Abstract
We have generated 36,991,173 17-base sequence “signatures” representing transcripts from the model plant Arabidopsis. These data were derived by massively parallel signature sequencing (MPSS) from 14 libraries and comprised 268,132 distinct sequences. Comparable data were also obtained with 20-base signatures. We developed a method for handling these data and for comparing these signatures to the annotated Arabidopsis genome. As part of this procedure, 858,019 potential or “genomic” signatures were extracted from the Arabidopsis genome and classified based on the position and orientation of the signatures relative to annotated genes. A comparison of genomic and expressed signatures matched 67,735 signatures predicted to be derived from distinct transcripts and expressed at significant levels. Expressed signatures were derived from the sense strand of at least 19,088 of 29,084 annotated genes. A comparison of the genomic and expression signatures demonstrated that ∼7.7% of genomic signatures were underrepresented in the expression data. These genomic signatures contained one of 20 four-base words that were consistently associated with reduced MPSS abundances. More than 89% of the sum of the expressed signature abundances matched the Arabidopsis genome, and many of the unmatched signatures found in high abundances were predicted to match to previously uncharacterized transcripts.
Continued improvements in plant molecular biology will depend on parallel improvements in our understanding of how genes are encoded in the genomic sequence, how they are regulated, and the range of transcripts possible for each gene. Two plant genomes are complete or nearly complete (Arabidopsis Genome Initiative 2000; Feng et al. 2002; Goff et al. 2002; Sasaki et al. 2002; Yu et al. 2002), and it is likely that additional genomes will become available in the foreseeable future. The next challenge for plant biologists is to completely define the characteristics of the “transcriptome” contained within these plant genomes. Computational approaches to identifying genes from finished genomes are improving, but can produce inconsistent results and still need to be trained with experimentally derived data (Andrews et al. 2000; de Souza et al. 2000; Guigo et al. 2000; Haas et al. 2002; Reese et al. 2000). As genomic sequencing becomes faster and more economical, it is critically important that methods, technologies, and resources be developed to experimentally detect every active gene, alternatively spliced and alternatively terminated or polyadenylated transcript.
Changes in gene expression are often indicative of changes in underlying biochemical processes. Presently, it is more tractable to measure transcript profiles than protein profiles. The most widely used method to analyze global patterns of gene expression is currently the DNA microarray (Schena et al. 1995, 1998; Duggan et al. 1999). Microarrays represent a powerful and relatively inexpensive approach to simultaneously measure the expression patterns of many genes in a genome. The expression level is determined by relative changes in the hybridization intensity of the sequences that are present in the array. However, because microarrays are hybridization-based, background may interfere with the detection of weakly expressed genes, and gene families may cross-hybridize, confounding measurements of some transcripts. One of the most intriguing applications of microarrays is the development of whole-genome tiling arrays for the characterization of transcriptional activity (Shoemaker et al. 2001; Kapranov et al. 2002; Rinn et al. 2003; Yamada et al. 2003). This approach reduces one of the biases common in microarrays by not limiting the probes to annotated genes.
In parallel to the remarkable advances in hybridization-based techniques like microarrays, technologies for measuring the quantitative levels of gene expression have made significant advances. Large-scale quantitative expression technologies involve the generation of short sequence tags from a given RNA sample, and use the abundance of these sequence tags to determine the relative abundance of each transcript. These methods are complementary to standard microarrays, because sequence tags are not preselected like the probes on most microarrays. Such random sequencing approaches can identify novel genes or noncoding RNA sequences. Several technologies have been widely used to generate sequence tags. One of the first such methods was the development of collections of single-pass sequencing reads (expressed sequence tags, or ESTs) that were used to estimate gene expression levels (Adams et al. 1995). More recently, shorter sequence tags have been used, such as the 14-base tags produced by the Serial Analysis of Gene Expression, or SAGE (Velculescu et al. 1995, 1997). These tags can be generated in large numbers for a given tissue to determine gene expression levels quantitatively. SAGE is one of the most popular of these quantitative methods to characterize gene expression because it can be performed in individual labs and generates data sets that have proven valuable for the annotation of complex genomes (Velculescu et al. 1999, 2000; Caron et al. 2001; Saha et al. 2002). The short length of SAGE tags makes it difficult to unambiguously assign tags within a genome, but recent modifications have adapted the method to generate 21-base tags that may occur uniquely in a complex genome (Saha et al. 2002). The number of signatures generated for most available SAGE libraries limits this method to the analysis of moderate-to-highly expressed transcripts (Ishii et al. 2000).
One of the most powerful new technologies for quantitative analysis of gene expression is Massively Parallel Signature Sequencing, or MPSS (Brenner et al. 2000a,b). MPSS involves the cloning of a cDNA library on beads and the acquisition of 17–20-nt “signatures” (tags) from these cDNAs using an unconventional sequencing method (summarized in Fig. 1). The abundance of the sequence signatures precisely reflects gene expression levels in the sampled tissue. With more than 1 million signatures obtained per library, the technology is sensitive to transcripts expressed at very low levels. Each signature is derived from the 3′-most DpnII (Sau3A) site 5′ to the poly(A) tail of a cDNA molecule. The sequencing process proceeds by the identification of sets of four bases by hybridization to labeled linker-probes, then removal of that set of four bases by a Type IIS restriction enzyme site contained in the linker, and then repetition of the process (Brenner et al. 2000a). These fluorescent reactions occur underneath an automated microscope and scanner while the beads are immobilized in a flow-cell, with no gels or capillaries. The procedure is completely parallel, facilitating large-scale sequencing, and 17–20 nt of high-quality sequence is routinely obtained per bead (Brenner et al. 2000a). Signatures or tags of this length have a high specificity and may match to just one position in a complex genome (Saha et al. 2002).
Figure 1.
Overview of MPSS methodology. A brief outline of the steps required for the processing of mRNA to obtain the MPSS signatures, in order from top to bottom. The methodology is described in more details in publications by Brenner et al. (2000a,b) or at http://www.lynxgen.com. The step indicated by an asterisk and in parentheses only takes place in the “signature” version of the MPSS method (Lynx Therapeutics, Inc.).
The high-quality sequence and annotation of the Arabidopsis genome makes it possible to assess the utility of tag-based expression analyses for this model plant. Expressed tags can be matched back to their genomic context to reveal the full set of transcripts and their precise expression levels (Meyers et al. 2004b). Parallel analyses of the tags and the genomic context may reveal systematic technological biases. With an understanding of the bias inherent in a specific transcriptional profiling technology, it may be possible to make corrections and arrive at a more accurate assessment of gene expression. In this report, we characterize the application and utility of MPSS in an analysis of gene expression in Arabidopsis. This bioinformatics analysis is based on the extraction of all potential signatures from the annotated genome and comparison of these signatures to the MPSS expression data.
RESULTS
MPSS Library Construction, Data Processing, and Signature Filtering
A total of 14 libraries were constructed using mRNA from diverse tissues, mutants, and treatments of Arabidopsis. These libraries were generated as part of specific experiments that are or will be described elsewhere. The libraries are listed in Table 1, and include untreated silique, callus, leaves, roots, and inflorescence, with all five sequenced using the “classic” method of MPSS (described in more detail in Meyers et al. 2004b). The leaf, root, and inflorescence libraries were also sequenced using the “signature” variation of MPSS. Briefly, the difference between the classic and signature MPSS methods is that in the former the entire 3′-DpnII-to-poly(A) fragment is cloned, whereas the latter takes advantage of the Type IIS enzyme MmeI to clone a fragment of only 21 to 22 bases that includes the DpnII site (Lynx Therapeutics, Inc.). The inflorescence was analyzed using the signature MPSS method using RNA from the four floral mutants agamous, apetala1-10, apetala3-6, and a superman/apetala1-10 double mutant (H. Ghazal, H. Sakai, and B.C. Meyers, unpubl.). Two signature MPSS libraries were constructed from leaves treated with salicylic acid; the tissue was collected 4 h or 52 h after treatment (B.C. Meyers, M. West, R.W. Michelmore, and D. St. Clair, unpubl.). All 14 libraries were from the Arabidopsis ecotype Col-0. Taken together, these libraries represent a range of tissues and treatments with which to assess the application of MPSS for transcriptional analysis in a complex eukaryotic genome.
Table 1.
