Abstract
Genome-wide association studies using large numbers of bi-allelic single nucleotide polymorphisms (SNPs) have been proposed as a potentially powerful method for identifying genes involved in common diseases. To assemble a SNP collection appropriate for large-scale association, we designed assays for 226,099 publicly available SNPs located primarily within known and predicted gene regions. Allele frequencies were estimated in a sample of 92 CEPH Caucasians using chip-based MALDI-TOF mass spectrometry with pooled DNA. Of the 204,200 designed assays that were functional, 125,799 SNPs were determined to be polymorphic (minor allele frequency >0.02), of which 101,729 map uniquely to the human genome. Many of the commonly available RefSNP annotations were predictive of polymorphic status and could be used to improve the selection of SNPs from the public domain for genetic research. The set of uniquely mapping, polymorphic SNPs is located within 10 kb of 66% of known and predicted genes annotated in LocusLink, which could prove useful for large-scale disease association studies.
Single nucleotide polymorphisms (SNPs) are the most abundant genetic variations in the human genome. They occur, on average, once every 300 base pairs of sequence with a minor allele frequency (MAF) greater than 1% (Kruglyak and Nickerson 2001; Stephens et al. 2001; Reich et al. 2003). The high abundance of SNPs and the availability of increasingly high-throughput and cost effective methods of measuring them encourage their use in many kinds of human genetic studies (Marnellos 2003). SNPs are of particular value in whole-genome association studies for identifying the genes involved in complex trait variation (Lander 1996; Risch and Merikangas 1996). The expected number of SNPs required for successful population-based association studies depends on the distribution of linkage disequilibrium (LD) across the genome in the populations of interest. The lengths of genomic segments in strong LD varies tremendously throughout the genome, ranging from less than 1 kilobase pairs (kb) to over 500 kb, with most blocks estimated to be less than 20 kb in Caucasian populations (Daly et al. 2001; Reich et al. 2001; Dawson et al. 2002; Gabriel et al. 2002; Clark et al. 2003). The international haplotype mapping (HapMap) project was initiated to characterize the patterns of LD throughout the human genome to help identify the most informative set of SNP markers for LD mapping (Gibbs et al. 2003).
To explore the potential of large-scale association studies, we set out to develop a suitable collection of approximately 100,000 SNPs. With the HapMap project far from completion, the SNPs could not be selected on the basis of LD patterns. Since a collection of 100,000 SNPs would be far too few to provide dense coverage throughout the genome, we primarily focused on SNPs located within and around known and predicted genes. Additionally, we sought SNPs with MAF greater than 5% in Caucasian populations that could be used in case-control type study designs, assuming that relatively common genetic variations are responsible for common diseases.
There are a large number of publicly available SNPs. The number of reported nonredundant SNPs in NCBI's dbSNP database at the time these analyses were initiated (refSNPs) exceeded four million (dbSNP build 114, April 2003, http://www.ncbi.nlm.nih.gov/SNP/). Most recently, the number of SNPs in the public domain stands at over nine million. Originally, these SNPs were primarily putative polymorphisms discovered by in silico datamining algorithms (Buetow et al. 1999; Irizarry et al. 2000; Marth et al. 2001) or shotgun sequencing (Altshuler et al. 2000). Most of them have not been confirmed independently. Previous studies suggested that 50% to 80% of the SNPs in dbSNP are polymorphic in any population (Marth et al. 2001; Carlson et al. 2003; Reich et al. 2003). To create a SNP resource useful for genetic studies we tested more than 200,000 publicly available SNPs located primarily within gene regions.
