Abstract
Type I chaperonins are molecular chaperones present in virtually all bacteria, some archaea and the plastids and mitochondria of eukaryotes. Sequences of cpn60 genes, encoding 60-kDa chaperonin protein subunits (CPN60, also known as GroEL or HSP60), are useful for phylogenetic studies and as targets for detection and identification of organisms. Conveniently, a 549–567-bp segment of the cpn60 coding region can be amplified with universal PCR primers. Here, we introduce cpnDB, a curated collection of cpn60 sequence data collected from public databases or generated by a network of collaborators exploiting the cpn60 target in clinical, phylogenetic, and microbial ecology studies. The growing database currently contains ∼2000 records covering over 240 genera of bacteria, eukaryotes, and archaea. The database also contains over 60 sequences for the archaeal Type II chaperonin (thermosome, a homolog of eukaryotic cytoplasmic chaperonin) from 19 archaeal genera. As the largest curated collection of sequences available for a protein-encoding gene, cpnDB provides a resource for researchers interested in exploiting the power of cpn60 as a diagnostic or as a target for phylogenetic or microbial ecology studies, as well as those interested in broader subjects such as lateral gene transfer and codon usage. We built cpnDB from open source tools and it is available at http://cpndb.cbr.nrc.ca.
The advent of genome-scale sequencing projects has led to the availability of full-genome sequences for a variety of organisms including eukaryotes, bacteria, and archaea. This data is an invaluable resource for studies of genome-scale evolutionary processes such as lateral gene transfer and organelle evolution, as well as providing fuel for debates surrounding topics such as the definition of “species,” particularly in the microbial world (Perna et al. 2001). NCBI currently lists 155 complete microbial genomes from 138 bacteria and 17 archaeal species. Obviously, this limited selection inadequately represents the vast microbial diversity present in the environment. As a result of this limitation, large collections of gene-specific sequence data, particularly from universal genes, are an important resource. Historically, small subunit ribosomal RNA (16S rRNA) sequences have been the primary resource for phylogenetic studies and for sequence-based taxonomy (Olsen et al. 1986; Woese et al. 1990; Cole et al. 2003). Given our current understanding of the dynamic nature of genomes and the impact of lateral gene transfer on genome evolution (Boucher et al. 2003), it is important that our view of taxonomy and phylogenetics be informed by more than one target.
A comparison of the sequences of the Escherichia coli groEL gene, which encodes a protein identified as being essential for the posttranslational assembly of bacteriophage particles and the Rubisco subunit-binding protein of higher plant chloroplasts, led to the discovery that these two proteins represent a ubiquitous protein family now known as the type I chaperonins (CPN60; Hemmingsen et al. 1988). “CPN60” is our preferred term for the type I chaperonins that are variously referred to in the literature as “GroEL,” “MopA,” and “Hsp60.” Among the bacterial and eukaryal organisms for which complete genome sequences are available, only the intracellular organisms Mycoplasma pulmonis and Ureaplasma urealyticum have been found to lack cpn60 genes. These organisms also lack other genes previously considered essential for prokaryotic life (Glass et al. 2000; Chambaud et al. 2001). Originally thought to be confined to the bacteria and eukaryotes, cpn60 genes have recently been identified in two members of the archaeal genus, Methanosarcina (Klunker et al. 2003).
Multiple functions have been ascribed to CPN60. Whereas the primary intracellular role of CPN60 is thought to be as a molecular chaperone in the processes of posttranslational protein folding and assembly of protein complexes (for review, see Saibil and Ranson 2002), CPN60 also appears to function as an intercellular signaling molecule (for review, see Maguire et al. 2002). Bacterial and mitochondrial CPN60 complexes consist of homo-14mers, whereas plastid CPN60 complexes contain two subunit types. Multiple cpn60 genes are rare in bacteria but commonplace in eukaryotes, particularly in plants where the genomes contain genes for the mitochondrial cpn60, as well as the two chloroplast cpn60 subunits. For example, the model plant Arabidopsis thaliana contains a total of nine cpn60 genes, three mitochondrial, two chloroplast cpn60-α, and four chloroplast cpn60-β subunit genes (Hill and Hemmingsen 2001).