Libraries and MPSS Signature Summary Statistics
| # | Library | Code | 2-step runs | 2-step signatures | 4-step runs | 4-step signatures | Total signatures | Total distinct signatures | Total distinct, significant, reliable | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A. 17-base MPSS signature data | |||||||||||
| 1 | Callus | CAFa | 2 | 1,014,442 | 2 | 945,097 | 1,959,539 | 40,901 | 20,081 | ||
| 2 | Inflorescence | INFa | 3 | 1,137,173 | 2 | 637,133 | 1,774,306 | 37,750 | 18,367 | ||
| 3 | Leaves | LEFa | 3 | 1,364,617 | 3 | 1,519,981 | 2,884,598 | 53,394 | 20,465 | ||
| 4 | Roots | ROFa | 5 | 2,340,375 | 3 | 1,302,257 | 3,642,632 | 48,100 | 20,713 | ||
| 5 | Silique | SIFa | 2 | 980,274 | 2 | 1,032,585 | 2,012,859 | 38,501 | 19,714 | ||
| 6 | apt1-10 Inflor. | AP1 | 2 | 1,290,799 | 2 | 1,673,925 | 2,964,724 | 61,270 | 22,454 | ||
| 7 | ap3-6 Inflor. | AP3 | 2 | 1,097,876 | 2 | 1,338,089 | 2,435,965 | 52,172 | 25,229 | ||
| 8 | agamous Inflor. | AGM | 2 | 1,058,093 | 2 | 1,517,577 | 2,575,670 | 41,091 | 17,488 | ||
| 9 | Inflorescence-2 | INS | 2 | 1,315,770 | 2 | 1,575,124 | 2,890,894 | 53,982 | 22,347 | ||
| 10 | Roots-2 | ROS | 2 | 1,247,698 | 2 | 1,210,738 | 2,458,436 | 43,728 | 22,274 | ||
| 11 | sup/ap1-10 Inflor. | SAP | 2 | 1,061,249 | 2 | 1,249,101 | 2,310,350 | 38,002 | 20,836 | ||
| 12 | SA 4 hr—leaf | S04 | 2 | 1,469,036 | 2 | 1,537,939 | 3,006,975 | 21,701 | 11,374 | ||
| 13 | SA 52 hr—leaf | S52 | 2 | 1,476,603 | 2 | 1,488,237 | 2,964,840 | 24,213 | 12,628 | ||
| 14 | Leaves-2 | LES | 2 | 1,562,225 | 2 | 1,547,160 | 3,109,385 | 44,086 | 22,525 | ||
| Total | 33 | 18,416,230 | 30 | 18,574,943 | 36,991,173 | 268,132b | 87,705b | ||||
| B. 20-base MPSS signature data | |||||||||||
| 1 | Callus | CAFa | 2 | 832,604 | 2 | 804,803 | 1,637,407 | 35,284 | 19,542 | ||
| 2 | Inflorescence | INFa | 3 | 933,710 | 2 | 522,137 | 1,455,847 | 31,561 | 17,271 | ||
| 3 | Leaves | LEFa | 3 | 1,144,721 | 3 | 1,313,015 | 2,457,736 | 46,113 | 18,320 | ||
| 4 | Roots | ROFa | 5 | 1,897,860 | 3 | 1,104,358 | 3,002,218 | 41,225 | 19,660 | ||
| 5 | Silique | SIFa | 2 | 808,348 | 2 | 865,560 | 1,673,908 | 32,474 | 17,614 | ||
| 6 | ap1-10 Inflor. | AP1 | 2 | 1,087,743 | 2 | 1,471,249 | 2,558,992 | 57,404 | 21,731 | ||
| 7 | ap3-6 Inflor. | AP3 | 2 | 892,242 | 2 | 1,158,405 | 2,050,647 | 48,644 | 22,050 | ||
| 8 | agamous Inflor. | AGM | 2 | 867,439 | 2 | 1,298,189 | 2,165,628 | 34,950 | 15,543 | ||
| 9 | Inflorescence-2 | INS | 2 | 1,119,870 | 2 | 1,396,268 | 2,516,138 | 50,839 | 23,059 | ||
| 10 | Roots-2 | ROS | 2 | 1,020,885 | 2 | 1,026,684 | 2,047,569 | 38,015 | 21,457 | ||
| 11 | sup/ap1-10 Inflor. | SAP | 2 | 835,574 | 2 | 1,038,723 | 1,874,297 | 33,026 | 19,615 | ||
| 12 | SA 4 hr—leaf | S04 | 2 | 1,277,278 | 2 | 1,349,668 | 2,626,946 | 18,787 | 10,557 | ||
| 13 | SA 52 hr—leaf | S52 | 2 | 1,276,762 | 2 | 1,308,033 | 2,584,795 | 21,381 | 11,962 | ||
| 14 | Leaves-2 | LES | 2 | 1,378,453 | 2 | 1,373,972 | 2,752,425 | 41,218 | 22,495 | ||
| Total | 33 | 15,373,489 | 30 | 16,031,064 | 31,404,553 | 238,311b | 84,262b | ||||
Sequenced by “classic” MPSS method.
Calculated as the union of the set of all libraries.
The libraries were constructed and sequenced at Lynx Therapeutics, Inc. (Hayward, CA) using previously published methods for MPSS (Brenner et al. 2000a,b). Sequencing runs were performed using two separate 4-nt sequencing frames offset by two bases (Brenner et al. 2000a); these sequencing reactions are hereafter called steppers, and the two reactions are referred to by the sequencing frame, 2-step or 4-step. These steppers and the sequencing frames are explained in more detail below.
Including the sequence GATC that is derived from the anchoring DpnII site, the standard length of the MPSS signatures was 17 nt; a continuation of each sequencing reaction generated an additional three bases of sequence information. Therefore, each library was obtained both as signatures of 17 bases and of 20 bases. Between 1.7 and 3.6 million 17-base signatures were obtained for each tissue (Table 1A). A small number of signatures with ambiguities resulting from low-quality sequence were removed; the numbers reported here only reflect high-quality sequencing reactions. The total number of runs varied for each library and each stepper, but between two and five runs, usually two, were performed for each stepper in each library (Table 1). The complete set of 63 sequencing runs comprised 36,991,173 17-base signatures, including 18,416,230 from the 33 2-step runs and 18,574,943 from the 30 4-step runs. Slightly lower numbers of signatures were obtained by MPSS for the 20-base signatures, with the 63 runs producing 31,404,553 sequences (Table 1B).
Within each library, distinct signatures were summed within a single sequencing run to determine an “abundance” value. The raw abundances were merged, first for runs within the steppers, and then across the two steppers to produce a single, normalized value for each observed signature. This process of merging the runs is described in more detail in the Methods section. The outcome was a single normalized value for each signature in units of Transcripts Per Million (TPM). The normalized value is largely independent of the total number of signatures that comprise the library size, and therefore this normalized value is comparable across libraries.
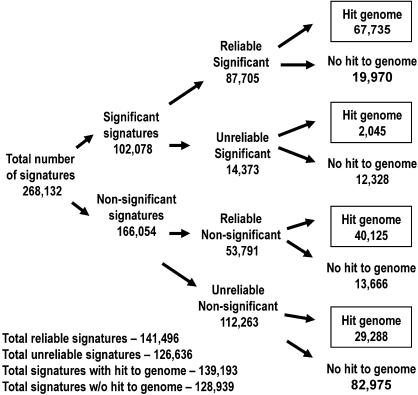
The distinct MPSS signatures from the 14 libraries were filtered using two criteria. We called these filters reliability and significance. The filters were designed to remove noisy sequences that may have resulted from erroneous processes or systematic errors. The filters are described in the Methods section. Briefly, the reliability filter removes signatures observed in only one sequencing run across all libraries (e.g., inconsistent expression). The significance filter removes signatures never observed above 3 TPM in any library (e.g., very low expression). The 36,991,173 17-base signatures in the 14 libraries represented 268,132 distinct signatures. Of these distinct signatures, 102,078 were significant, 141,496 were reliable, and 87,705 were both significant and reliable (Fig. 2). Most of our analyses focused on the 87,705 significant and reliable signatures, because these signatures comprised 97.5% of the sum of the abundances across the libraries (18,187,589 of 18,654,747 total TPM). The unreliable signatures represented only 1.7% of the sum of the abundances, and these were removed for the majority of our analyses. Unreliable signatures were matched to the genome at a very low rate and are probably the result of sequencing errors (see below).
Figure 2.
Filter results for 14 MPSS libraries. A total of 268,132 distinct 17-base expressed signatures from 14 Arabidopsis MPSS libraries were processed according to three filters, “significance,” “reliability,” and “genomic match.” The numbers indicate distinct signatures that are separated by each filter or set of filters. Signatures that do not match to the genome correspond to the “Class 0” signatures discussed in the text; those that match the genome correspond to Classes 1 to 7.
Extraction of Signatures From Genomic Sequence
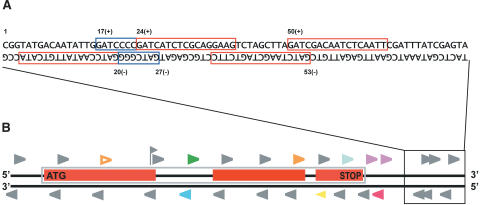
We developed a script to extract potential MPSS signatures from genomic DNA sequence; we refer to these signatures as genomic signatures, in contrast to the expressed or MPSS signatures. A Web-based version of this script can be used on our Web page (http://mpss.udel.edu/at/query.php). This script finds all occurrences of GATC in a given sequence, reports or stores each 17- or 20-base signature, and reports the coordinates of the first base of this signature (Fig. 3). Because signatures will occur in pairs on opposite strands (GATC is a palindrome), the script determines the sequence of the genomic signatures that occur on both the sense and the anti-sense strands (Fig. 3). We used this script to extract a total of 851,212 genomic signatures and their coordinates (chromosome, position, and strand) from the TIGR genomic sequence (117,277,014 bp; version 3 from July 2002; http://www.tigr.org), reflecting a total of 425,606 DpnII (Sau3A) sites. These genomic signatures were stored for comparison to the annotation of genes and for matching against expressed signatures.
Figure 3.
Extraction and classification of signatures from genomic sequence. (A) Example of signatures extracted from genomic DNA sequence. Red and blue boxes indicate signatures identified in the genomic sequence; a noncomplementary signature on each strand is identified from each DpnII site because of the palindromic nature of the site. Numbers above and below the sequence indicate the nucleotide position and strand information stored for each signature. Blue boxes indicate overlapping genomic signatures; the more 5′ signature can either be grouped with the 3′ signature and contain a second occurrence of GATC or be separated and consist of a signature of <17 bases or 20 bases. (B) Horizontal black lines indicate the two strands of DNA. Red boxes indicate exons of a gene on the top strand; the blue box enclosing the exons denotes the extent of the entire gene. Arrowheads indicate the positions of signatures found in the sequence. Signatures duplicated in the genome are indicated using hollow arrowheads; filled arrowheads indicate signatures unique in the genome. The format of the diagram is the same as used in the viewer on our Web page (http://mpss.udel.edu/at). Expressed signatures are indicated in color, whereas nonexpressed genomic signatures are shown in gray. The color of the triangle indicates the signature “class,” and the colors are used as follows: (orange) Class 1—in an exon, same strand as ORF; (purple) Class 2—within 500 bp after stop codon, same strand as ORF; (yellow) Class 3—antisense of ORF; (red) Class 4—in genome but not Class 1, 2, 3, 5, or 6; (green) Class 5—entirely within intron, same strand; (blue) Class 6—entirely within intron, antisense; (light green flag) Class 7—signature includes an exon/intron boundary and is spliced. Not shown are Class 0 signatures that are identified by MPSS but do not match to the genome.
We used the exon coordinates from the Arabidopsis annotation to identify signatures spanning introns, generating an additional set of genomic signatures. In addition to the 851,212 signatures extracted directly from the Arabidopsis chromosomal sequence, 6807 signatures were derived from annotated gene sequences (TIGR version 3.0) and included 13 bases of sequence flanking both sides of an intron, plus the GATC. Including these spliced signatures, our database was comprised of 858,019 genomic signatures.