RESULTS
From November 1999 through September 2001, we collected 226,099 putative SNPs, primarily ascertained from in silico expressed sequence tag (EST) comparison projects (Buetow et al. 1999; Irizarry et al. 2000), SNPs from NCBI's database that mapped to RefSeq and UniGene sequences, and a small portion through external collaborators. To confirm the existence of these putative SNPs, primer extension assays (MassEXTEND) were designed and applied to a pool of 92 individual Caucasian DNA samples for each SNP. Extension products were analyzed by chip-based mass spectrometry. The areas under the two mass spectrum peaks corresponding to the expected mass values of the two alleles were calculated. The ratio of each peak area to the total area of both peaks was used to estimate the relative allele frequency in the sample pool (Buetow et al. 2001; Bansal et al. 2002; Mohlke et al. 2002; Shifman et al. 2002). A SNP was considered confirmed and polymorphic if the estimated allele frequency was at least 0.02, and the estimated frequency was greater than twice the measurement standard deviation.
Of the 226,099 SNP assays designed and tested, 204,200 (90%) were functional, producing at least one of the two expected extension products based on the SNP definition. Out of these functional assays, 126,391 SNPs (62%) were identified as polymorphic in this Caucasian sample. To improve our ability to select additional polymorphic SNPs amenable to assay design from the public domain, we investigated the relationships between the standard RefSNP annotations and functional and polymorphic status (Table 1). For this comparison, we further subdivided the polymorphic SNPs into those with frequencies equal to or less than 0.05 (N = 12,169) and those greater than 0.05 (N = 114,222). While the strengths of associations between polymorphic status and RefSNP attributes vary, all are statistically significant (P-value < 10–6), owing to the large sample size. For the few SNPs with frequency information (14%), those reporting high heterozygosity were much more likely to be higher frequency in our sample. The two strongest predictors of polymorphic status available for nearly all SNPs were NCBI validation status and the number of submitters reporting the SNP (Submitter Count). We observed that 85% of SNPs that were reported as “validated” by NCBI were identified as polymorphic in this sample. Polymorphic SNPs were also more likely to have longer sequences for their submission (Length), be drawn from more recent RefSNP submissions (RS Build), be derived from genomic DNA (MolType), be mapped within introns (SNP Type), and map exactly one time to the genome (RS Mapping).
Table 1.
Comparison of SNP Annotations Between Failed and Functional Assays and Between Polymorphic Classes
|
Functionalc
|
|||||
|---|---|---|---|---|---|
| Na | Failed (N = 21,899) | Non-Polymorphic (N = 77,809) | MAFd≤0.05 (N = 12,169) | MAF >0.05 (N = 114,222) | |
| Heterozygosity | 26,248 | 0.19/0.39/0.49 | 0.01/0.13/0.42 | 0.10/0.24/0.43 | 0.32/0.44/0.49 |
| Length | 193,940 | 394/419/551b | 356/401/534 | 401/436/561 | 401/447/548 |
| RS Build | 193,940 | 87/92/108b | 86/92/106 | 87/92/108 | 88/100/111 |
| NCBI validation | 193,941 | ||||
| No | 73% (11,077)b | 86% (55,541) | 72% (7,641) | 57% (59,340) | |
| Yes | 27% (4,157) | 14% (8,683) | 28% (2,929) | 43% (44,573) | |
| MolType | 193,940 | ||||
| cDNA | 13% (1,924)b | 18% (11,754) | 9% (948) | 5% (5,397) | |
| Genomic | 87% (13,310) | 82% (52,470) | 91% (9622) | 95% (98,515) | |