The universal nature of cpn60 genes makes them attractive targets for phylogenetic studies (Viale and Arakaki 1994; Viale et al. 1994; Viale 1995; Bush and Everett 2001; Jian et al. 2001), as well as clinical tools for detection and identification of organisms (Goh et al. 1997, 1998, 2000; Dale et al. 1998; Kwok et al. 2002; Kwok and Chow 2003; Lew et al. 2003). An analysis of the cpn60 sequences from a variety of bacterial and eukaryotic species led to the design of universal, degenerate PCR primers, which can be applied for the amplification of a 549- to 567-bp region of cpn60 corresponding to nucleotides 274–828 of the E. coli cpn60 sequence from virtually any genome (Goh et al. 1996). The utility of this cpn60 “universal target” (UT) for discriminating closely related bacterial species has been established, and it has been demonstrated that the cpn60 UT region generally provides more discriminating and phylogenetically informative data than the 16S rDNA target (Marston et al. 1999; Brousseau et al. 2001).
The ability to amplify the cpn60 UT from any genomic template has also facilitated the study of complex microbial communities, in which the UT region is amplified from a complex template and libraries of cloned UT sequences are created and sequenced (Hill et al. 2002). A number of the characteristics of the cpn60 gene and of the UT region offer significant advantages over 16S rDNA for studies of complex microbial populations and for quantitative assays. As protein-coding genes, cpn60 sequences are less constrained from sequence variation than are structural RNA-encoding genes. Furthermore, sequence variation extends quite uniformly throughout the cpn60 coding region, whereas variable regions of 16S rRNA genes are dispersed between regions of highly conserved sequence. Highly stable secondary structure that is associated with 16S rRNA is not present in cpn60 genes or transcripts. Generally, cpn60 genes are single copy in prokaryotic genomes, and the relatively small size of the UT facilitates high-throughput sequencing approaches.
Our ongoing efforts to exploit cpn60 as a target for phylogenetic studies, microbial detection and identification, and microbial ecology have led us to gather and curate a large collection of cpn60 (Type I chaperonin) sequence data, as well as sequence from the archaeal thermosome (Type II chaperonin), a homolog of cpn60. To share this resource with the scientific community, we have designed and implemented a Web interface for cpnDB, a curated collection of cpn60 sequence data that is available at http://cpndb.cbr.nrc.ca.
RESULTS
cpnDB Contents
cpnDB currently contains ∼2000 records, approximately one third of which have full-length cpn60 gene sequence data associated with them. The remaining two-thirds of the records contain exclusively UT sequence data. Organisms represented in cpnDB include eukaryotes, bacteria, and archaea, and most of the major taxonomic groups defined by the 16S rRNA “backbone tree” are represented. Table 1 summarizes the database contents by major taxonomic group and number of genera in each group. Taxonomic lineages associated with each record are derived from the full lineages provided by the NCBI taxonomy database for each organism. Currently, the primary focus of cpnDB is on cpn60 sequences (type I chaperonin), although it may be expanded in the future to include eukaryotic type II chaperonins in addition to the archaeal type II chaperonins currently included. The cpn60 universal PCR primers do not amplify any part of the type II chaperonin genes.
Table 1.