The 858,019 genomic signatures were clustered to identify duplications and to determine the uniqueness of each signature. Clustering of the 17-base signatures identified 753,894 distinct genomic signatures (Table 2). Of these, 703,161 genomic signatures were “unique,” and have only one “hit” in the genome. Duplicated signatures (hits ≥2) were found in 154,858 different locations in the Arabidopsis genome and comprised 50,733 distinct signatures (Table 2). Of the duplicated signatures, 37,315 distinct genomic signatures were found in two locations, 6484 were found in three locations, and the remaining 6934 distinct signatures in four or more locations. The most highly duplicated genomic signatures, such as GATCATACCAGCACTAA, found 281 times, were derived from repetitive regions such as ribosomal DNA, retrotransposons, or transposons (data not shown). Some signatures with multiple hits may also be derived from duplicated genes. A comparison to randomly generated DNA sequence suggests that most signatures with hits >1 result from duplication events (see Supplemental material). We also extracted and analyzed 20-base genomic signatures to facilitate the use of 20-base MPSS expression data (Table 2). Increasing the signature length to 20 from 17 bases will improve the specificity of the MPSS data for many genes, but even within a relatively compact genome like Arabidopsis, genomic duplications may confound the assignment of expression data for a subset of genes.
Table 2.
Duplications of Genomic Signatures
|
17-base signatures
|
20-base signatures
|
|||||||
|---|---|---|---|---|---|---|---|---|
| “Hits”a | Total locationsb | % of total | # distinctc | % of distinct | Total locationsb | % of total | # distinct | % of distinct |
| 1 | 703,161 | 81.95 | 703,161 | 93.27 | 750,792 | 87.50 | 750,792 | 95.91 |
| 2 | 74,630 | 8.70 | 37,315 | 4.95 | 43,664 | 5.09 | 21,832 | 2.79 |
| 3 | 19,452 | 2.27 | 6,484 | 0.86 | 13,437 | 1.57 | 4,479 | 0.57 |
| 4 | 9,528 | 1.11 | 2,382 | 0.32 | 7,700 | 0.9 | 1,925 | 0.25 |
| 5 | 6,545 | 0.76 | 1,309 | 0.17 | 5,345 | 0.62 | 1,069 | 0.14 |
| 6 | 4,344 | 0.51 | 724 | 0.10 | 3,576 | 0.42 | 596 | 0.08 |
| 7 | 3,066 | 0.36 | 438 | 0.06 | 2,401 | 0.28 | 343 | 0.04 |
| 8 | 2,480 | 0.29 | 310 | 0.04 | 2,024 | 0.24 | 253 | 0.03 |
| 9 | 2,106 | 0.25 | 234 | 0.03 | 1,845 | 0.22 | 205 | 0.03 |
| 10 | 1,980 | 0.23 | 198 | 0.03 | 1,770 | 0.21 | 177 | 0.02 |
| 11-20 | 12,932 | 1.51 | 893 | 0.12 | 11,521 | 1.34 | 801 | 0.1 |
| 21-30 | 5,622 | 0.66 | 229 | 0.03 | 4,806 | 0.56 | 195 | 0.02 |
| 31-50 | 4,853 | 0.57 | 127 | 0.02 | 3,796 | 0.44 | 99 | 0.01 |
| >50 | 7,320 | 0.85 | 90 | 0.01 | 5,342 | 0.62 | 68 | 0.01 |
| Total | 858,019 | 753,894 | 858,019 | 782,834 | ||||
“Hits” refers to the total number of occurrences in the genome.
Includes both spliced and unspliced versions of the 6807 signatures that span the intron/exon boundaries (See Table 3).
“Distinct” refers to the number of different sequences found within the set.
Classification of Signatures From Genomic Sequence
We assigned a “class” to each genomic signature based on the position of the signature relative to annotated genes (Table 3). All 858,019 signatures extracted from the Arabidopsis genome were assigned a class. The classification system was similar to that used for SAGE data in the Saccharomyces Genome Database (Ball et al. 2001). Each signature was compared to the annotation and assigned a class depending on the position and strand relative to the exons of annotated genes (classes are described in the Fig. 3B legend). Because a small number of genes are annotated as overlapping, a small number of signatures met the criteria for more than one class. We retained at most two classifications for these signatures; the “secondary classification” is described in Supplemental material. Using the TIGR annotation (version 3.0), we identified 173 genes that did not contain Class 1, 2, 5, or 7 genomic signatures, and are therefore not expected to be detectable by MPSS.
Table 3.
Classification of 17-Base Genomic and Expressed Signatures
|
Secondary class
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Class | Position | Arabidopsis genomea | 3 | 6 | # with hits = 1 | % with hits = 1b | Grouped by genec | Mean # per gene | # in MPSS librariesd | MPSS data, grouped by gened |
| 1 | Within exon, same strand | 203,174 | 204 | 149 | 168,024 | 82.7 | 28,134 | 7.22 | 29,352 | 16,312 |
| 2 | Within 500 bp potential 3′UTR | 44,536 | 7,047 | 733 | 37,831 | 84.9 | 21,735 | 2.05 | 6,844 | 5,862 |
| 3 | Antisense to exon | 197,083 | — | — | 163,052 | 82.7 | 27,813 | 7.09 | 16,897 | 9,474 |
| 4 | Unannotated | 288,142 | — | — | 223,609 | 77.6 | — | — | 5,622 | — |
| 5 | Within intron, sense strand | 60,430 | 153 | 107 | 53,639 | 88.8 | 17,003 | 3.55 | 1,476 | 1,317 |
| 6 | Within intron, antisense strand | 57,847 | — | — | 51,354 | 88.8 | 16,715 | 3.46 | 685 | 650 |
| 7 | Spans an exon/intron splice site | 6,807 | — | — | 5,652 | 83.0 | 5,369 | 1.27 | 955 | 920 |
| Total | 858,019 | 7,404 | 989 | 703,161 | 82.0 | 28,912 | 61,831 | 19,820 | ||
| 0 | Non genomic match | — | — | — | — | 19,970 | ||||
| 1/2/5/7 | Normal coding gene | 314,947 | — | — | 28,911 | 38,627 | 19,088 | |||
Number of signatures occurring at distinct positions in TIGR annotation version 3.0.
Calculated as the number of signatures in each class with hits = 1 divided by the total number of signatures in that class.
This value is the union of the set of gene identifiers matched by all genomic signatures in each class.
From 14 libraries, considering only “significant” and “reliable” expressed signatures for which hits = 1, and excluding 37,192 “nonsignificant” hits to the genome. Calculated as the union of the set of all libraries.
Matching of Expressed Signatures to Genomic Signatures
We compared the set of expressed MPSS signatures with the set of genomic signatures to make signature-to-gene assignments. Among the 17-base signatures identified for the 14 libraries, the 268,132 distinct, expressed signatures were compared with 753,894 distinct genomic signatures, producing 139,193 hits to the genome and leaving 128,939 unmatched signatures. This comparison matched 67,735 of 87,705 (77.2%) “significant” and “reliable” expressed signatures to genomic signatures, and these were considered the most dependable representation of “real” transcripts. Among these 67,735 matched signatures were 38,627 that uniquely corresponded to the sense strand of 19,088 annotated genes (Table 3). Predicted antisense transcripts were identified by a large number of signatures that matched uniquely in the Arabidopsis genome; 17,582 Class 3 or 6 signatures were found in the 14 libraries. The set of 5622 expressed Class 4 signatures that uniquely mapped to unannotated regions of the genome (Table 3) demonstrates that the transcriptional activity of a large amount of the Arabidopsis genome remains to be characterized (Meyers et al. 2004b). This assessment of transcriptional activity was performed using only the significant and reliable signatures that mapped to the genome; the 40,125 reliable but nonsignificant signatures were not considered but may be derived from weakly expressed but real transcripts (see below).
Expressed signatures that did not match to any of the Class 1 to 7 genomic signatures were designated as Class 0. In the 14 MPSS libraries, this included 128,939 signatures, of which 19,970 were significant and reliable (Fig. 2). There are several reasons why a signature might not match to the genome. Sequencing errors during MPSS could generate novel signatures that differ by one or several bases from genomic signatures. We address this possibility in more detail below. It is also possible that no sequencing errors occurred but the signature spanned a splice site that has not yet been annotated. This novel splice site, when annotated, would generate a new Class 7 signature instead of the current unmatched Class 0 signature. One further possibility is that the unmatched signatures occurred within an existing gap in the genomic sequence, such as from the poorly covered centromeric regions.
The Sources of Class 0 MPSS Signatures
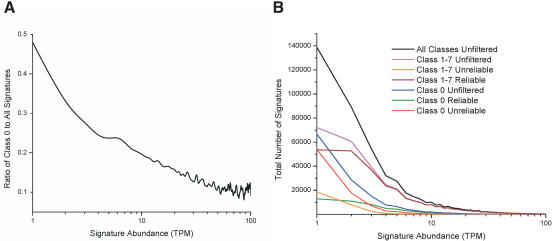
We investigated the source of the 128,939 distinct, expressed signatures that did not match to the Arabidopsis genome. Using the sum of the normalized abundances values for all distinct transcripts across the 14 libraries in our database, we determined that these Class 0 signatures represented 9.0% of the normalized MPSS expression data, based on summed signature abundances (1,680,361 TPM divided by 18,654,747 TPM). In contrast, the number of distinct Class 0 signatures comprised 48.1% of the total number of distinct expressed signatures (128,939 of 268,132). This comparison suggested that most Class 0 signatures were present at very low abundances. This was more apparent when we calculated the proportion of distinct signatures at each expression level that were Class 0 (Fig. 4A). Nearly twice the proportion of Class 0 signatures was found at 1 TPM than at 3 TPM. At 1 TPM, almost 50% of the distinct signatures did not match to the genome (Fig. 4A). At 8 TPM, the fraction of Class 0 signatures dropped below 20%, and at higher abundance levels the percentage rarely dropped below 10% (Fig. 4A). Next, we determined the proportion of Class 0 signatures that were unreliable. The “reliability” filter identifies as unreliable those signatures that were observed in only one of 63 sequencing runs in the data set but does not stipulate any specific expression level. This sporadic appearance may signify a sequencing error. The Class 0 signatures at all abundance levels were predominantly identified as unreliable (Fig. 4B). In contrast, the majority of signatures mapped to the genome (Class 1 to 7) were reliable, even at low expression levels (Fig. 4B). Therefore, many Class 0 signatures are observed in only one run and at low abundances; these characteristics are consistent with the product of random sequencing errors. The corollary to this observation is that Class 0 signatures found at higher levels and occurring more consistently across the libraries may represent real but previously uncharacterized splicing events.
Figure 4.