| SNP Type | 193,932 | ||||
| Not annotated | 34% (5,219)b | 30% (19,462) | 32% (3,385) | 31% (32,131) | |
| Intron | 30% (4,608) | 31% (19,962) | 34% (3,614) | 38% (39,046) | |
| Locus region | 15% (2,227) | 14% (9,301) | 15% (1,571) | 15% (15,491) | |
| mRNA UTR | 13% (2,002) | 15% (9,720) | 13% (1,331) | 11% (11,718) | |
| Coding | 0% (30) | 0% (130) | 0% (15) | 0% (94) | |
| Coding nonsynon | 4% (609) | 5% (3,344) | 3% (358) | 3% (2,663) | |
| Coding synon | 3% (425) | 3% (1,792) | 2% (243) | 2% (2,287) | |
| Exception | 1% (110) | 1% (496) | 1% (53) | 0% (465) | |
| Splice site | 0% (4) | 0% (13) | 0% (0) | 0% (13) | |
| Submitter count | 193,941 | ||||
| 1 | 48% (7,382) | 62% (39,822) | 49% (5,203) | 38% (39,873) | |
| 2 | 33% (5,056) | 28% (17,869) | 35% (3,694) | 38% (39,572) | |
| 3 | 14% (2,148) | 9% (5,527) | 12% (1,321) | 18% (18,321) | |
| >3 | 4% (648) | 1% (1,006) | 4% (352) | 6% (6,147) | |
| RS Mapping | 193,941 | ||||
| 0 | 8% (1,178)b | 6% (3,885) | 5% (544) | 2% (2,315) | |
| 1 | 90% (13,746) | 92% (59,280) | 93% (9,819) | 95% (99,185) | |
| >1 | 2% (310) | 2% (1,059) | 2% (207) | 2% (2,413) | |
| eXTEND mapping | 226,059 | ||||
| 0 | 29% (6,281)b | 9% (7,006) | 7% (860) | 7% (7,455) | |
| 1 | 57% (12,532) | 73% (56,566) | 72% (8,782) | 81% (92,947) | |
| >1 | 14% (3,085) | 18% (14,208) | 21% (2,526) | 12% (13,811) | |
| TSC MAF range | 7,997 | ||||
| [0, 0.025] | 23% (120) | 74% (1,325) | 29% (129) | 5% (239) | |
| [0.025, 0.05] | 6% (31) | 7% (133) | 22% (99) | 3% (163) | |
| [0.05, 0.075] | 3% (16) | 3% (59) | 10% (43) | 3% (141) | |
| [0.075, 0.1] | 5% (28) | 2% (33) | 10% (46) | 5% (282) | |
| >0.1 | 63% (331) | 13% (234) | 29% (130) | 84% (4,415) | |
The first eight annotation categories were drawn from NCBI dbSNP for the 193,940 SNPs that overlap with SNPs in this study.
Quantitative variables are summarized as 1st quartile/median/3rd quartile. Categorical variables are summarized by column percent (count).
N: Number of valid nonmissing observations for each variable.
Comparisons between failed and functional assays significant at α = 0.05. All significant results have P-values < 10-6.
All comparisons among functional assay groups are statistically significant with P-values < 10-6.
MAF: Minor allele frequency.
We developed an algorithm called eXTEND based on NCBI's ePCR program (Schuler 1997) to test for possible coamplification of homologous regions, improve the success rate of SNP assay designs, and map the confirmed SNPs to the human genome. The algorithm estimates the specificity of a MassEXTEND SNP assay by analyzing in silico the annealing specificity of the two amplification primers and one extension primer used in each reaction. The GoldenPath genome assembly from the University of California, Santa Cruz (version hg15, based on NCBI's Genome Build 33, April 2003) was used for eXTEND to map all SNP assays. The distribution of mapping results among all assay categories is presented in Table 1. Of the 204,200 functional assays, 188,840 putative SNPs could be mapped to the human genome and 158,295 assays mapped uniquely. The remaining 30,545 assays were ambiguous, mapping to the genome more than once. Assays that did not map were somewhat more likely to be non-polymorphic. With each release of the human genome assembly, more of the previously unmapped SNPs can be mapped, and many of the assays that previously mapped multiple times have reduced numbers of mapping positions.
