with input library sequences, affinity-purified (with recombinant CTCF) CTCF target sites, and the selection of clones with enhancer-blocking properties. (E) Presents scatter plot analyses of insulator strength, determined from the microarray analysis, and in vitro binding patterns, broken down into different sequence categories of the CTCF library. (F) Shows a scatter plot analysis between the insulator/in vitro binding ratios and DNA content of the corresponding spots of the microarrays, as determined by oligo hybridization.
Using the toxin-A approach, we assayed the insulator properties of our entire CTCF target-site library by inserting this into the multiple cloning site of the pREPtoxA plasmid. After transfection into JEG-3 cells and hygromycin selection, as outlined in Figure 4C, the emerging clones (usually more than 20-fold over background per transfection event) were pooled from several (usually 4–5) transfection experiments, and the library inserts were amplified from total DNA preparations. By labeling the input material with Cy3 and the functionally selected sequences with Cy5, we could readily visualize changes in the representation of sequences as a result of the insulator assay (Fig. 4D). To confirm these data, several clones of the CTCF target-site library which showed high insulator function were individually subjected to the toxin-A assay. Figure 4B shows that they all indeed were able to block the communication between the enhancer and the promoter of the reporter gene, albeit less efficiently than the H19 ICR. Conversely, two other members of the library with no insulator function were unable to prevent activation of the toxin-A gene (Fig. 4B).
To examine the relationship between target-site affinity and insulator function, we generated scatterplots by comparing quantitative information from the insulator trap assay with the in vitro and in vivo CTCF binding patterns. Figure 4E (and data not shown) shows that the insulator strength shows strongest correlation with the intrinsic in vitro binding affinity to CTCF. Because this relationship was best visualized in the log scale, it is possible that stronger in vitro binding/insulator function correlates with the presence of multiple CTCF target sites that attract CTCF in a cooperative manner. Conversely, the moderate agreement between in vivo binding and insulator strength parameters (data not shown) suggests that endogenous sequences flanking the CTCF target sites dictate chromatin conformations that serve to limit CTCF availability or affinity for some sites. Indeed, the patterns of CTCF target-site occupancy are dramatically modified during development (R. Mukhopadhyay, J. Whitehead, M. Lezcano, W.-Q. Yu, A. Mattsson, and R. Ohlsson, unpubl.). The correlation between CTCF affinity and insulator strength was further underscored by the fact that the relationship was essentially identical within each subclass of library sequences, including single copy sequences and LTRs (data not shown). By generating scatterplots between the insulator/in vitro binding data and the DNA content, as determined by the oligo hybridization approach, we were also able to rule out that varying DNA amounts on the microarrays skewed the data inappropriately (Fig. 4F). We conclude that the CTCF-dependent chromatin insulator function operates in an analog (continuous) mode, potentially prompting a redefinition of the chromatin insulator concept.
Cross-Referencing CTCF Target Sites with CpG Methylation Status
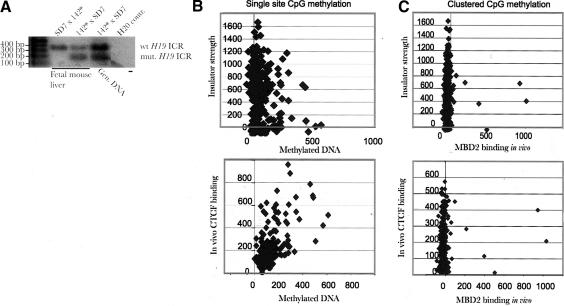
We and others showed previously that CTCF target sites within the maternal H19 ICR allele protect against de novo methylation (Pant et al. 2003; Schoenherr et al. 2003). To examine the generality of this feature, we cross-referenced our CTCF target-site library with DNA methylation marks. The probe hybridized to the microarrays was derived from immunopurified sequences using a specific antibody against 5-methylcytidine. Figure 5A shows that this antibody is highly specific, because it pulls down only the paternal H19 ICR allele when the maternal allele is unmethylated (of the wild type). Conversely, both parental alleles are pulled down by the anti-methylcytidine antibody when the maternal H19 ICR allele carries mutated CTCF binding sites (Fig. 5A). This result is expected due to massive de novo methylation of the maternal ICR allele when mutated, as determined by bisulfite sequencing (Pant et al. 2003). Figure 5B shows a scatterplot analysis revealing that a fraction of the CTCF binding sequences is at least partially methylated at single CpGs. The possibility that a small level of background DNA methylation both within and at sites flanking the CTCF target-site sequence could reflect plasticity of the methylation status at single CpGs is supported by the observation that the insulator strength and the in vivo CTCF target-site occupancy in mouse fetal liver are generally devoid of the methyl-binding protein MBD2 (Fig. 5C), which interacts with clusters of methylated CpGs (Ballestar and Wolffe 2001).