MPSS expression levels and reliability of signatures not matched to the genome. (A) The ratio was calculated as the number of distinct Class 0 signatures to the number of all expressed signatures, with no filters applied. The number of signatures in each group was calculated by joining data across all 14 libraries. The x-axis indicates each level of abundance, in TPM, for the signatures that were compared. The y-axis indicates the ratio of the two frequency counts. The x-axis is shown in logarithmic scale because the majority of the signatures are found at low abundance levels; likewise, this axis ends at 100 TPM because only a small percentage of the signatures are found above this level. (B) For each abundance level indicated on the x-axis, the number of distinct signatures is indicated on the y-axis. The data from all 14 libraries were considered together. The black line indicates the abundance levels for all 268,132 distinct signatures (no filters applied). The following colored lines correspond to each group of Class 1 to 7 signatures: (cyan line) unfiltered; (orange line) “unreliable” signatures; (brown line) “reliable” signatures. The following colored lines correspond to each group of Class 0 signatures: (blue line) unfiltered; (red line) “unreliable” signatures; (green line) “reliable” signatures.
To assess sequencing errors, the unmatched Class 0 signatures were compared with significantly expressed genomic signatures, allowing one polymorphic base in a match. The first four bases of every signature are GATC and are invariant; the remaining 13 positions of the 17-base MPSS signatures could contain errors. For this comparison, we derived a set of signatures one base different (OBD) from the 126,598 17-base Class 0 signatures not perfectly matched to the genomic or plastid sequences (see Supplemental material). To generate the OBD signatures, each position was changed to one of three other bases, so that a total of 3 × 13 = 39 OBD signatures were derived from each Class 0 signature (see Supplemental material). Out of 67,735 significant and reliable Class 1 to 7 genome-matched, expressed signatures, 28,414 (41.2%) were matched to OBD signatures corresponding to 73,815 of the Class 0 signatures (58.3%; see Supplemental material). These Class 0 signatures may have resulted from single-base-sequencing errors in the OBD-matching, expressed genomic signatures. The higher-abundance Class 0 signatures that were not matched in the OBD comparisons may be further enriched for those derived from as-yet-uncharacterized transcripts or splicing events.
Next, we used the abundance of OBD-matching Class 0 and 1 to 7 signatures to estimate the error rate for MPSS. Our estimate for 17-base MPSS signatures was ∼3.2% per signature, which corresponds to an error rate of 0.25% for each of the 13 sequenced bases (GATC excluded). For this calculation, we determined the sum of the abundance of OBD matches to expressed Class 1 to 7 signatures. A subset of the expressed, genome-matched signatures was used for this calculation. The purpose of selecting this subset was to avoid basing the calculation of the error rate on any falsely matching OBD and genomic signatures. We used the 18,192 Class 1 to 7 signatures that were unique in the genome (hits = 1) that were significant and reliable, and that were at least two bases different from any other signature in the list. We then removed signatures that had any single OBD-matching signature for which the sum of the abundances across the 14 libraries was >25% of that of the matched Class 1 to 7 signature. This step was based on the assumption that for random sequencing errors, the 39 possible OBD signatures should be more or less equally represented. A total of 4,370 Class 1 to 7 signatures met this criterion and matched OBD Class 0 signatures that were present at levels ∼10-fold higher than expected for random sequencing errors. The OBD-matching Class 0 signatures could represent Class 7 signatures from uncharacterized splicing events that were coincidently one base different from the genome-matched signatures. The final list of 13,822 signatures was sorted based on the sum of abundances for each signature across all 14 libraries. The error rate was then calculated as 3.52%, 3.19%, or 2.68% using the top 1000, 2000, or 5000 most abundantly expressed signatures, respectively. The higher-abundance signatures were used because they were sampled in large numbers, providing a better estimate of error. The median estimated error rate, 3.19%, precisely matches the error rate for MPSS determined by sequencing Escherichia coli genomic DNA (S. Luo and C. Haudenschild, unpubl.), and corresponds to an error rate of ∼0.25% for each of the 13 variable bases in an MPSS signature.
Sequence-Specific Effects on MPSS Signatures
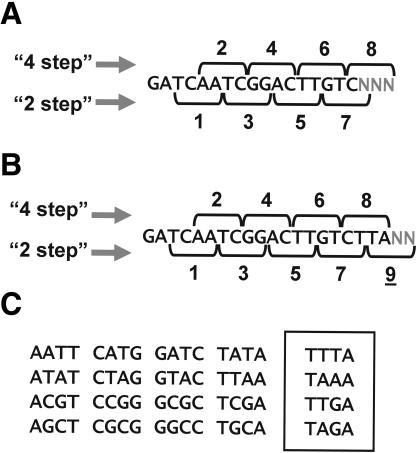
We analyzed the frequency of four-base words in the genomic and expressed signatures to determine if any sequence-specific biases resulted in underrepresentation of certain signatures in the MPSS data. This analysis is based on the four-base words that are sequenced during the MPSS procedure in which digestion with a Type II enzyme exposes the four-base sequencing frame (Brenner et al. 2000a). The MPSS sequencing takes place in two sets of frames (e.g., 2-step and 4-step reactions; Fig. 5A,B). These steppers represent a type of replication because the sequencing reactions for the same library are performed independently (Brenner et al. 2000a). A given signature should have a similar abundance in both steppers for a given library. If the result of the 2-step and 4-step reactions differs for the same signature in a library, this difference could be attributed to sequence-specific effects. Such effects are possible because different four-base words are observed in the two sequencing frames for each signature (Fig. 5A,B). Any word-specific bias may affect one stepper for a given signature but not the other. A possible source of a word-specific bias could be the Type IIS restriction enzyme BbvI that is used to “expose” the four-base words as single-stranded sequence (Brenner et al. 2000a); if this enzyme does not cut each site with equal efficiency, the presence of the disfavored words may result in decreased sequencing efficiency for signatures containing those words.
Figure 5.
Sequencing frames and “bad” words in MPSS signatures. MPSS uses two sequencing reactions that are performed in reading frames shifted by two bases. These two frames are the 2-step or 4-step frames. Sequencing proceeds in sets of four bases (“words”). (A) The eight sequencing frames are indicated for the 17-base signatures, with the 2-step frames indicated below the example signature and the 4-step frames indicated above. The numbering 1 to 8 for the 17-base signatures is only used to illustrate the total number of words that are sequenced. The first sequencing reaction in the 2-step reaction always begins with TC. The three Ns in gray at the end of the 17-base signature indicate the bases that are not sequenced but that can affect the performance of the seventh or eighth word. (B) The 20-base signatures require an extra word (underlined “9”) that includes the two unsequenced bases indicated by the gray letter N. (C) The 20 “bad words” that underperform in MPSS sequencing. All 16 four-base palindromic words are listed at left; these words underperform in the MPSS sequencing reaction when they occur in the frames indicated for the 17- or 20-base signatures (see text). The four words in the box also underperform in the MPSS sequencing reaction, but these are not palindromic.
Using all significant and reliable, expressed signatures, we identified signatures with significantly different abundances in the 2-step or 4-step runs for all 14 libraries (P < 0.05 when H0 = no difference using a Z-test). For this calculation, we used the raw normalized abundance for each stepper, described as “t_norm” and “f_norm” in the Methods section. The signatures were separated into one of three “bins” based on whether the t_norm value was equal to, lower than, or higher than the f_norm value. This partitioning step is described in more detail in the Methods section. Within the groups of signatures with either higher 2-step or higher 4-step abundances, the number of occurrences of each of 256 possible four-base words was determined. These occurrences were counted for 2-step frames compared with those in 4-step frames for either 17- or 20-base signatures (Fig. 5A,B). Additional details of the steps involved in this analysis are provided in the Methods section. The result of this analysis allowed us to correlate word occurrence with bias in the success rate of the 2- and 4-step MPSS reactions.
These comparisons identified 20 four-base words that were underrepresented in one of the two MPSS steppers or sequencing frames (Table 4). All 16 “palindromic” words were among the 20 words that were most underrepresented in the expression data; the word “GATC” was extremely rare in the expressed signatures because it is the recognition site for DpnII, the MPSS anchoring enzyme. In addition to the palindromes, the words TTTA, TAAA, TTGA, and TAGA were also associated with a reduction in the normalized abundance of the signature, but less strongly than the palindromic words (Table 4). The 20 most underrepresented words were the same when the calculation was performed using the 17-base signatures (Table 4; Supplemental Figs. S4–S6) and the 20-base signatures (Supplemental Figs. S4–S6), although the ranking of underrepresentation was slightly different. We refer to these 20 words that underperform in MPSS reactions as “bad words” (Fig. 5C). In a randomly composed genome, ∼7.7% of all potential 17-base MPSS signatures will be poorly measured by this technology because of occurrences of these 20 bad words in both sequencing frames (see Methods for calculation). Several overrepresented words were also identified (Table 4), but the reason for overrepresentation of a four-base word was not apparent; we chose instead to focus on an analysis of the underrepresented words. Palindromic sequences may be underrepresented in the four-base sequencing frames in which the palindrome occurs because of intermolecular interactions of complementary single-stranded four-base overhangs on the microbead. These interactions would effectively block MPSS analysis in that sequencing frame by preventing the addition of the adapters used in sequencing.
Table 4.