Taxonomic Lineages of cpnDB Sequence Records and Numbers of Records and Genera
| Lineage | No. of records | No. of genera |
|---|---|---|
| TYPE I CHAPERONIN | ||
| Archaea; Euryarachaeota | 2 | 1 |
| Bacteria; Actinobacteria | 130 | 16 |
| Bacteria; Aquificae | 1 | 1 |
| Bacteria; Bacteroidetes/Chlorobi group; Bacteroidetes | 25 | 11 |
| Bacteria; Bacteroidetes/Chlorobi group; Chlorobi | 1 | 1 |
| Bacteria; Chlamydiae/Verrucomicrobia group; Chlamydiae | 27 | 3 |
| Bacteria; Cyanobacteria | 22 | 8 |
| Bacteria; Deinococcus-Thermus | 2 | 2 |
| Bacteria; Fibrobacteres/Acidobacteria group | 1 | 1 |
| Bacteria; Firmicutes; Bacilli | 340 | 22 |
| Bacteria; Firmicutes; Clostridia | 22 | 9 |
| Bacteria; Firmicutes; Mollicutes | 11 | 3 |
| Bacteria; Fusobacteria | 3 | 1 |
| Bacteria; Planetomycetes | 1 | 1 |
| Bacteria; Proteobacteria; Alphaproteobacteria | 187 | 28 |
| Bacteria; Proteobacteria; Betaproteobacteria | 39 | 15 |
| Bacteria; Proteobacteria; delta/epsilon subdivisions | 60 | 8 |
| Bacteria; Proteobacteria; Gammaproteobacteria | 264 | 50 |
| Bacteria; Spirochaetes | 8 | 3 |
| Bacteria; Thermotogae | 2 | 1 |
| Eukaryota; Alveolata | 9 | 4 |
| Eukaryota; Cryptophyta | 2 | 2 |
| Eukaryota; Diplomonadida group | 2 | 2 |
| Eukaryota; Entamoebidae | 5 | 1 |
| Eukaryota; Euglenozoa | 7 | 3 |
| Eukaryota; Fungi/Metazoa group; Fungi | 12 | 10 |
| Eukaryota; Fungi/Metazoa group; Metazoa | 29 | 22 |
| Eukaryota; Viridiplantae | 29 | 11 |
| Eukaryota; Rhodophyta | 4 | 3 |
| Eukaryota; Others | 7 | 6 |
| Various (clones from microbial ecology studies) | 595 | n/a |
| TYPE II CHAPERONIN | ||
| Archaea; Crenarchaeota | 20 | 6 |
| Archaea; Euryarchaeota | 44 | 12 |
| Archaea; Nanoarchaeota | 1 | 1 |
Although multiple cpn60 genes are common in complex eukaryotes, we have found only a few examples of multiple cpn60 genes in bacteria. Table 2 lists the 16 bacterial species with multiple cpn60 genes for which complete genome sequences are available to date.
Table 2.
Occurrence of Multiple cpn60 Genes in Bacteria for Which Full Genome Sequences Are Available
| Organism | Taxon | Genome accession(s) | No. of cpn60 genes |
|---|---|---|---|
| Chlamydia muridarum | Chlamydiales | NC_002620 | 3 |
| Chlamydia caviae | Chlamydiales | NC_003361 | 3 |
| Chlamydophila pneumoniae J138 | Chlamydiales | NC_002491 | 3 |
| Chlamydophila pneumoniae TW183 | Chlamydiales | NC_005043 | 3 |
| Chromobacterium violaceum | Betaproteobacteria; Chromobacterium group | NC_005085 | 2 |
| Corynebacterium diphtheriae biotype gravis | Actinobacteria; Corynebacteriaceae | NC_002935 | 2 |
| Corynebacterium efficiens | Actinobacteria; Corynebacteriaceae | NC_004369 | 2 |
| Mycobacterium bovis subsp. bovis | Actinobacteria; Mycobacteriaceae | NC_002945 | 2 |
| Mycobacterium tuberculosis | Actinobacteria; Mycobacteriaceae | NC_002755 | 2 |
| Nostoc punctiforme | Cyanobacteria; Nostocaceae | NZ_AAAY02000020 | 3 |
| NZ_AAAY02000051 | |||
| NZ_AAAY02000174 | |||
| Streptomyces avermitilis | Actinobacteria; Streptomycetaceae | NC_003155 | 2 |
| Thermosynechococcus elongatus | Cyanobacteria; Chroococcales | NC_004113 | 2 |
| Vibrio vulnificus | Gammaproteobacteria; Vibrionaceae | NC_005139 (chromosome I) | 2 |
| NC_005140 (chromosome II) | |||
| Mesorhizobium loti | Alphaproteobacteria; Rhizobiales; Phyllobacteriaceae | NC_002679 | 5 |
| Bradyrhizobium japonicum | Alphaproteobacteria; Rhizobiales; Bradyrhizobiaceae | NC_004463 | 7 |
| Rhodopseudomonas palustris | Alphaproteobacteria; Rhizobiales; Bradyrhizobiaceae | NC_005296 | 2 |
cpn60 UT Versus Full-Length Gene Sequence
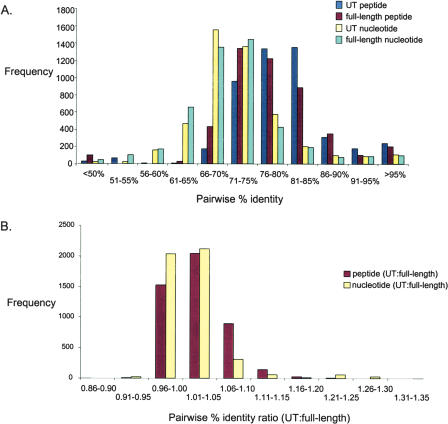