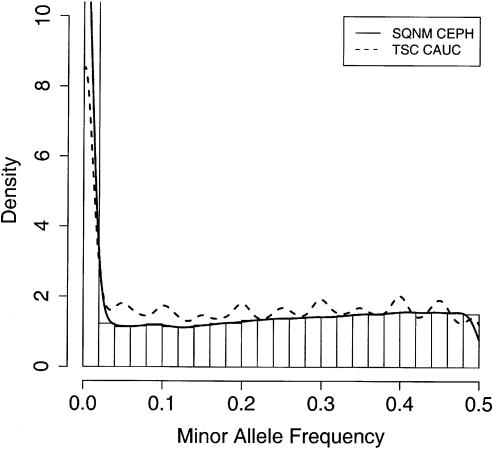
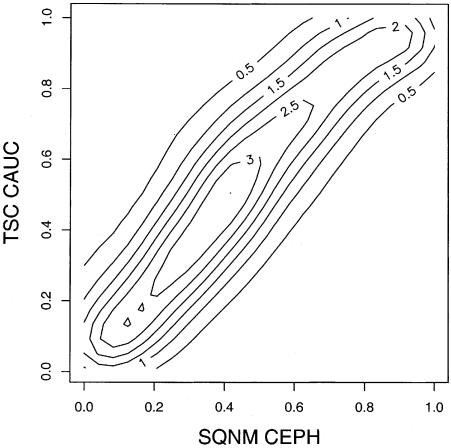
The distribution of MAFs for the 158,295 functional assays that mapped uniquely to the human genome is shown in Figure 1. The shape of this distribution shows a larger proportion of high frequency than low frequency SNPs. This distribution is compared in Figure 1 to the distribution for 61,173 SNPs with Caucasian frequencies available from The SNP Consortium (TSC). Ignoring the excess of SNPs with frequencies at 0.05 intervals due to rounding in the TSC data set, the distribution is more uniform than we observed. The overabundance of high frequency SNPs can be partially explained by the tendency of the pool-based approach used in this study to overestimate the low mass extension product compared to the high mass extension product (Jurinke et al. 2003). For example, the median of the distribution of high mass allele frequencies for the polymorphic SNPs is 0.42, a significant departure from the expected value of 0.5 assuming an equal probability of the assay design assigning the minor allele to the high or low mass extension products. Regardless of this bias in the allele frequency measurement, there was general agreement in allele frequency estimates from 5,644 polymorphic SNPs (MAF > 0.05) that were in common between the current study and TSC SNPs (Fig. 2; Spearman correlation ρ = 0.79). At the lower frequency range, 81% of SNPs determined to be non-polymorphic had corresponding TSC MAF ≤ 0.05 (Table 1).
Figure 1.
SNP allele frequency distribution. Minor allele frequency (MAF) distribution of 158,295 functional SNP assays that map uniquely to the human genome from this study (SQNM CEPH; histogram and solid line) compared to the distribution of 61,173 SNPs with Caucasian frequencies estimated by TSC (TSC CAUC; dashed line). The distributions are presented as densities, such that the area under each curve sums to one. The y-axis is truncated at 10, although the left-most histogram bar for SQNM CEPH extends to 17.
Figure 2.
Correlation between frequencies from this study and matched Caucasian TSC SNP allele frequencies. The relationship between allele frequencies for 5,644 SNPs measured in the current study (SQNM CEPH) and matched Caucasian frequencies in the TSC collection (TSC CAUC) is compared with a bivariate contour plot. Density lines are plotted every 0.5 units, ranging from 0.5 for the outermost line to the highest density of 3.0 for the innermost line. The allele presented was arbitrarily standardized to the MassEXTEND high mass allele from each assay in this study. SNPs with minor allele frequencies <0.05 in both SQNM CEPH and TSC CAUC samples were excluded from this plot. Lower frequency SNPs are compared at the bottom of Table 1.