Abundance of Four-Base Words in Expressed Signatures
| Rank (by ratio)a | Wordb | N2 (2-step)c | N4 (4-step) | N2:N4 Adj. ratiod | ||||
|---|---|---|---|---|---|---|---|---|
| A. Word use in 17-base signatures for which the 2-step abundance was significantly greater than 4-step abundance. This subset of expressed signatures is described as “Bin 2” in the Methods section. | ||||||||
| 1 | GGCC | 1 | 232 | 0.0043 | ||||
| 2 | TTAA | 6 | 846 | 0.0071 | ||||
| 3 | TATA | 3 | 415 | 0.0072 | ||||
| 4 | TCGA | 7 | 757 | 0.0092 | ||||
| 5 | AGCT | 11 | 1120 | 0.0098 | ||||
| 6 | CGCG | 2 | 155 | 0.0129 | ||||
| 7 | TGCA | 13 | 819 | 0.0159 | ||||
| 8 | CATG | 15 | 747 | 0.0201 | ||||
| 9 | ATAT | 18 | 762 | 0.0236 | ||||
| 10 | GCGC | 3 | 106 | 0.0283 | ||||
| 11 | CCGG | 13 | 401 | 0.0324 | ||||
| 12 | AATT | 33 | 887 | 0.0372 | ||||
| 13 | GTAC | 12 | 317 | 0.0379 | ||||
| 14 | ACGT | 12 | 291 | 0.0412 | ||||
| 15 | CTAG | 30 | 341 | 0.0880 | ||||
| 16 | TTTA | 111 | 788 | 0.1409 | ||||
| 17 | TAAA | 127 | 705 | 0.1801 | ||||
| 18 | TTGA | 202 | 1034 | 0.1954 | ||||
| 19 | TAGA | 81 | 412 | 0.1966 | ||||
| 20 | TCAC | 138 | 436 | 0.3165 | ||||
| 21 | TCGG | 89 | 249 | 0.3574 | ||||
| 22 | TCAA | 311 | 799 | 0.3892 | ||||
| 23 | TAAG | 171 | 403 | 0.4243 | ||||
| 24 | TGAC | 154 | 352 | 0.4375 | ||||
| 25 | TCCG | 98 | 221 | 0.4434 | ||||
| [see Supplemental Figures S4 to S6 for complete tables] | ||||||||
| 246 | GAAT | 484 | 172 | 2.8140 | ||||
| 247 | CGTA | 186 | 65 | 2.8615 | ||||
| 248 | GCTT | 490 | 171 | 2.8655 | ||||
| 249 | GATA | 385 | 131 | 2.9389 | ||||
| 250 | ACAT | 488 | 159 | 3.0692 | ||||
| 251 | GCTA | 307 | 85 | 3.6118 | ||||
| 252 | ACTC | 495 | 136 | 3.6397 | ||||
| 253 | ACTT | 600 | 162 | 3.7037 | ||||
| 254 | CCTA | 265 | 70 | 3.7857 | ||||
| 255 | ACTA | 365 | 89 | 4.1011 | ||||
| B. Word use in 17-base signatures for which the 4-step abundance was significantly greater than 2-step abundance. | ||||||||
| 1 | GCGC | 189 | 1 | 0.0053 | ||||
| 2 | TTAA | 837 | 10 | 0.0119 | ||||
| 3 | AGCT | 1359 | 21 | 0.0155 | ||||
| 4 | TATA | 407 | 9 | 0.0221 | ||||
| 5 | GGCC | 321 | 9 | 0.0280 | ||||
| 6 | TCGA | 782 | 22 | 0.0281 | ||||
| 7 | TGCA | 886 | 27 | 0.0305 | ||||
| 8 | CCGG | 449 | 19 | 0.0423 | ||||
| 9 | CATG | 729 | 31 | 0.0425 | ||||
| 10 | ATAT | 726 | 33 | 0.0455 | ||||
| 11 | GTAC | 405 | 21 | 0.0519 | ||||
| 12 | CGCG | 196 | 11 | 0.0561 | ||||
| 13 | ACGT | 366 | 21 | 0.0574 | ||||
| 14 | AATT | 879 | 52 | 0.0592 | ||||
| 15 | CTAG | 396 | 32 | 0.0808 | ||||
| 16 | TAAA | 750 | 111 | 0.1480 | ||||
| 17 | TTTA | 718 | 112 | 0.1560 | ||||
| 18 | TAGA | 546 | 95 | 0.1740 | ||||
| 19 | TTGA | 1100 | 213 | 0.1936 | ||||
| 20 | TCAC | 502 | 209 | 0.4163 | ||||
| 21 | TGGC | 381 | 161 | 0.4226 | ||||
| 22 | AGCC | 380 | 166 | 0.4368 | ||||
| 23 | TGAC | 414 | 184 | 0.4444 | ||||
| 24 | TTAG | 447 | 200 | 0.4474 | ||||
| 25 | GCGG | 184 | 83 | 0.4511 | ||||
| [see Supplemental Figures S7 to S9 for complete tables] | ||||||||
| 246 | AAAG | 443 | 1151 | 2.5982 | ||||
| 247 | GCTT | 227 | 594 | 2.6167 | ||||
| 248 | CATT | 229 | 605 | 2.6419 | ||||
| 249 | CATA | 157 | 429 | 2.7325 | ||||
| 250 | CCTT | 179 | 504 | 2.8156 | ||||
| 251 | ACTC | 185 | 523 | 2.8270 | ||||
| 252 | ACAT | 183 | 647 | 3.5355 | ||||
| 253 | ACTT | 205 | 744 | 3.6293 | ||||
| 254 | ACTA | 111 | 444 | 4.0000 | ||||
| 255 | CCTA | 60 | 265 | 4.4167 | ||||
For brevity, only the first 25 and last 10 rows of 255 four-base words are shown; GATC was not considered because it is rarely observed among expressed signatures. For the complete set of data corresponding to this subset of signatures, the other “bins”, and the 20-base expressed signatures, see Supplemental Figures S4-S9.
Palindromic words are shown in bold; other “bad” words are indicated in italics. Frame 1 (see Fig. 5A) was not considered because only the 16 words initiating with “TC” can be observed in this frame.
“N” indicates the frequency of occurrence of the word among the frames of the expressed signatures for either of the indicated steppers. The frequency of the words was calculated with all expressed signatures considered equally, independent of the expression abundance.
“Adj. ratio” indicates that the ratio was adjusted to account for the different number of frames in the 2- and 4-step reactions for which the word frequencies were counted; for the 17-base expressed signatures, words in 2-step frames 3 and 5 were counted and 4-step frames 2, 4, and 6 (Fig. 5A). Therefore, frequency counts for the 4-step words were adjusted by 2/3 prior to calculating the ratio.
Online Resources for Arabidopsis MPSS Data
We have developed a publicly available, Web-based interface to our database (http://mpss.udel.edu/at; Meyers et al. 2004a). This interface includes a graphical interface and permits simple queries based on gene or genomic sequence, MPSS or genomic signature sequence, gene identifier, or chromosomal region. MPSS expression data are available for both 17- and 20-base signatures for the 14 Arabidopsis libraries described above. The Web site also analyzes the Arabidopsis genomic and expressed signatures for the presence of bad words.
DISCUSSION
We have described our methods for normalizing and filtering MPSS expression data, for matching MPSS expression data with the genomic sequence and annotation data, and for detecting sequence-specific biases in MPSS. These methods would work in any genome, but the high level of finishing work that has been performed on the Arabidopsis genome has facilitated our analyses. Although our analysis mapped to the genome >90% of the MPSS expression data, additional improvements to the annotation and addition of new cDNA data will incrementally improve the number and accuracy of the MPSS signatures mapped to transcripts. Improvements in signature mapping will refine the quantitative estimates of expression levels for certain genes including those in which all matching signatures may not have yet been mapped. Our analyses of MPSS data also suggest that a subset of unmatched (Class 0) signatures is enriched for and derived from as-yet-uncharacterized transcripts.
Development of Specialized Methods for Working With MPSS Data
We designed several filters to remove inconsistent or low-abundance signatures, but these filters may have been overly stringent. Most of our analyses focused on the 87,705 reliable and significant signatures that matched the genomic sequence and comprised 97.5% of the MPSS expression data. The number of distinct “unreliable” signatures was high, but the total abundance of these signatures was low. We demonstrated that the majority of these unreliable signatures may result from sequencing errors. Of the 53,791 distinct, expressed signatures that were considered reliable but not significant, 40,125 matched to the Arabidopsis chromosomal sequences. These were signatures observed in multiple runs and possibly in multiple libraries but never observed at a level higher than 3 TPM. It is possible that normal mRNA transcripts are expressed at levels below 4 TPM and our filter for significance was too stringent. To determine if our significance filter will miss some real transcripts, we calculated the number of libraries in which significantly expressed signatures (e.g., >3 TPM in at least one library) that matched to the genome were found at 1, 2, or 3 TPM. These low abundances were observed 81,155 times for 40,590 distinct signatures across the 14 libraries. This suggests that some transcripts may be expressed at low levels in certain cells or tissues; these levels could be further diluted by sampling tissues rather than specific cell types. These transcripts might be observed at a “significant” level if we sampled specific cells by using a method such as laser-capture microscopy (Asano et al. 2002; Kerk et al. 2003) or protoplast sorting (Galbraith 2003).
New libraries from diverse tissues and treatments may also render a new set of significant signatures by revealing specific conditions under which certain transcripts are expressed at higher levels. Because our libraries represented a limited number of treatments or tissues, we may not have sampled the specific conditions under which some transcripts may be expressed at high levels. The signatures uniquely expressed at significant levels in novel libraries could include many of the 40,125 reliable but not significant genome-matched signatures identified in our 14 libraries. These data would improve our ability to distinguish the signal (signatures from real transcripts) from the noise (artifactual or erroneous signatures). The addition of more libraries to our database should therefore improve our recognition of valid signatures. The number of transcripts encoded in a genome must be finite, and with deep-enough sampling, the number of “significant” signatures will eventually approach the total number of distinct transcripts that are possible from a given genome.
The “reliability” filter will be useful until the number of signatures resulting from errors reaches a low degree of saturation. If each of the 87,705 distinct 17-base “reliable and significant” signatures generated at least one copy of each of the 39 possible one-base-different signatures (39 = 3 different bases × 13 variable sites), a total of 3,420,495 OBD signatures could be observed at “unreliable” levels. However, if sequencing errors occur at a low but consistent rate, and the same transcript occurs at high levels in several libraries, some erroneous signatures may be observed in multiple runs. These erroneous signatures would escape the “reliability” filter. An alternative to this filter may be possible when the complete set of transcripts from a genome is known; in this case, unmatched signatures could be removed, assuming that errors affect all signatures at the same rate and there is no benefit to retaining the erroneous data. Our analyses indicate that many splicing events remain uncharacterized, suggesting that significant efforts in targeted cDNA sequencing will be required before Arabidopsis or any other plant transcriptome is completely characterized.
MPSS as a Technology for Gene Expression Analysis
“Bad” Words That Affect MPSS
We identified artifacts that for technical reasons confound the measurement of gene expression by MPSS in certain sequence-specific cases. These artifacts were identified by comparing the extracted genomic signatures to the expressed signatures. The principal problem results when certain four-base words occur in-frame with the “stepper” or sequencing frame of the sequencing reaction. The occurrence of any of 20 of the 256 possible words resulted in underrepresentation of that signature in the set of expressed signatures, but only for the specific stepper in which the word occurred. Sixteen of the 20 words were palindromic sequences, a result consistent with intermolecular interactions on the surface of the MPSS microbead that could block the sequencing reaction. The remaining four words were TAAA, ATTT, TTGA, and TAGA. The reason for decreased sequencing success rates for these four words is not clear. It is possible that the Type IIS restriction enzyme used in the sequencing reaction (BbvI; Brenner et al. 2000a) cuts poorly at these sequences. In addition, certain words were overrepresented in the poorly sequencing stepper, but there was no clear explanation for this effect. It is difficult to attribute overrepresentation of certain words to biased digestion by BbvI. Successful MPSS sequencing of an entire signature depends on the efficiency of four BbvI digestion reactions (e.g., the frames in Fig. 5). An average level of efficiency of digestion for all four words is all that is required to obtain the sequence, and an increased efficiency for one of the words will not affect the overall sequencing success rate for that signature.