To determine whether relationships between cpn60 genes for any two organisms are reliably reflected in the UT region, we examined the pairwise percent identities between UT nucleotide sequences and full-length cpn60 gene sequences, and between UT peptide sequences and full-length CPN60 protein sequences for 97 Gammaproteobacteria sequences representing 32 genera (Fig. 1A). For each pair of sequences (4656 pairs), we determined the ratio of the UT nucleotide sequence identity to the full-length nucleotide sequence identity and the ratio of the UT peptide sequence identity to the full-length sequence identity. For example, if the UT nucleotide sequence identity between two sequences is 78%, and the full-length nucleotide sequence identity for the same pair is 76%, then the ratio would be 1.03. We found that the majority of ratios for the gammaproteobacteria (3569 of 4656 peptide ratios; 4161 of 4656 nucleotide ratios) are between 0.96 and 1.10, indicating that the level of difference between the UT regions of any two organisms in this group is representative of the level of difference between the full-length cpn60 sequences (Fig. 1B).
Figure 1.
(A) Distribution of pairwise sequence identities for 97 gammaproteobacteria-derived cpn60 UT nucleotide sequences, full-length nucleotide sequences, UT peptide, and full-length peptide sequences (representing 32 genera). (B) Distribution of ratios of peptide (UT:full-length CPN60) and nucleotide (UT:full-length cpn60) pairwise sequence identities.
Sequence Diversity Within cpnDB
Figure 2 shows the cumulative frequency distribution for pairwise percent nucleotide and peptide identities and pairwise peptide similarities between cpn60 UT sequences across a data set composed of one representative of each of 247 eukaryotic and bacterial genera represented in cpnDB. Pairwise UT nucleotide identities in this data set range from 26.4% to 100%, with a median value of 55.4% identity and a mean of 56.5% identity. The peptide identity range extends from 15.4% to 100%, with a median of 53.5% and a mean of 54.6%. Pairwise peptide percent similarity ranges from 33.6% to 100%, with a median value of 74.3% similarity and a mean of 74.5%.
Figure 2.
Cumulative frequency distribution plot of pairwise UT nucleotide sequence identity and pairwise UT peptide sequence identity and similarity for representatives of each of 247 bacterial and eukaryotic genera.
Web Interface
cpnDB was constructed with MySQL, and the Web interface was implemented with PHP. Database contents can be searched using text or sequence queries. The text search window (Fig. 3) allows searching by elements of the organism name as well as specific strain identifiers such as American Type Culture Collection numbers. A keyword query searches all text fields. The number of records retrieved can be restricted and records can be sorted in a number of ways. It is also possible to view only recent deposits to the database or to browse the entire selection by general taxonomic class and alphabetical listing of organisms.
Figure 3.
The cpnDB text query page. A screenshot shows different fields by which a user can effectively search the database.
Search results are presented in table form as shown in Figure 4. In this case, a search for genus Achromobacter yielded two records. Full records can be viewed by selecting the database ID number, or added to a download cart for later retrieval. Individual records (Fig. 5) include deposit date, organism identification, taxonomic lineage, unique GenBank identifiers, source of the data, and available nucleotide and peptide sequence data. Links to the NCBI taxonomy database entries, nucleotide, and peptide are provided for retrieval of full GenBank records and links to associated resources provided by NCBI. For nonreference sequences, which include clinical isolates, field isolates, and sequences derived from environmental samples in microbial population studies, the record also includes a description of the nearest reference sequence neighbor in the database. For example, Figure 5 shows the record for a cloned cpn60 sequence derived from a study of pig feces microbial flora. In this case, the library sequence (001_g11, GenBank accession AF436914) is shown to be 72.939% identical to the reference nucleotide sequence from Bacteroides uniformis. The date of the most recent search of the reference database is indicated and links to the FASTA and BLASTp results are provided. All nonreference sequences are searched against the reference data set following each deposit of new reference data.