Of 226,099 putative SNPs tested, 21,899 reactions (9.7%) did not result in a functional assay. The majority of such reaction failures could be attributed to one of four causes: (1) inaccurate sequence information in the region of the SNP for those that could not be mapped, (2) non-functional PCR and/or Mass EXTEND primers, (3) random processing failures, or (4) genomic regions that are difficult to amplify (e.g., GC-rich). We found that for 29% of the failed assays, the oligos did not map to the genome by eXTEND analysis, compared to 9% of nonpolymorphic assays and 7% of polymorphic assays (Table 1). We also found that failed assays were more likely to have shorter sequences for their submission, be drawn from earlier RefSNP submissions, not have been reported as validated by NCBI, be derived from cDNA submissions, not have a SNP type annotation, and map less often to the genome. Results of a mass spectrometry analysis of oligonucleotides used in failed assays ranged from the expected reagent, to incomplete synthesis, reagent with salt adducts, and in the most extreme cases, no product. Based on these observations, we developed oligonucleotide quality control software (Spectro-CHECK) for mass spectrometric monitoring of reagent quality. With these procedures in place the average failure rate for new SNP assays has been successfully reduced to 6%.
DISCUSSION
In this study we estimated the allele frequencies in 92 Caucasian subjects for 204,200 SNPs derived from public sources available from 1999 to 2001. Of 158,295 SNPs that map uniquely, 64% were confirmed polymorphic (MAF > 0.02). We compared our confirmation rates of polymorphic SNPs to four published studies using Caucasian samples. The studies by Marth et al. (2001), Gabriel et al. (2002), Reich et al. (2003), and Carlson et al. (2003) reported polymorphic rates of approximately 80%, 76%, 83%, and 51%, respectively. The differences in confirmation rates between these studies likely stem from the different technologies and methodologies that were used to test SNPs and the different protocols and thresholds that were applied to determine false-positive SNPs and frequencies of true SNPs. For example, most studies considered a SNP to be polymorphic if it was identified at least once. However, since we used a quantitative assessment of allele frequencies we selected a cutoff of 2%, near the technology measurement limit. We also compared the proportion of common SNPs estimated in each study to our results. The study by Marth et al. (2001) found that around 52% to 54% of TSC SNPs had frequencies of at least 20%, which is slightly higher than the 44% we determined in our analysis. The studies by Gabriel et al. (2002), Reich et al. (2003), and Carlson et al. (2003) reported approximately 58%, 71%, and 41% of public SNPs with frequencies greater than 10%, respectively. This range includes our estimate of 56% of SNPs that are similarly common. By comparison, the various sources of Caucasian (most of CEPH origin) allele frequencies in the TSC data evaluated in this study gave estimates of 66 to 79% of SNPs with MAF > 0.05.
Apart from measurement methods, there were other notable differences between our study and those cited. Our study tested a larger number of SNPs than the previous studies, the largest of which (Gabriel et al. 2002) examined 4,532 SNPs. It was also broader in its coverage of various classes of public dbSNPs: For instance the ratio of random read SNPs (dbSNP submitter TSC-CSHL) versus BAC-overlap SNPs (dbSNP submitters SC_JIM and KWOK) in our study was similar to the corresponding ratio in dbSNP (even for dbSNP builds more recent than our study, like build 116), whereas Gabriel et al. (2002) examined only TSC SNPs. And in our study, even though we were mostly interested in broader gene regions, we did not concentrate exclusively on genes, as in some of the other studies that resulted in higher confirmation rates (Carlson et al. 2003; Reich et al. 2003). Had we concentrated only on SNPs located within or close to LocusLink genes, our confirmation rate would have been higher as well. Finally, the SNPs for these studies may have been collected from dbSNP more recently than our set, thus benefiting from an improvement in the quality of SNPs in the public domain. With the experience gathered over the years by the SNP discovery community, it is likely that newer SNPs in dbSNP have been discovered and filtered by improved techniques, resulting in a higher confirmation rate.
An important consideration for all researchers using SNPs for genetic research is the selection of informative SNPs for the study in question. In the absence of thoroughly validated allele frequencies for the ethnicity of interest, we found that the standard NCBI annotations can improve the selection of polymorphic SNPs (Table 1). For example, restricting our data only to those SNPs with Length > 447, RS Build > 100, NCBI Validation = “YES”, and Submitter Count > 1 results in 14,640 SNPs, 80% of which have MAF greater than 0.1. This compares to 53% of all SNPs tested (including zero and multiple mapping SNPs), representing a substantial improvement for selecting common SNPs. The single most useful factor in this selection is NCBI Validation. Approximately 75% of SNPs annotated with “YES” in our sample are common in Caucasians. Ignoring NCBI Validation status results in a more modest improvement from 53% to 69% of common SNPs.