Figure 5.
Cross-referencing methylation status with CTCF occupancy. (A) shows that an antibody against 5-methylcytidine immunopurifies only the methylated paternal H19 ICR allele if the maternally inherited allele is of the wild type. Conversely, when the mutated H19 ICR allele is inherited maternally (labeled 142* and unable to interact with CTCF in vivo while displaying massive de novo methylation; Pant et al. 2003), both alleles are brought down as determined by using PCR primers spanning CTCF target site #3 and a diagnostic EcoRV site (Pant et al. 2003). (B,C) Scatterplot analyses comparing CTCF in vivo occupancy/insulator strength vs. single CpG methylation (B, using an antibody against methylated cytidine) and clustered (C, using an antibody against MBD2) CpG methylation states in mouse fetal liver.
DISCUSSION
We report here the uncovering of a CTCF-organized network that coordinates the epigenetic states of numerous target sites throughout the genome both by performing as a chromatin insulator and by reading or maintaining methylation-free domains. This additional level of connectivity between previously unlinked pathways is of major importance, as it demonstrates new interactions between loci with pivotal functions, such as metabolism, growth, neurogenesis, and cell signaling. The absence of previously known CTCF target sites in our library suggests that our screening has covered only a fraction of potential CTCF target sites. A conservative assessment of the total number of CTCF target sites, based on ChIP-on-chip analysis of the entire human chromosome 22, suggests a distribution of high-affinity CTCF target sites on average every 400 kb (W.-Q.Yu, J. Dumanski, and R. Ohlsson, unpubl.). Assuming that this number is representative for all chromosomes of the mouse, our library covers ∼5%–7% of all potential high-affinity CTCF target sites.
The apparent discrepancy between in vivo and in vitro binding affinities, as seen in Figure 1F, could originate from several factors. The in vivo binding affinity as measured here will reflect the proportion of cells within the heterogeneous liver tissue utilising a particular target site. A sequence with a strong intrinsic affinity for CTCF (high in vitro binding score) may be only functionally occupied by CTCF in a subset of cells, and therefore present at low concentration in the probe used for the in vivo assay, resulting in a low in vivo binding score. Our preliminary results indeed reveal the existence of lineage-specific patterns of occupancy of CTCF binding sites (R. Mukhopadhyay, J. Whitehead, M. Lezcano, W.-Q. Yu, A. Mattsson, and R. Ohlsson, unpubl.), which is in keeping with chromatin conformation-based restrictions in the availability of CTCF target sites in these instances. The chromatin-specific parameter controlling CTCF target-site occupancy may involve nucleosome positioning, because CTCF is unable to interact with its target site if this is covered by a nucleosome (Kanduri et al. 2002). In vivo, CTCF might be outcompeted from a subset of its target sites which overlap with cis elements interacting with other trans-acting factors. The context-dependent combinatorial use of specific zinc fingers at particular target sites and hence the affinity of this interaction may not be consistently recapitulated in our in vitro binding studies. Finally, binding affinity may also be modulated by posttranslational modification of CTCF (Klenova et al. 2001). We therefore cannot rule out the possibility that the conformation assumed by the recombinant CTCF in an in vitro assay may not necessarily reflect the natural context of the DNA binding interaction at all target sites.
Our results here provide new insights into the mode of insulator function: Considerable differences in insulator strength are proportional to the binding efficiency of CTCF. When combined with chromatin modifications to increase or decrease availability of CTCF target sites, a picture of a rheostat or analog mode of insulator function is emerging. These data support an increasing recognition of the role of stochastic events in gene expression. Insulator function could perhaps be characterized in terms of enhancer or silencer signals leaking through an insulator with probability inversely proportional to the binding efficiency. Because the insulator strength correlated with binding efficiency in the log scale, it is conceivable that some insert clones contain multiple binding sequences that interact in a cooperative manner. Precedence for such a scenario has recently been uncovered: The mutation of only one of the four CTCF binding sites in the H19 ICR leads to robust activation of the maternal Igf2 allele and complex patterns of de novo methylation when maternally inherited (Pant et al. 2004). These results suggest that there is a need for all four CTCF target sites to cooperate to both efficiently insulate the maternal Igf2 allele from downstream enhancers and to maintain the methylation privilege of the maternal H19 ICR allele.