With an understanding of the bad words, it is possible to identify transcripts that could be underestimated by MPSS, but it is difficult to determine the extent of the missing expression data. Whole-genome analyses using the MPSS data will be affected by noise resulting from underestimated expression for genes containing the 7.71% of Arabidopsis signatures containing any one of the 20 bad words in both MPSS sequencing frames. One approach to reduce the frequency and importance of palindromes would be to replace BbvI with a Type IIS enzyme that creates five-base words for the MPSS sequencing process (five-base words would not be palindromic). Another approach would be to simultaneously obtain and then compare MPSS and microarray data to identify genes consistently underestimated by MPSS. Or, additional MPSS reactions using different anchoring enzymes (such as NlaIII instead of DpnII) could be performed and the data compared; the resulting signatures, derived from different sites within the same transcripts, are unlikely to contain the same bad words.
The Merits of Longer MPSS Signatures
Is it beneficial to increase the length of MPSS signatures from 17 to 20 bases? The answer to this question lies in the relative merits of increased specificity versus a reduction in sequencing success caused by a greater chance of a bad word occurring in the longer signatures. Extracting 20-base signatures instead of 17-base signatures increased the number of signatures occurring uniquely in the Arabidopsis genome by 47,631. This represents 5.5% of the 858,019 potential signatures in the genome that are duplicated when only 17 bases long but unique when sequenced to 20 bases. Although this would increase the number of distinct signatures, this could potentially decrease the abundance below the significant threshold for certain signatures. Another decrease in the number of 20-base signatures measured by MPSS results from sequencing failures due to the additional frames that could expose bad words. Owing to the additional frame in the 2-step and 4-step sequencing reaction for 20-base MPSS signatures (Fig. 5B), we can predict that with 20 bad words, 9.3% of distinct signatures will sequence poorly, an increase from the 7.7% calculated for 17-base signatures. Therefore, 1.6% of the signatures were predicted to be blocked for 20-base MPSS sequencing but not for 17-base sequencing. Across the 14 libraries, the total number of distinct, significant, and reliable expressed signatures was 4.1% lower for the 20-base than the 17-base MPSS signatures (Table 1). The difference between the observed and predicted reduction in signatures in the 20-base libraries probably results from a combination of length-dependent effects and could include issues that we have not yet characterized.
For the 20-base signatures, the increased specificity and decreased sequencing success rate are independent and unrelated for any given signature and transcript. These independent effects make it difficult to state categorically whether 17- or 20-base signatures are better. If a gene of interest is not sequenced because of a bad word in frame 9 (Fig. 5B), then the 20-base signature is uninformative; but if a duplicated 17-base signature can be uniquely distinguished only by the 18-th, 19-th, or 20-th bases, the 20-base MPSS signatures may be essential. This choice between specificity and bad words is more problematic when considering the source of data to use for genome-wide analyses, because the two issues will introduce different types of noise. If the problem of sequence-specific artifacts resulting from certain words were eliminated from the MPSS procedure, the longer signature length would be better.
The Influence of Sequencing Errors in MPSS
Our estimate for the error rate of MPSS is ∼0.25% per base. With this error rate, the 13 bases sequenced by MPSS (e.g., excluding GATC) will result in ∼3.25% of expressed signatures containing a single base error. Chen et al. (2002) determined an overall error rate of ∼2%, or ∼0.2% per base, from synthetic SAGE experiments. Other estimates for the error rate of SAGE tags range as high as 10% per tag (1% per base) because the tags represent single-pass sequences (Lash et al. 2000). Therefore, the error rate of MPSS is only slightly higher than the error rate of SAGE. Our analyses suggest that the majority of the erroneous MPSS signatures were captured and removed by our “reliability” and “significance” filters.
The Class 0 signatures that passed our filters were expressed at higher abundances and may be derived from previously unidentified Arabidopsis transcripts. These sequences represent unrecognized Class 7 signatures derived from splicing events not yet annotated or observed in characterized full-length cDNA clones. We were using version 3.0 of the Arabidopsis annotation, and future versions are likely to incorporate many more cDNA sequences that should improve the annotation of 3′-UTR splicing and increase the number of known Class 7 signatures (Haas et al. 2003; Wortman et al. 2003). The detection of Class 0 signatures that correspond to splicing events in as-yet-uncharacterized transcripts is consistent with an experimental demonstration that most unmatched SAGE tags are derived from real transcripts that have not been previously identified (Chen et al. 2002). Chen et al. (2002) demonstrated that >70% of SAGE tags that did not match to human transcripts were derived from previously uncharacterized transcripts. PCR-based strategies such as RACE (Frohman et al. 1988) or the so-called GLGI procedure (Chen et al. 2002) could be used to isolate the corresponding full-length transcripts using primers based on the MPSS signatures. The identification and validation of these transcripts will be facilitated by the quantitative expression data and tissue specificity that is already known from the MPSS libraries. The resulting sequence data would provide additional transcriptional information with which to improve the annotation of the Arabidopsis genome.
The Impact of Duplicated Signatures
One complication in the use of expression data for whole-genome studies results from genomic duplications that produce signatures that do not match uniquely in the genome. The duplications in the Arabidopsis genome have been documented extensively (Arabidopsis Genome Initiative 2000; Blanc et al. 2000; Vision et al. 2000; Simillion et al. 2002). In the case of MPSS, the number of “hits” that we calculated for the genomic signatures reflected this complexity. Approximately 18% of the 17-base genomic signatures were found in more than one location in the genome. Increasing the length of the signatures from 17 to 20 bases decreased the total number of duplicated signatures in the Arabidopsis genome by approximately one-third, from 18% to 12.5%. To determine the source of expression for a MPSS signature duplicated in the genome, it may be possible to compare microarray and MPSS data for the same tissue. This marriage of technologies would complement the weaknesses inherent in each, although it would also depend on the specificity and sensitivity of hybridization to the array.
The utility of MPSS and related tag-based methods like SAGE will depend on the degree and patterns of duplication and redundancy in the genome under analysis. These methods will be most useful if duplications occur mainly in the nongenic, transcriptionally inactive regions of the genome. In Arabidopsis, the proportion of duplicated signatures was slightly lower for signatures associated with genes than for those in intergenic regions (e.g., Class 4 signatures versus any other in Table 3). And many of the most duplicated genomic signatures mapped to repetitive intergenic sequences like retrotransposons or transposons (data not shown). In the cereals, genome expansion has occurred primarily through increased numbers of LTR retrotransposons (Kumar and Bennetzen 1999). A large proportion of the maize genome is comprised of several highly duplicated families of retrotransposons that are weakly if at all expressed (Meyers et al. 2001). The genic regions may be less repetitive and MPSS signatures may match expressed genes with a higher degree of specificity than may be predicted by the genome size. On the other hand, polyploid genomes or genomes containing a high degree of genic repeats may be less amenable to the use of MPSS, particularly if the duplicated genes have not substantially diverged. When MPSS is used in less-duplicated genomes, comparisons across closely related genomes may be highly informative. The sensitivity of this technology to single nucleotide polymorphisms may reveal allele-specific effects that are quantified by measuring the expression of orthologous genes detected by distinct signatures. Such measurements could be made by comparing genetically distinct individuals, subspecies, or inbred lines and their hybrids.
MPSS Versus Other Global Measurements of Gene Expression
The sampling depth of MPSS is greater than most other gene expression technologies. The 17-base signatures for our 14 libraries represented a total of 36,991,173 transcripts, or ∼2.64 million transcripts per library. The set of 36,991,173 signatures is nearly twice the total number of ESTs or SAGE tags present in GenBank for all organisms (in November 2003). The average sampling depth of SAGE is <1% of that of our MPSS libraries (http://www.ncbi.nlm.nih.gov/SAGE/). Although SAGE also provides a quantitative estimate of expression (Velculescu et al. 1995), the 14-base tags will be more ambiguous because of higher numbers of “hits” than MPSS. LongSAGE tags, however, are 21 or 22 bases in length and as a result have a high degree of specificity, similar to that of MPSS signatures (Saha et al. 2002). Neither SAGE nor LongSAGE is subject to the issues of the “bad words” described here for MPSS. A significant disadvantage of any variant of SAGE remains the cost on a per-tag basis, which is still much higher than that of MPSS. This prohibitive cost of sampling hundreds of thousands of SAGE tags for a single library is probably the reason for the relatively small average number of tags for the SAGE libraries in GenBank.
Because tag-based technologies are open-ended and do not use preselected targets for expression analysis, novel transcripts or variants of known transcripts can be identified in completely sequenced genomes. The direct comparison of SAGE tags, MPSS signatures, or ESTs to genomic sequence data facilitates and improves annotation by providing experimental data for transcripts missed by prediction-based annotation (Andrews et al. 2000; de Souza et al. 2000; Guigo et al. 2000; Reese et al. 2000; Haas et al. 2002; Saha et al. 2002; Meyers et al. 2004b). These tags can also identify transcripts that were missed because of gaps, partial genome sequences, or incomplete annotation. Improvements to microarrays in the density of oligonucleotides and the flexibility of design have facilitated the development of whole-genome tiling arrays for the characterization of transcriptional activity (Shoemaker et al. 2001; Kapranov et al. 2002; Rinn et al. 2003; Yamada et al. 2003). This approach makes it possible to use microarrays as an open-ended technology by not restricting probe sets to annotated genes.
Based on complementary strengths and weaknesses, cDNA sequences, MPSS signatures, and microarrays each have an appropriate role in the development of genomic resources for a particular organism. An appropriate course for the development of genomic resources in a poorly characterized genome may start with ESTs and full-length sequencing of cDNA clones, followed by MPSS profiling of diverse tissues and treatments, and finally by PCR-based transcript identification using MPSS signatures. These efforts would complete a gene inventory in the organism, and provide the sequences for functional studies and microarray design. In genomes that are only partially sequenced, or for which only ESTs are available, profiling by MPSS combined with significant cDNA amplification and sequencing will define both the quantitative and qualitative transcriptional complexity. The filtered MPSS signatures create a “unigene” set that describes the transcript inventory for a tissue; MPSS signatures unmatched by cDNA sequences represent transcripts in the inventory that remain to be characterized. After generating a substantial set of quantitatively and qualitatively validated transcripts, microarrays could be used as a relatively inexpensive means to identify transcriptional changes under specific biological conditions.