Figure 4.
Search results table. A text search of the database contents results in a tabular display of matching records. Users can retrieve full record details by clicking on the record ID number or add selected records to a download cart.
Figure 5.
cpnDB record details. Full records include the type of record (e.g., reference, clinical isolate, field isolate, microbial population study clone), name of the organism, taxonomic lineage, links to full GenBank records, source information, and for nonreference sequences, a description of the most similar reference sequence along with links to the most recent search results.
Sequence-based searches can be conducted using nucleotide or peptide queries. In addition to standard FASTA and BLASTp searches, implementation of a modified version of BIBI (http://pbi1.univ-lyon1.fr/bibi/; Devulder et al. 2003) with a nucleotide query results in production of a FASTA results table as well as a CLUSTALW multiple-sequence alignment of sequences included in the FASTA output and a CLUSTALW distance tree. The output also includes a brief description of the query, including size and G+C content. BIBI was modified to use the FASTA search protocol rather than BLASTn. The multiple sequence alignment and tree are viewed and manipulated through java applets Jalview (http://www.jalview.org/) and ATV (http://www.genetics.wustl.edu/eddy/atv; Zmasek and Eddy 2001). All data files produced, including sequences in FASTA format, the CLUSTALW alignment, and treefile are also available for download in text format.
Documentation for all applications implemented is available through the cpnDB Web interface. Descriptions and protocols for the application of the universal cpn60 primers and related literature references are also presented.
DISCUSSION
16S rRNA genes have long been the standard for molecular systematic studies as well as the rapidly expanding fields of molecular diagnostics and microbial ecology. These genes are universal, the multiple copies of 16S rRNA genes per genome make it an abundant, easily detectable target, and universal primers have been developed to amplify specific fragments for sequence analysis. Potential pitfalls of 16S rRNA as a target for organism detection and identification are related to the fact that there is often insufficient discriminating sequence information within the 16S rRNA target to distinguish between closely related species and strains. Also, the multiple variable copy numbers per genome complicate quantitative assays on the basis of this target. The structure of the gene, with alternating regions of variable and conserved sequence, facilitate the formation of chimeric PCR products, especially when amplifying from a complex template (Wang and Wang 1997).
Universal, protein-encoding genes offer alternatives to 16S rRNA with some particular advantages. As a consequence of the degeneracy of the genetic code, protein-encoding genes can diverge and evolve more rapidly than genes encoding structural RNA molecules, where even minor changes to the nucleotide sequence can have catastrophic effects. This results in the observation that protein-encoding gene sequences can be used to discriminate between species, or even strains within a species. Genes present in a single copy in microbial genomes, although more difficult to detect, offer superior targets for application quantitative methods such as quantitative, real-time PCR, while eliminating potential sequencing artifacts produced when multiple gene copies are not identical.
Other protein-encoding genes beside cpn60 have proven useful for phylogenetic and diagnostic purposes, including rpoB (Adekambi et al. 2003; Khamis et al. 2003), pmoA (Bourne et al. 2001), and gyrB (Yamamoto and Harayama 1995; Kasai et al. 2000). A database of gyrB sequences has been published (Kasai et al. 1998), but it is limited to bacterial sequences, and it is unclear whether it is currently being maintained and updated.
We have found that the cpn60 UT region is reasonably representative of the entire open reading frame in terms of phylogenetically informative sequence variation (Fig. 1), and that it usually provides more discriminating information than corresponding 16S rRNA sequences (Brousseau et al. 2001). Within a population of one representative of each of 247 eukaryotic and bacterial genera, we observe pairwise nucleotide sequence identities over a wide range (26%–100%, Fig. 2) in a normal distribution. These observations, combined with the accessibility of the UT sequence for any given organism through use of universal, degenerate primers, has resulted in the large accumulation of exclusively UT sequence data for many of the organisms represented in cpnDB. Surprisingly, we identified two pairs of identical nucleotide UT sequences in this intergenic comparison. In one case, the pair is composed of two synonyms for the same organism [Mobiluncus curtsii (AY123679) and Falcivibrio vaginalis (AY123678)]. The other case occurred between the sequences for Alcaligenes faecalis subsp. faecalis (AY123668) and Eikenella corrodens (AY123719), both betaproteobacteria. In this case, the identity could be actual or could be the result of source strain misidentification or misannotation.