Although there was a certain degree of plasticity of methylation of single CpGs, as determined by using the antibody against methyl-cytidine, there was a general lack of overall methylation as determined by using the antibody against MBD2. Because this protein interacts with methylated CpGs only when these are clustered, it could be argued that there is a trivial explanation of methylation-free states of CTCF target sites genome-wide, that is, absence of CpGs. However, the inserts and their immediate flanks (up to 1 kb) of the single copy category contain on average 21 CpGs (ranging from five to 74 CpGs). The exceptions, with a relatively high degree of methylation despite relatively strong insulator activity and in vivo binding to CTCF, could be explained by invoking the possibility that these clones reside in imprinting control regions in which one of the alleles is methylated, while the unmethylated allele is binding CTCF, as in the case of the H19 ICR (Kanduri et al. 2000a,b). Indeed, two of the library CTCF target sites mapped within known imprinted domains (Grb10 and Snrpn), whereas five other members of the CTCF target-site library could be identified in the EICO library of candidate imprinted genes (Nikaido et al. 2004), strengthening a link between CTCF and genomic imprinting.
A surprising result of our analyses was that the majority of sequences could not be identified using any of the established databases, such as Celera and NCBI. In light of the fact that 99% of the euchromatin, but very little of the heterochromatin, has been sequenced (The Mouse Genome Sequencing Consortium 2002), these clones might belong to heterochromatic domains. Support for this deduction comes from our observation here that the distribution of CTCF is essentially identical to that of a marker of heterochromatin, HP1β, in cultured mouse embryo fibroblasts. Moreover, preliminary immunostaining observations show that the bulk of CTCF associated with mitotic chromosomes maps to centromeres, which constitute the main heterochromatic compartment (V. Lobanenkov, unpubl.). Finally, Lsh, a chromatin remodeling factor associated primarily with heterochromatin (Yan et al. 2003), extensively interacts with CTCF target sites of our library (J. Whitehead, P. Mariano, M. Lezcano, C. Kanduri, W.-Q. Yu, M. Parrinen, K. Muegge, E. Klenova, V. Lobanenkov, and R. Ohlsson, unpubl.). We propose therefore that a significant proportion of CTCF target sites belong to the heterochromatic compartment and that the main properties of CTCF target sites are shared between heterochromatic and euchromatic domains. The implication of this statement, that CTCF might organize active expression domains within heterochromatin, is supported by our preliminary observation that a CTCF binding site at the Xist promoter is occupied only on the active Xist allele of the inactive, heterochromatinized X chromosome in female mouse placenta (E. Pugacheva, V.K. Tiwari, A.A. Vostrov, W.W. Quitschke, D.I. Loukinov, R. Ohlsson, and V.V. Lobanenkov, unpubl.).
The genome-wide distribution of insulation and methylation protection features reported here and previously associated with only differentially methylated imprinting control regions supports a prior proposal that the imprinting phenomenon has evolved from unusual combinations of common epigenetic determinants (Horsthemke et al. 1999). In this regard, the identification of repeat elements in many CTCF target sites suggests the possibility that the transposition of a subset of repeats that displays cooperating CTCF target sites can modify expression domains by insulation if inserted strategically between enhancers or silencers and promoters. These considerations might be profoundly influenced by the emergence of BORIS, which is a mammalian paralog of CTCF with extensive similarities in the central zinc finger binding domain and which is exclusively expressed during male germline development (Loukinov et al. 2002). Because BORIS is frequently activated in human cancer cells (Klenova et al. 2002), we infer that its pathological interaction with CTCF target sites breaks down the CTCF network with ensuing aberrant epigenetic states and unscheduled expression at vital loci identified in this report.