Microarray data, although less expensive to generate, may be less transferable across laboratories because the data are quantitative only when compared with other samples. Moreover, microarrays may also be subject to platform-specific or user-specific biases, although careful documentation according to MIAME protocols (Brazma et al. 2001) and storage of raw image files for reanalysis may extend the life span of these data. Nevertheless, there are likely significant effects in microarray data, such as oligonucleotide position within a transcript, composition, or placement within the array (Qian et al. 2003) that have yet to be fully characterized through empirical analysis for each platform and each organism. The effect of cross-hybridization is often disregarded in microarray analyses because it is not easily measured or known. As we have demonstrated, method-specific and sequence-specific biases in MPSS can be measured and characterized by comparing across sequencing runs or by comparing the expressed signatures with the genomic signatures. Although there may be additional issues with MPSS that we have yet to identify, we believe that we have characterized the most prevalent problems inherent in this technology. The type of empirical analysis we have presented here for MPSS will be necessary to characterize the strengths and weaknesses of every technology used to measure gene expression. Full examination and empirical analysis of these issues may suggest ways to improve existing gene expression technologies.
METHODS
Plant Materials and RNA
For roots and leaves, plants were grown in 16 h of light for 21 d, grown under sterile conditions in vermiculite or perlite moistened with half-strength MS salts. The entire plant was harvested ∼2 h after dark and separated into roots and leaves. For flowers and siliques, plants were grown in soil in a growth chamber at 22°C with 16 h of light for 5 wk. The immature inflorescence was harvested ∼2 h after dark for wild-type and floral mutants. Floral tissues included inflorescence meristem and early stage floral buds (up to Stage 11/12). Siliques were harvested ∼24 to 48 h after fertilization, when petals have begun to detach (Stage 16–17). Callus was initiated from Col-0 seeds grown on media containing half-strength MS salts, 3% sucrose in the presence of 2,4D (0.5 mg/L), IAA (2 mg/L), and kinetin (0.1 mg/L); the tissue was grown ∼3 mo in dark at 22°C and transferred to fresh plates every ∼10–14 d. For salicylic acid treatments, the plants were grown in soil at 20°C with 8 h of light for 54 d. Half-strength Hoagland's solution was used to bottom-water the plants. Plants during daylight were sprayed to run-off with 0.3 mM SA in 0.02% Silwet (Lehle Seeds). Rosettes were cut above the soil line and harvested 4 h or 52 h posttreatment and still in the light.
Total RNA was prepared using TRIzol reagent (Invitrogen). mRNA was obtained using the Poly(A) Purist kit from Ambion, following the manufacturer's protocol.
MPSS Protocol
MPSS was performed as previously described (Brenner et al. 2000a) for libraries #1 to #5 (Table 1); this approach is called the classic methodology. Libraries #6 to #14 (Table 1) were sequenced using the MPSS “signature cloning” method (Lynx Therapeutics, Inc.). Briefly, the difference in the classic and signature cloning methods is that for the classic method, the entire fragment from the DpnII site to the poly(A) tail is cloned into the vector, and this fragment is loaded onto the beads for sequencing, as previously described. In the signature cloning method, an MmeI enzyme recognition site is added during cloning to the 5′-end of the fragment, adjacent to the DpnII site; MmeI is a Type IIS restriction enzyme that cuts 21 or 22 bp from the recognition site. After cloning, the fragment is cut with this enzyme to remove the cDNA 3′ to the 21- or 22-base fragment containing the signature that will be sequenced. In this way, signature cloning produces an “iso-length” library of 21 or 22 bases, removing any bias during the bead loading or sequencing reactions that may result from different DpnII-to-poly(A) fragment sizes.
MPSS Expression Data Processing
To determine the normalized abundance value for each signature in the library, we first merged 2-step runs and separately merged 4-step runs to create a raw abundance count for each stepper. The raw abundance count was calculated as the average from all 2-step or from all 4-step runs for a given signature. The average total number of signatures sequenced in all 2-step or for all 4-step runs was also calculated within the library. The final stage to merge the data within the steppers was to calculate the normalized abundance for each stepper. The normalized abundance is the raw abundance count divided by the average total number of signatures for both of the steppers in the library, multiplied by 106 to obtain a “transcripts per million” value. The calculation can be simplified to the following equations:
 |
where t_norm is the 2-step normalized value and f_norm is the 4-step normalized value.
Next, an overall normalized abundance was obtained by merging the separate 2-step and 4-step normalized abundances. For each signature, the normalized abundance of either the 2-step or 4-step runs was selected. This merge was simply a selection of the stepper for which the sum across all libraries of the normalized abundances for a given signature was higher. The higher of the two steppers was selected for each signature instead of averaging the two steppers because some sequence-specific artifacts can underestimate the abundance in one or both steppers (e.g., the “bad words” described in Results). In the rare cases in which the normalized abundances were precisely equal for the two steppers across all libraries (<0.1% of the total number of signatures), the stepper that was higher in a greater number of libraries was chosen; if this number was equal, the steppers were essentially indistinguishable and the 2-step value was arbitrarily chosen.
For each signature, the normalized abundance for the chosen stepper in each library was stored as the “norm_taken” value. This norm_taken value was used as the final value for further analyses and library-to-library comparisons. Because norm_taken for a signature in a single library is derived from a comparison of all libraries, the addition of new libraries can affect the norm_taken value for some signatures in existing libraries. However, this should occur only when there are few existing libraries, when the steppers are extremely similar across all libraries, or when the new library contains a much higher abundance for a given signature than is found in the existing libraries. In the first and last cases, there was likely not much support for either stepper, and in the middle case, the choice of one stepper over the other is largely irrelevant because the differences are insignificant.
Once the runs are merged, filters were applied to identify signatures that were unusually rare and could have resulted from technical artifacts. The filter for “reliability” determines if a given signature is found in more than one of the runs in all libraries; the goal is to exclude signatures that may have resulted from technical problems in a single run. Signatures found in only a single run, regardless of the abundance in that run, were flagged as “unreliable.” The filter for “significance” determines if a given signature is found in any Arabidopsis library at ≥4 PPM. The goal of this filter is to distinguish signatures that were consistently present at levels that might be interpreted as background (e.g., low levels of OBD signatures resulting from sequencing errors, as described in the Results section) from those that were potentially derived from a real transcript expressed at a significant level in at least one of the sampled conditions. These filters were applied by analyzing all libraries in the database, and the filters were reapplied with the addition of each new library to our database. Therefore, as with the norm_taken value described above, it is possible for the addition of a new library to alter the composition of the set of filtered signatures in an existing library. The filtered results from previous library additions and calculations are stored as different “builds” of our database so that it is possible to reconstruct analyses performed on earlier versions of the data set.
Calculation of Four-Base Word Usage in Genomic and Expressed Signatures
Each 17-base genomic signature was divided into eight frames of four bases that correspond to the MPSS sequencing process (Fig. 5A). Four different frames are used for each of the 2-steppers. The four-base words in each frame were counted across different sets of signatures. For the analyses of words in 17-base genomic signatures, bases 18 to 20 in the seventh and eighth frames were required (Fig. 5B). Similarly, word analysis of the 20-base genomic signatures required an additional two bases of sequence data to determine the word in the ninth frame (Fig. 5B). In each case, if we were comparing the genomic and expressed signatures, we limited our analyses to signatures occurring uniquely in the genome to avoid false matches.
We were most interested in the expressed signatures in which the 2-step and 4-step normalized abundances varied (see description above for t_norm and f_norm). To detect differential representation of the four-base words, we created three bins representing signatures in which (1) the 2-step abundance was equal to the 4-step abundance, (2) the 2-step abundance was significantly higher than the 4-step abundance (with P > 0.05), or (3) the 4-step abundance was significantly higher than the 2-step abundance (with P > 0.05). In the 2-step sequencing reaction, the words in frame 1 (Fig. 5A,B) always start with TC; this frame was excluded from our analysis because the overabundance of “TCNN” words skewed the results (data not shown). The fraction of the total comprised by each four-base word was calculated and then compared for the 2-step and 4-step frames (Table 4). For the 17-base signatures, the frequency of different words in frames 3 and 5 of expressed signatures was compared with the frequency of words in frames 2, 4, and 6 (Fig. 5A). For the 20-base signatures, the frequency of different words in frames 3, 5, and 7 of expressed signatures was compared with the frequency of words in frames 2, 4, 6, and 8 (Fig. 5B). Therefore, one fewer 2-step frame than 4-step frame was used to calculate the word frequency (two frames vs. three for the 17-base signatures, and four frames vs. five for the 20-base signatures). Therefore, we adjusted the final ratio of 2-step words to 4-step words to account for this difference in the number of frames that were analyzed, although the ratio adjustment merely scaled the data and did not affect the rankings of the words. These calculations used only 255 of the 256 possible four base words in the genomic and expressed signatures; the word “GATC” was removed because it was not detected within expressed signatures for the reasons described above. This analysis was performed for the 17-base and 20-base genomic and expressed signatures (Supplemental Figs. S4–S9).
We estimated the likelihood that a signature would have one of these 20 bad words in both 2-step and 4-step sequencing frames. These signatures are likely to be underrepresented in the MPSS data, as described in the Results section. The theoretical probability P of a genomic signature having one or more four-base palindrome in both sequencing frames is defined by the following equation considering 16 palindromic words:
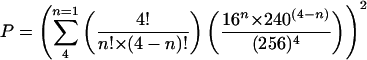 |
The calculation of the probability was performed with the assumptions that (1) each stepper sequences from four frames (see Fig. 5A), and (2) the genomic sequence is essentially random. If we consider only the 16 palindromic words, then 5.17% of all genomic signatures are predicted to be affected in the MPSS reactions. If the 20 bad words that we identified as underperforming in MPSS are considered, the percentage of genomic signatures containing one or more four-base palindromes in both sequencing frames is 7.71%.