A summary of each of the 226,086 tested SNPs along with the allele frequency estimates is available as Supplemental material (Table S1). Allele frequencies for polymorphic SNPs have been submitted to the NCBI dbSNP repository. Such public information may prove useful to develop SNP maps of various sizes targeting gene regions of the human genome. Until the haplotype map is completed (Gibbs et al. 2003), the use of gene-based SNP panels such as the one presented here offer a feasible alternative for large-scale association.
METHODS
Construction of DNA Pool and SNP Confirmation
Unrelated Caucasian DNA samples were purchased from Coriell. Ninety-two (92) DNA samples were measured and pooled in equimolar amounts to generate a single DNA pool for SNP confirmation and allele frequency estimation (Buetow et al. 2001). Oligonucleotides were purchased from Integrated DNA Technology, Operon, and Metabion. For each reaction, 25 ng of pooled DNA was used. PCR and MassEXTEND reactions were conducted using standard conditions as previously described (Buetow et al. 2001). Reaction products were dispensed independently onto four SpectroCHIPs and analyzed on a mass spectrometer (Sequenom). Spectra were then analyzed using SpectroTYPER software that includes quantitative peak area calculation and baseline correction. The peak areas for both alleles were used to calculate the allele frequencies (Buetow et al. 2001; Bansal et al. 2002; Mohlke et al. 2002).
SNP Mapping
Assays were mapped to the genome using eXTEND, a modified version of the ePCR program (Schuler 1997). ePCR identifies potential amplicons for given PCR primer pairs; similarly, eXTEND identifies potential MassEXTEND sites on the genome using the PCR primer pair and the corresponding MassEXTEND primer of each assay. Each primer was annealed in silico to the genome allowing up to two mismatches over its length. PCR primers were then mapped in all combinations of order, orientation, and spacing that could produce an amplicon less than 1000 bp and a MassEXTEND reaction product within the amplicon. Using eXTEND we obtained genome positions for the majority of SNP assays. Assays that did not map with this procedure were subjected to two additional passes of eXTEND with relaxed annealing parameters for primers to increase mapping efficiency. The GoldenPath genome assembly from the University of California, Santa Cruz (version hg15, based on NCBI's Genome Build 33, April 2003) was used for mapping.
Public SNP Annotations
For comparing our SNPs with the public domain SNPs, we used the nonredundant set in NCBI's dbSNP database (refSNP, build 114, April 28 2003) and the annotations that NCBI was providing for that set with respect to Genome Build 33 (Sherry et al. 2001). Uniquely mapping Sequenom SNPs were matched to uniquely mapping refSNPs that mapped to the same position on the genome. The set of LocusLink genes (Pruitt and Maglott 2001) considered in this study consisted of 35,817 known and predicted genes for which NCBI was providing positions on the genome (NCBI Genome Build 33, April 2003) on the Mapview FTP site (ftp://ftp.ncbi.nih.gov/refseq/H_sapiens/H_sapiens/maps/mapview/BUILD.33/seq_gene.md.gz).