METHODS
Creation of the CTCF Library
Fetal mouse liver cells (E16) were mechanically dispersed and formaldehyde-crosslinked for chromatin immunopurification (ChIP) as described (Kuo and Allis 1999). The immunopurified material (ChIP DNA), using the C-terminal CTCF antibody (Upstate Biotechnology), was endfilled, phosphorylated, and ligated to T7 and T3 linkers. Using the same primers the DNA was amplified and cloned into pGemT-Easy vector to generate the initial library of 2200 clones. The inserts of the library were size-selected (100–300 bp) and cloned back into pGemT-Easy to make a secondary library. Following transformation into XL1 Blue cells, 1128 clones, consisting of 266 unique sequences, were isolated. The plasmid DNA was prepared using the Montage Plasmid Miniprep96kit (Millipore) and inserts were PCR amplified, purified (Millipore PCR purification kit), precipitated, and dissolved in a 50% DMSO/nitrocellulose printing solution. The DNA was spotted on poly-L-lysine-coated glass slides (in-house coated) to generate the CTCF target-site library microarray using a Cartesian Technologies Prosys 5510A printer with Telechem Stealth SMP 3B pins, at 24°C and relative humidity 45% during printing. Slides also contained several positive and negative controls. Each clone, including controls, was spotted eight times. The slides were UV cross-linked at 450 mJ and stored away from light.
Sequencing and Analysis of Clones
The clones were sequenced using Applied Biosystem's BigDye Terminator cycle sequencing kit and run on an ABI377 sequencer. The bioinformatic analysis was done using genome information from Celera Discovery System, NCBI Mouse Genome Resource, and Ensembl Mouse Genome Server, as well as the Gene Ontology database (www.geneontology.org). Analysis was aided by tools from RepeatMasker (A. Smit and P. Green, http://ftp.genome.washington.edu/RM/RepeatMasker.html) and the EMBOSS sequence analysis suite (NewCpGSeek, SeqMatchAll; http://bioinfo.pbi.nrc.ca:8090/EMBOSS/index.html). Band-shift analysis was carried out as described (Filippova et al. 1996).
Immunopurification of Methylated Sequences
Ten μg of mouse fetal liver DNA resulting from a cross between mice carrying a mutant H19 ICR allele paternally or maternally (the 142* strain) and SD7 mice as described (Pant et al. 2003) was sonicated by three pulses at 30% power, 10 sec per pulse using a Branson digital sonifier, to yield fragments on average 200–300 bp. Following denaturation, the DNA was precleared with protein A 4 Fast FlowSepharose (Amersham Biosciences) and incubated overnight at 4°C with 2.5 μg of 5-methylcytidine monoclonal antibody (Eurogentec). The complex was purified using protein-A Sepharose (Pharmacia) and washed as described for the ChIP protocol (see below). The immunopurified samples were analyzed for allelic distribution at CTCF target site #3 in the mouse H19 ICR by PCR amplification using forward primer 5′CT CAGTGGTCGATAT3′ and reverse primer 5′TGAGTCAAGTTC TCT3′ for CTCF target sites with a PCR cycle 95°C 5 min, 25× (95°C 40 sec, 54°C 30 sec, 72°C 30 sec), 72°C 7 min. The parental origin of the H19 ICR alleles was confirmed by EcoRV digestion of the PCR samples, as this site was specific for the mutant allele (Pant et al. 2003). The immunopurified samples were amplified and hybridized to the CTCF target-site microarrays as outlined below.
Probe Preparation and Hybridization
ChIP DNA was prepared from fetal mouse liver by using either an affinity-purified rabbit antibody against the N-terminal portion of CTCF or a rabbit antibody against MBD2 (kind gift from Dr. A. Wolffe, Sangamo, Inc.). The ChIP DNA samples were amplified by a two-step PCR method. The first amplification was done using a random-specific primer SR1 (5′GCCGTCGAC GAATTCNNNNNNNNN3′) with PCR cycle 94°C 5 min, 5× (94°C 30 sec, 15°C 60 min, 20°C 45 min, 25°C 30 min, 30°C 30 min, 35°C 20 min, 40°C 1 min, 45°C 30 sec, 50°C 30 sec, 55°C 20 sec, 60°C 20 sec), 60°C 7 min. The resulting PCR product was used as the starting material for the second PCR reaction with specific primer SR2 (5′GCCGTCGACGAATTC3′), with PCR cycle 94°C 5 min, 23× (94°C 30 sec, 50°C 55 sec, 72°C 1 min), 72°C 7 min. The probe was prepared using CyScribe Post-Labelling kit (Cy Dye Post-labelling Reactive Dye Pack, Amersham Biosciences) and also by direct PCR labeling. The primers used for the direct PCR labelling reaction were SR2 and a random primer (N)10, with Cy5 or Cy3 fluorophores using 94°C 2 min, 23 × (94°C 20 sec, 50°C 40 sec, 72°C 1 min), 72°C 7 min. Probes prepared by Cy dye post-labeling and direct PCR labeling were pooled and precipitated with 100 μg of Cot-1 DNA (Clontech). The labeled DNA was dissolved in water and hybridization solution (GlassHyb Hybridization solution, Clontech), denatured, and incubated at 45°C for 2 h. The slides were prehybridized in solution containing 3.5 × SSC, 0.1% SDS, 0.1% BSA, and 1.3 mg/mL herring sperm ssDNA at 55°C for 1 h. Hybridization and washing were done according to the GlassHyb Hybridization Solution kit User Manual (Clontech). The slides were scanned using ScanArray 4000, and analysis was done with QuantArray version 3.0 (Packard Biosciences). Background subtraction and normalization were done, which included the subtraction of local background and also of the signal obtained in the negative controls. The oligo hybridization assay to quantitate DNA in each spot was carried out using the 9mer hybridization protocol (Operon, QIAGEN).