Acknowledgments
We thank Pam Green for helpful comments and critical reading of the manuscript. This work was supported by a grant to B.C.M. from the NSF Plant Genome Research Program (DBI-0110528).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2275604.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Adams, M.D., Kerlavage, A.R., Fleischmann, R.D., Fuldner, R.A., Bult, C.J., Lee, N.H., Kirkness, E.F., Weinstock, K.G., Gocayne, J.D., White, O., et al. 1995. Initial assessment of human gene diversity and expression patterns based upon 83 million nucleotides of cDNA sequence. Nature 377: 3–174. [PubMed] [Google Scholar]
- Andrews, J., Bouffard, G.G., Cheadle, C., Lu, J., Becker, K.G., and Oliver, B. 2000. Gene discovery using computational and microarray analysis of transcription in the Drosophila melanogaster testis. Genome Res. 10: 2030–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Asano, T., Masumura, T., Kusano, H., Kikuchi, S., Kurita, A., Shimada, H., and Kadowaki, K. 2002. Construction of a specialized cDNA library from plant cells isolated by laser capture microdissection: Toward comprehensive analysis of the genes expressed in the rice phloem. Plant J. 32: 401–408. [DOI] [PubMed] [Google Scholar]
- Ball, C.A., Jin, H., Sherlock, G., Weng, S., Matese, J.C., Andrada, R., Binkley, G., Dolinski, K., Dwight, S.S., Harris, M.A., et al. 2001. Saccharomyces Genome Database provides tools to survey gene expression and functional analysis data. Nucleic Acids Res. 29: 80–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc, G., Barakat, A., Guyot, R., Cooke, R., and Delseny, M. 2000. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12: 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma, A., Hingamp, P., Quackenbush, J., Sherlock, G., Spellman, P., Stoeckert, C., Aach, J., Ansorge, W., Ball, C.A., Causton, H.C., et al. 2001. Minimum information about a microarray experiment (MIAME)—Toward standards for microarray data. Nat. Genet. 29: 365–371. [DOI] [PubMed] [Google Scholar]
- Brenner, S., Johnson, M., Bridgham, J., Golda, G., Lloyd, D.H., Johnson, D., Luo, S., McCurdy, S., Foy, M., Ewan, M., et al. 2000a. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 18: 630–634. [DOI] [PubMed] [Google Scholar]
- Brenner, S., Williams, S.R., Vermaas, E.H., Storck, T., Moon, K., McCollum, C., Mao, J.I., Luo, S., Kirchner, J.J., Eletr, S., et al. 2000b. In vitro cloning of complex mixtures of DNA on microbeads: Physical separation of differentially expressed cDNAs. Proc. Natl. Acad. Sci. 97: 1665–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron, H., van Schaik, B., van der Mee, M., Baas, F., Riggins, G., van Sluis, P., Hermus, M.C., van Asperen, R., Boon, K., Voute, P.A., et al. 2001. The human transcriptome map: Clustering of highly expressed genes in chromosomal domains. Science 291: 1289–1292. [DOI] [PubMed] [Google Scholar]
- Chen, J., Sun, M., Lee, S., Zhou, G., Rowley, J.D., and Wang, S.M. 2002. Identifying novel transcripts and novel genes in the human genome by using novel SAGE tags. Proc. Natl. Acad. Sci. 99: 12257–12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza, S.J., Camargo, A.A., Briones, M.R., Costa, F.F., Nagai, M.A., Verjovski-Almeida, S., Zago, M.A., Andrade, L.E., Carrer, H., El-Dorry, H.F., et al. 2000. Identification of human chromosome 22 transcribed sequences with ORF expressed sequence tags. Proc. Natl. Acad. Sci. 97: 12690–12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan, D.J., Bittner, M., Chen, Y., Meltzer, P., and Trent, J.M. 1999. Expression profiling using cDNA microarrays. Nat. Genet. 21: 10–14. [DOI] [PubMed] [Google Scholar]
- Feng, Q., Zhang, Y., Hao, P., Wang, S., Fu, G., Huang, Y., Li, Y., Zhu, J., Liu, Y., Hu, X., et al. 2002. Sequence and analysis of rice chromosome 4. Nature 420: 316–320. [DOI] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. 1988. Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. 85: 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith, D.W. 2003. Global analysis of cell type-specific gene expression. Compar. Funct. Genomics 4: 208–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, S.A., Ricke, D., Lan, T.H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100. [DOI] [PubMed] [Google Scholar]
- Guigo, R., Agarwal, P., Abril, J.F., Burset, M., and Fickett, J.W. 2000. An assessment of gene prediction accuracy in large DNA sequences. Genome Res. 10: 1631–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J., Volfovsky, N., Town, C.D., Troukhan, M., Alexandrov, N., Feldmann, K.A., Flavell, R.B., White, O., and Salzberg, S.L. 2002. Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol. 3: RESEARCH0029.0021–0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J., Delcher, A.L., Mount, S.M., Wortman, J.R., Smith Jr., R.K., Hannick, L.I., Maiti, R., Ronning, C.M., Rusch, D.B., Town, C.D., et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31: 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii, M., Hashimoto, S., Tsutsumi, S., Wada, Y., Matsushima, K., Kodama, T., and Aburatani, H. 2000. Direct comparison of GeneChip and SAGE on the quantitative accuracy in transcript profiling analysis. Genomics 68: 136–143. [DOI] [PubMed] [Google Scholar]
- Kapranov, P., Cawley, S.E., Drenkow, J., Bekiranov, S., Strausberg, R.L., Fodor, S.P., and Gingeras, T.R. 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916–919. [DOI] [PubMed] [Google Scholar]
- Kerk, N.M., Ceserani, T., Tausta, S.L., Sussex, I.M., and Nelson, T.M. 2003. Laser capture microdissection of cells from plant tissues. Plant Physiol. 132: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A. and Bennetzen, J.L. 1999. Plant retrotransposons. Annu. Rev. Genet. 33: 479–532. [DOI] [PubMed] [Google Scholar]
- Lash, A.E., Tolstoshev, C.M., Wagner, L., Schuler, G.D., Strausberg, R.L., Riggins, G.J., and Altschul, S.F. 2000. SAGEmap: A public gene expression resource. Genome Res. 10: 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Tingey, S.V., and Morgante, M. 2001. Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11: 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Lee, D.K., Vu, T.H., Tej, S.S., Edberg, S.B., Matvienko, M., and Tindell, L.D. 2004a. Arabidopsis MPSS: An online resource for quantitative expression analysis. Plant Physiol. (in press). [DOI] [PMC free article] [PubMed]
- Meyers, B.C., Vu, T.H., Tej, S.S., Ghazal, H., Matvienko, M., Agrawal, V., Ning, J., and Haudenschild, C. 2004b. Analysis of the transcriptional complexity of Arabidopsis by massively parallel signature sequencing. Nat. Biotech. (in press). [DOI] [PubMed]
- Qian, J., Kluger, Y., Yu, H., and Gerstein, M. 2003. Identification and correction of spurious spatial correlations in microarray data. Biotechniques 35: 42–48. [DOI] [PubMed] [Google Scholar]
- Reese, M.G., Hartzell, G., Harris, N.L., Ohler, U., Abril, J.F., and Lewis, S.E. 2000. Genome annotation assessment in Drosophila melanogaster. Genome Res. 10: 483–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn, J.L., Euskirchen, G., Bertone, P., Martone, R., Luscombe, N.M., Hartman, S., Harrison, P.M., Nelson, F.K., Miller, P., Gerstein, M., et al. 2003. The transcriptional activity of human Chromosome 22. Genes & Dev. 17: 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha, S., Sparks, A.B., Rago, C., Akmaev, V., Wang, C.J., Vogelstein, B., Kinzler, K.W., and Velculescu, V.E. 2002. Using the transcriptome to annotate the genome. Nat. Biotechnol. 20: 508–512. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., Matsumoto, T., Yamamoto, K., Sakata, K., Baba, T., Katayose, Y., Wu, J., Niimura, Y., Cheng, Z., Nagamura, Y., et al. 2002. The genome sequence and structure of rice chromosome 1. Nature 420: 312–316. [DOI] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470. [DOI] [PubMed] [Google Scholar]
- Schena, M., Heller, R.A., Theriault, T.P., Konrad, K., Lachenmeier, E., and Davis, R.W. 1998. Microarrays: Biotechnology's discovery platform for functional genomics. Trends Biotechnol. 16: 301–306. [DOI] [PubMed] [Google Scholar]
- Shoemaker, D.D., Schadt, E.E., Armour, C.D., He, Y.D., Garrett-Engele, P., McDonagh, P.D., Loerch, P.M., Leonardson, A., Lum, P.Y., Cavet, G., et al. 2001. Experimental annotation of the human genome using microarray technology. Nature 409: 922–927. [DOI] [PubMed] [Google Scholar]
- Simillion, C., Vandepoele, K., Van Montagu, M.C., Zabeau, M., and Van De Peer, Y. 2002. The hidden duplication past of Arabidopsis thaliana. Proc. Natl. Acad. Sci. 99: 13627–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu, V.E., Zhang, L., Vogelstein, B., and Kinzler, K.W. 1995. Serial analysis of gene expression. Science 270: 484–487. [DOI] [PubMed] [Google Scholar]
- Velculescu, V.E., Zhang, L., Zhou, W., Vogelstein, J., Basrai, M.A., Bassett Jr., D.E., Hieter, P., Vogelstein, B., and Kinzler, K.W. 1997. Characterization of the yeast transcriptome. Cell 88: 243–251. [DOI] [PubMed] [Google Scholar]
- Velculescu, V.E., Madden, S.L., Zhang, L., Lash, A.E., Yu, J., Rago, C., Lal, A., Wang, C.J., Beaudry, G.A., Ciriello, K.M., et al. 1999. Analysis of human transcriptomes. Nat. Genet. 23: 387–388. [DOI] [PubMed] [Google Scholar]
- Velculescu, V.E., Vogelstein, B., and Kinzler, K.W. 2000. Analysing uncharted transcriptomes with SAGE. Trends Genet. 16: 423–425. [DOI] [PubMed] [Google Scholar]
- Vision, T.J., Brown, D.G., and Tanksley, S.D. 2000. The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117. [DOI] [PubMed] [Google Scholar]
- Wortman, J.R., Haas, B.J., Hannick, L.I., Smith Jr., R.K., Maiti, R., Ronning, C.M., Chan, A.P., Yu, C., Ayele, M., Whitelaw, C.A., et al. 2003. Annotation of the Arabidopsis genome. Plant Physiol. 132: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, K., Lim, J., Dale, J.M., Chen, H., Shinn, P., Palm, C.J., Southwick, A.M., Wu, H.C., Kim, C., Nguyen, M., et al. 2003. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846. [DOI] [PubMed] [Google Scholar]
- Yu, J., Hu, S., Wang, J., Wong, G.K., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://mpss.udel.edu/at/query.php; MPSS signature extraction script.
- http://mpss.udel.edu/at; MPSS database.
- http://www.lynxgen.com; MPSS methodology.
- http://www.ncbi.nlm.nih.gov/SAGE/; SAGE database.
- http://www.tigr.org; TIGR.