The sequence data accumulated in cpnDB is derived from public repositories or has been generated by a collaborative network of clinicians, phylogeneticists, and microbial ecologists exploiting the cpn60 and archaeal chaperonin targets in their work. All of the sequence data in cpnDB is also present in the public sequence repositories. However, the advantages to a curated collection of sequences are obvious. We do not have any immediate plans to accept direct sequence submissions to cpnDB, but instead encourage interested parties to submit their data to the public sequence databases (EMBL, GenBank, or DDBJ), as surveillance of these resources for new cpn60 sequence data is ongoing.
To the best of our knowledge, cpnDB is the largest curated collection of gene-specific sequence data for a protein-encoding gene, and as such, it is a valuable resource for phylogenetic studies, clinical applications, and microbial ecology investigations.
METHODS
Data Collection
Surveillance of the NCBI GenBank sequence database is managed with the Pubcrawler service (http://www.pubcrawler.ie/; Hokamp and Wolfe 1999). Weekly Pubcrawler reports include lists of new or updated cpn60 records in GenBank. Each record is evaluated by the curator as described below before deposit into cpnDB. GenBank flatfiles are retrieved in batch from NCBI, and relevant information concerning the identification of the source organism and its taxonomic lineage are extracted along with the sequence data and links to the unique GenBank identifiers (gi and accession).
Curation and Annotation
Only complete and unambiguous cpn60 sequence data is deposited in cpnDB. To be considered complete, the sequence must, at a minimum, encompass the universal target region of cpn60. We do not incorporate sequence data that fails to cover this region or that contain ambiguous nucleotides. For archaeal type II chaperonin sequences, only full-length sequences are included. We also make every effort to completely annotate the record regarding the source organism, including strain name and culture collection synonyms. Type strains and strain synonyms are also indicated where possible, on the basis of information retrieved from a number of other resources, including the American Type Culture Collection catalog (http://www.atcc.org/) and the List of Bacterial Names with Standing in Nomenclature (http://www.bacterio.cict.fr/). Taxonomic lineages are derived from the NCBI taxonomy database (http://www.ncbi.nlm.nih.gov/Taxonomy/). Complete annotation often involves consulting the primary literature describing the sequence and its source, as this information is often not completely described in the GenBank annotation.
Updates
Sequences from nonreference sources (including those derived from clinical isolates, field isolates, and microbial population studies) are compared with the reference data set upon deposit. Best FASTA and BLASTp scores are reported in the record, along with the date of the most recent search and links to the most closely related reference records and search results. Each time the reference data set is updated, all nonreference sequences are automatically recompared with the new reference data, and fields describing nearest database neighbors are refreshed.
Acknowledgments
Funding for the development of cpnDB was provided by the Canadian Biotechnology Strategy and the National Research Council Genomics and Health Initiative. The authors gratefully acknowledge the support and contributions of the Canadian Bioinformatics Resource (http://cbr-rbc.nrc-cnrc.gc.ca/).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2649204.