Multiplex PCR Analysis
To independently verify the presence or absence of individual clones in the ChIP material, and to test for bias in the random amplification step used during probe preparation, a multiplex PCR screen was used. Primers were designed within the cloned fragments, and groups of three to four loci, plus the H19 ICR as a control, were amplified simultaneously from a dilution series of genomic DNA, as well as the original and random-amplified ChIP material and serum controls. Amplified fragments were resolved on 10% acrylamide gels, stained with SYBR Green, and visualized on a Fuji FLA3000 phosphorimager. The amplification shown in Figure 1D was carried out at 94°C 3 min, 30× (94°C 30 sec, 57°C 30 sec, 72°C 40 sec), 72°C 3 min, using primers: 29F 5′gtctgcagaa gcacttgaag3′, 29R 5′ccatcttctggtgcatcatc3′, 163F 5′gtatgcagagactt ggagac3′, 163R 5′agccgcatcagtcattagtc3′, 363F 5′tcctggatgttgagaa cacg3′, 363R 5′aaacctctagcctggagaag3′, H19F 5′cggactcccaaatcaac aag3′, and H19R 5′gcaatccgttttaggactgc3′.
Array-Based Binding Assay
The in vitro binding reaction between inserts from the library and recombinant Pichia CTCF (Quitschke et al. 2000) was performed in binding buffer (Filippova et al. 1996) at room temperature, and the complexes were recovered using the CTCF antibody (against the N-terminal portion) and protein A 4 Fast Flow Sepharose beads. The purified DNA was PCR-amplified and labeled using T7 and SP6 primers, followed by hybridization to the CTCF target-site library microarray as described above.
Episomal Insulator Assay
Two positive clones based on hybridization and multiplex PCR results were selected and cloned into the episomal vector pREPH19B at the Kpn1 and Xho1 sites. These were transfected into the JEG-3 cell line. After 9 d, DNA and RNA were prepared using the Wizard Genomic DNA Purification kit (Promega) and RNeasy Minikit (QIAGEN), respectively. The RNase protection expression analysis was performed as described (Kanduri et al. 2000a).
Insulator Trap Assay
A 670-bp Diphtheria Toxin A chain gene segment (derived from pIBI30-DT-A, a kind gift from Dr. Ian Maxwell, Univ. of Colorado, Colorado.) was inserted into pGEM-H19 containing the H19 promoter (–166 to +336 relative to the H19 transcriptional start site) using the restriction sites PstI and SalI. The whole cassette containing the H19 promoter and the DT-A reporter gene was first restricted with ApaI, blunt-ended, and restricted with XbaI. This was inserted into the XbaI site of episomal vector pREPH19B (Kanduri et al. 2000a), replacing the existing H19 minigene from the vector to generate the pREPtox plasmid. The control plasmids were generated by a similar strategy. The negative control plasmids were pREPtoxO, which lacks the SV40 enhancer, and pREPtoxA, which includes the enhancer but lacks the H19 ICR. The positive control was taken as the plasmid containing both H19 ICR and the enhancer. The multiple cloning site was used to insert the entire CTCF library with the aid of the following primers:
Xho1-T7: 5′CCGCTCGAGCGGTAATACGACTCACTATAGGG 3′; Kpn1-Sp6: 3′TAAGATATCACAGTGGATTTAGCCCCATGGGGC 5′; Kpn1-T7: 5′CGGGGTACCCCGTAATACGACTCACTATAGGG 3′; Xho1-Sp6: 3′TAAGATATCACAGTGGATTTAGGCGAGCTCGCC 5′.