For SNP frequency comparisons we used the data gathered by the allele frequency/genotype project of The SNP Consortium, as provided on their site (http://snp.cshl.org). There were 61,266 SNPs with refSNP identifiers and valid frequency results that we examined in this work. The SNPs were biallelic and for each SNP we put together the frequencies of the two alleles, as estimated for the Caucasian samples. A TSC-validated SNP and a Sequenom SNP were matched if both had been matched to the same refSNP. There was not enough information in the TSC downloaded data to unambiguously match alleles in TSC–Sequenom pairs. We only considered pairs for which: (1) TSC allele 1 was identical to Sequenom allele 1 and TSC allele 2 identical to Sequenom allele 2, in which case we matched TSC allele 1 frequency to Sequenom allele 1 frequency; or (2) TSC allele 1 was identical to Sequenom allele 2 and TSC allele 2 identical to Sequenom allele 1, in which case we matched TSC allele 1 frequency to Sequenom allele 2 frequency; or (3) there was only one TSC allele available (which was true for SNPs found to be nonpolymorphic) and this was identical to Sequenom allele 1 or 2. We considered 7,997 such pairs, associated with 7,026 distinct refSNP identifiers (some refSNPs in this set had more than one corresponding TSC–Sequenom pairs).
Statistical Methods
Univariate allele frequency densities were estimated using the gaussian kernel density estimation function available in version 1.8.1 of R (R Development Core Team 2004). The smoothing bandwidth was set at 0.015. The bivariate allele frequencies density was estimated using the kde2d two-dimensional kernel density estimation function available in the MASS package for R (Venables and Ripley 2002). A value of 0.25 was used as the smoothing bandwidth. The resulting density was displayed as a contour plot.
Acknowledgments
We are grateful to Dr. Eric Lai (GlaxoSmithKline, Inc., USA) and Dr. Ohara Osamu (Kazusa DNA Research Institute, Japan) for contributing SNPs to this project.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2421604.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: E. Lai, and O. Osamu.]
References
- Altshuler, D., Pollara, V.J., Cowles, C.R., Van Etten, W.J., Baldwin, J., Linton, L., and Lander, E.S. 2000. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407: 513–516. [DOI] [PubMed] [Google Scholar]
- Bansal, A., van den Boom, D., Kammerer, S., Honisch, C., Adam, G., Cantor, C.R., Kleyn, P., and Braun, A. 2002. Association testing by DNA pooling: An effective initial screen. Proc. Natl. Acad. Sci. 99: 16871–16874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buetow, K.H., Edmonson, M.N., and Cassidy, A.B. 1999. Reliable identification of large numbers of candidate SNPs from public EST data. Nat. Genet. 21: 323–325. [DOI] [PubMed] [Google Scholar]
- Buetow, K.H., Edmonson, M., MacDonald, R., Clifford, R., Yip, P., Kelley, J., Little, D.P., Strausberg, R., Koester, H., Cantor, C.R., et al. 2001. High-throughput development and characterization of a genomewide collection of gene-based single nucleotide polymorphism markers by chip-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Proc. Natl. Acad. Sci. 98: 581–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson, C.S., Eberle, M.A., Rieder, M.J., Smith, J.D., Kruglyak, L., and Nickerson, D.A. 2003. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat. Genet. 33: 518–521. [DOI] [PubMed] [Google Scholar]
- Clark, A.G., Nielsen, R., Signorovitch, J., Matise, T.C., Glanowski, S., Heil, J., Winn-Deen, E.S., Holden, A.L., and Lai, E. 2003. Linkage disequilibrium and inference of ancestral recombination in 538 single-nucleotide polymorphism clusters across the human genome. Am. J. Hum. Genet. 73: 285–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, M.J., Rioux, J.D., Schaffner, S.F., Hudson, T.J., and Lander, E.S. 2001. High-resolution haplotype structure in the human genome. Nat. Genet. 29: 229–232. [DOI] [PubMed] [Google Scholar]
- Dawson, E., Abecasis, G.R., Bumpstead, S., Chen, Y., Hunt, S., Beare, D.M., Pabial, J., Dibling, T., Tinsley, E., Kirby, S., et al. 2002. A first-generation linkage disequilibrium map of human chromosome 22. Nature 418: 544–548. [DOI] [PubMed] [Google Scholar]
- Gabriel, S.B., Schaffner, S.F., Nguyen, H., Moore, J.M., Roy, J., Blumenstiel, B., Higgins, J., DeFelice, M., Lochner, A., Faggart, M., et al. 2002. The structure of haplotype blocks in the human genome. Science 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- Gibbs, R.A., Belmont, J.W., Hardenbol, P., Willis, T.D., Yu, F., Yang, H., Ch'ang, L.Y., Huang, W., Liu, B., Shen, Y., et al. 2003. The international HapMap project. Nature 426: 789–796. [DOI] [PubMed] [Google Scholar]
- Irizarry, K., Kustanovich, V., Li, C., Brown, N., Nelson, S., Wong, W., and Lee, C.J. 2000. Genome-wide analysis of single-nucleotide polymorphisms in human expressed sequences. Nat. Genet. 26: 233–236. [DOI] [PubMed] [Google Scholar]
- Jurinke, C., Oeth, P., and van den Boom, D. 2003. MALDI-TOF mass spectrometry: A versatile tool for high-performance DNA analysis. Mol. Biotechnol. 25: 147–164. [DOI] [PubMed] [Google Scholar]
- Kruglyak, L. and Nickerson, D.A. 2001. Variation is the spice of life. Nat. Genet. 27: 234–236. [DOI] [PubMed] [Google Scholar]
- Lander, E.S. 1996. The new genomics: Global views of biology. Science 274: 536–539. [DOI] [PubMed] [Google Scholar]
- Marnellos, G. 2003. High-throughput SNP analysis for genetic association studies. Curr. Opin. Drug. Discov. Devel. 6: 317–321. [PubMed] [Google Scholar]
- Marth, G., Yeh, R., Minton, M., Donaldson, R., Li, Q., Duan, S., Davenport, R., Miller, R.D., and Kwok, P.Y. 2001. Single-nucleotide polymorphisms in the public domain: How useful are they? Nat. Genet. 27: 371–372. [DOI] [PubMed] [Google Scholar]
- Mohlke, K.L., Erdos, M.R., Scott, L.J., Fingerlin, T.E., Jackson, A.U., Silander, K., Hollstein, P., Boehnke, M., and Collins, F.S. 2002. High-throughput screening for evidence of association by using mass spectrometry genotyping on DNA pools. Proc. Natl. Acad. Sci. 99: 16928–16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt, K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2004. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- Reich, D.E., Cargill, M., Bolk, S., Ireland, J., Sabeti, P.C., Richter, D.J., Lavery, T., Kouyoumjian, R., Farhadian, S.F., Ward, R., et al. 2001. Linkage disequilibrium in the human genome. Nature 411: 199–204. [DOI] [PubMed] [Google Scholar]
- Reich, D.E., Gabriel, S.B., and Altshuler, D. 2003. Quality and completeness of SNP databases. Nat. Genet. 33: 457–458. [DOI] [PubMed] [Google Scholar]
- Risch, N. and Merikangas, K. 1996. The future of genetic studies of complex human diseases. Science 273: 1516–1517. [DOI] [PubMed] [Google Scholar]
- Schuler, G.D. 1997. Sequence mapping by electronic PCR. Genome Res. 7: 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry, S.T., Ward, M.H., Kholodov, M., Baker, J., Phan, L., Smigielski, E.M., and Sirotkin, K. 2001. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shifman, S., Pisante-Shalom, A., Yakir, B., and Darvasi, A. 2002. Quantitative technologies for allele frequency estimation of SNPs in DNA pools. Mol. Cell. Probes 16: 429–434. [DOI] [PubMed] [Google Scholar]
- Stephens, J.C., Schneider, J.A., Tanguay, D.A., Choi, J., Acharya, T., Stanley, S.E., Jiang, R., Messer, C.J., Chew, A., Han, J.H., et al. 2001. Haplotype variation and linkage disequilibrium in 313 human genes. Science 293: 489–493. [DOI] [PubMed] [Google Scholar]
- Venables, W.N. and Ripley, B.D. 2002. Modern applied statistics with S. Springer, New York.
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/SNP/; NCBI dbSNP home page.
- http://snp.cshl.org; The SNP Consortium home page.