The entire ligation mixture was transfected into the JEG-3 cell line. The clones were selected against hygromycin (150 μg/mL) for 2–3 wks. Genomic DNA was prepared from the surviving clones using the Wizard Genomic DNA Purification kit. The inserts were PCR-amplified using the above primers, labeled, and hybridized to the CTCF target-site library microarray as described above.
Colony Count Assay
The above-mentioned plasmids were used to generate individual clone constructs. Representative positive and negative clones scored in the toxin assay for insulator trap function were chosen and inserted individually into the multiple cloning site of the pREPtox vector. These newly generated plasmids containing the sequence from individual clones were then transfected into the JEG-3 cell line and selected against hygromycin (150 μg/mL) for 3 wks. The colonies obtained were then washed and fixed with paraformaldehyde (4% in PBS) and stained with hematoxylin. Following the final wash to remove the excess stain, the colonies were counted.
Immunohistochemistry of CTCF and HP1β Distribution
Murine adult lung fibroblasts were fixed in acetone for 10 min. Double-immunofluorescence staining was done using goat anti-HP1β (1:50 dilution, Santa Cruz Biotechnology) and affinity-purified rabbit anti-CTCF (1:50 dilution). Detection was performed sequentially using biotinylated anti-goat secondary antibody (1:200 dilution) made in horse (Vector) with the following avidin-FITC (Vector) conjugation for anti-HP1β; and with goat anti-rabbit secondary antibodies Cy-3 conjugated for the detection of CTCF. DAPI counterstaining allowed visualization of the cell nuclei. Images were captured using a Leica DMIRE2 fluorescence microscope equipped with the cooled CCD camera Evolution QE1 (Media Cybernetics) using IPLab Image software (Scanalytics).
Acknowledgments
We thank Dr. Anders Isaksson and gratefully acknowledge the Wallenberg microarray platform at the Rudbeck laboratory. This work was supported by the Swedish Science Research Council (VR, to R.O.), the Juvenile Diabetes Research Foundation International (JDRF, to R.O.), the Swedish Cancer Research Foundation (CF, to R.O.), the Swedish Pediatric Cancer Foundation (BCF, to R.O.), the Wallenberg and Lundberg Foundations (to R.O.), Stiftelsen Wenner-Grenska Samfundet (to R.O.), and intramural research funding from NIAID NIH (to V.V.L.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2408304. Article published online ahead of print in July 2004.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. AY457177–AY457567.]
References
- Ballestar, E. and Wolffe, A. 2001. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur. J. Biochem. 268: 1–6. [DOI] [PubMed] [Google Scholar]
- Bell, A.C. and Felsenfeld, G. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485. [DOI] [PubMed] [Google Scholar]
- Bell, A.C., West, A.G., and Felsenfeld, G. 2001. Insulators and boundaries: Versatile regulatory elements in the eukaryotic genome. Science 291: 447–450. [DOI] [PubMed] [Google Scholar]
- Filippova, G., Fagerlie, S., Klenova, E., Myers, C., Dehner, Y., Goodwin, G., Neiman, P., Collins, S., and Lobanenkov, V. 1996. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell Biol. 16: 2802–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2000. Gene ontology: Tool for the unification of biology. Nature Genet. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hark, A.T., Schoenherr, C.J., Katz, D.J., Ingram, R.S., Levorse, J.M., and Tilghman, S.M. 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489. [DOI] [PubMed] [Google Scholar]
- Holmgren, C., Kanduri, K., Dell, G., Ward, A., Mukhopadhya, R., Kanduri, M., Lobanenkov, V., and Ohlsson, R. 2001. CpG methylation regulates the Igf2/H19 insulator. Curr. Biol. 11: 1128–1130. [DOI] [PubMed] [Google Scholar]
- Horsthemke, B., Surani, M.A., James, T.C., and Ohlsson, R. 1999. The mechanisms of genomic imprinting. In Genomic imprinting: An interdisciplinary approach (ed. R. Ohlsson), pp. 91–118. Springer-Verlag, Berlin. [DOI] [PubMed]
- James, T.C. and Elgin, S.C. 1986. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell Biol. 6: 3862–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri, C., Holmgren, C., Franklin, G., Pilartz, M., Ullerås, E., Kanduri, M., Liu, L., Ginjala, V., Ulleras, E., Mattsson, R., et al. 2000a. The 5'-flank of the murine H19 gene in an unusual chromatin conformation unidirectionally blocks enhancer-promoter communication. Curr. Biol. 10: 449–457. [DOI] [PubMed] [Google Scholar]
- Kanduri, C., Pant, V., Loukinov, D., Pugacheva, E., Qi, C.-F., Wolffe, A., Ohlsson, R., and Lobanenkov, A. 2000b. Functional interaction of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10: 853–856. [DOI] [PubMed] [Google Scholar]
- Kanduri, C., Fitzpatrick, G., Mukhopadfhyay, R., Kanduri, M., Lobanenkov, V., Higgins, M., and Ohlsson, R. 2002. A differentially methylated imprinting control region within the Kcnq1 locus harbours a methylation-sensitive chromatin insulator. J. Biol. Chem. 277: 18106–18110. [DOI] [PubMed] [Google Scholar]
- Klenova, E., Chernukhin, I., El-Kady, A., Lee, R., Pugacheva, E., Loukinov, D., Goodwin, G., Delgado, D., Filippova, G., Leon, J., et al. 2001. Functional phosphorylation sites in the C-terminal region of the multivalent multifunctional transcriptional factor CTCF. Mol. Cell Biol. 21: 2221–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenova, E., Morse, H., Ohlsson, R., and Lobanenkov, V.V. 2002. The novel Boris + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Sem. Cancer Biol. 12: 399–414. [DOI] [PubMed] [Google Scholar]
- Kuo, M. and Allis, C. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein: DNA associations in a chromatin environment. Methods 19: 425–433. [DOI] [PubMed] [Google Scholar]
- Loukinov, D., Pugacheva, E., Vatolin, S., Pack, S., Moon, H., Chernukhin, I., Mannan, P., Larsson, E., Kanduri, C., Vostrov, A., et al. 2002. BORIS, a novel male germline-specific protein associated with epigenetic reprogramming events, shares the same 11 Zn-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc. Natl. Acad. Sci. 99: 6806–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. [DOI] [PubMed] [Google Scholar]
- Nikaido, I., Saito, C., Wakamoto, A., Tomaru, Y., Arakawa, T., Hayashizaki, Y., and Okazaki, Y. 2004. EICO (Expression-based Imprint Candidate Organizer): Finding disease-related imprinted genes. Nucleic Acids Res. 32: D548–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson, R., Renkawitz, R., and Lobanenkov, V. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17: 520–527. [DOI] [PubMed] [Google Scholar]
- Pant, V., Mariano, P., Kanduri, C., Mattsson, A., Lobanenkov, V., Heuchel, R., and Ohlsson, R. 2003. The nucleotides responsible for the direct physical contact between the chromatin insulator protein CTCF and the H19 imprinting control region manifest parent of origin-specific long-distance insulation and methylation-free domains. Genes & Dev. 17: 586–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant, V., Kurukuti, S., Pugacheva, E., Shamsuddin, S., Mariano, P., Renkawitz, R., Klenova, E., Lobanenkov, V., and Ohlsson, R. 2004. Mutation of a single CTCF target site within the H19 imprinting control region leads to loss of Igf2 imprinting and complex patterns of de novo methylation upon maternal inheritance. Mol. Cell Biol. 8: 3497–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quitschke, W.W., Taheny, M.J., Fochtmann, L.J., and Vostrov, A.A. 2000. Differential effect of zinc finger deletions on the binding of CTCF to the promoter of the amyloid precursor protein gene. Nucleic Acids Res. 28: 3370–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr, C., Levorse, J., and Tilghman, S. 2003. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 33: 66–69. [DOI] [PubMed] [Google Scholar]
- Yan, Q., Cho, E., Lockett, S., and Muegge, K. 2003. Association of Lsh, a regulator of DNA methylation, with pericentromeric heterochromatin is dependent on intact heterochromatin. Mol. Cell Biol. 23: 8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker site.
- www.geneontology.org; Gene Ontology database.
- http://bioinfo.pbi.nrc.ca:8090/EMBOSS/index.html; the EMBOSS sequence analysis suite.