References
- Adekambi, T., Colson, P., and Drancourt, M. 2003. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J. Clin. Microbiol. 41: 5699–5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher, Y., Douady, C.J., Papke, R.T., Walsh, D.A., Boudreau, M.E., Nesbo, C.L., Case, R.J., and Doolittle, W.F. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37: 283–328. [DOI] [PubMed] [Google Scholar]
- Bourne, D.G., McDonald, I.R., and Murrell, J.C. 2001. Comparison of pmoA PCR primer sets as tools for investigating methanotroph diversity in three Danish soils. Appl. Environ. Microbiol. 67: 3802–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brousseau, R., Hill, J.E., Prefontaine, G., Goh, S.H., Harel, J., and Hemmingsen, S.M. 2001. Streptococcus suis serotypes characterized by analysis of chaperonin 60 gene sequences. Appl. Environ. Microbiol. 67: 4828–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush, R.M. and Everett, K.D. 2001. Molecular evolution of the Chlamydiaceae. Int. J. Syst. Evol. Microbiol. 51: 203–220. [DOI] [PubMed] [Google Scholar]
- Chambaud, I., Heilig, R., Ferris, S., Barbe, V., Samson, D., Galisson, F., Moszer, I., Dybvig, K., Wroblewski, H., Viari, A., et al. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29: 2145–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole, J.R., Chai, B., Marsh, T.L., Farris, R.J., Wang, Q., Kulam, S.A., Chandra, S., McGarrell, D.M., Schmidt, T.M., Garrity, G.M., et al. 2003. The Ribosomal Database Project (RDP-II): Previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31: 442–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, C.J., Moses, E.K., Ong, C.C., Morrow, C.J., Reed, M.B., Hasse, D., and Strugnell, R.A. 1998. Identification and sequencing of the groE operon and flanking genes of Lawsonia intracellularis: Use in phylogeny. Microbiology 144: 2073–2084. [DOI] [PubMed] [Google Scholar]
- Devulder, G., Perriere, G., Baty, F., and Flandrois, J.P. 2003. BIBI, a bioinformatics bacterial identification tool. J. Clin. Microbiol. 41: 1785–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass, J.I., Lefkowitz, E.J., Glass, J.S., Heiner, C.R., Chen, E.Y., and Cassell, G.H. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407: 757–762. [DOI] [PubMed] [Google Scholar]
- Goh, S.H., Potter, S., Wood, J.O., Hemmingsen, S.M., Reynolds, R.P., and Chow, A.W. 1996. HSP60 gene sequences as universal targets for microbial species identification: Studies with coagulase-negative staphylococci. J. Clinic. Microbiol. 34: 818–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, S.H., Santucci, Z., Kloos, W.E., Faltyn, M., George, C.G., Driedger, D., and Hemmingsen, S.M. 1997. Identification of Staphylococcus species and subspecies using the Chaperonin-60 gene identification method and reverse checkerboard hybridization. J. Clin. Microbiol. 35: 3116–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, S.H., Driedger, D., Gillett, S., Low, D.E., Hemmingsen, S.M., Amos, M., Chan, D., Lovgren, M., Willey, B.M., Shaw, C., et al. 1998. Streptococcus iniae, a human and animal pathogen: Specific identification by the Chaperonin-60 gene identification method. J. Clin. Microbiol. 36: 2164–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, S.H., Facklam, R.R., Chang, M., Hill, J.E., Tyrrell, G.J., Burns, E.C., Chan, D., He, C., Rahim, T., Shaw, C., et al. 2000. Identification of enterococcus species and phenotypically similar lactococcus and vagococcus species by reverse checkerboard hybridization to chaperonin 60 gene sequences. J. Clin. Microbiol. 38: 3953–3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmingsen, S.M., Woolford, C., van der Vies, S.M., Tilly, K., Dennis, D.T., Georgopoulos, C.P., Hendrix, R.W., and Ellis, R.J. 1988. Homologous plant and bacterial proteins chaperone oligomeric protein assembly. Nature 333: 330–334. [DOI] [PubMed] [Google Scholar]
- Hill, J.E. and Hemmingsen, S.M. 2001. Arabidopsis thaliana type I and II chaperonins. Cell Stress & Chaperones 6: 190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, J.E., Seipp, R.P., Betts, M., Hawkins, L., Van Kessel, A.G., Crosby, W.L., and Hemmingsen, S.M. 2002. Extensive profiling of a complex microbial community by high-throughput sequencing. Appl. Environ. Microbiol. 68: 3055–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokamp, K. and Wolfe, K. 1999. What's new in the library? What's new in GenBank? Let PubCrawler tell you. Trends Genet. 15: 471–472. [DOI] [PubMed] [Google Scholar]
- Jian, W., Zhu, L., and Dong, X. 2001. New approach to phylogenetic analysis of the genus Bifidobacterium based on partial HSP60 gene sequences. Int. J. Syst. Evol. Microbiol. 51: 1633–1638. [DOI] [PubMed] [Google Scholar]
- Kasai, H., Watanabe, K., Gasteiger, E., Bairoch, A., Isono, K., Yamamoto, S., and Harayama, S. 1998. Construction of the gyrB database for the identification and classification of bacteria. Genome Inform. Ser. Workshop Genome Inform. 9: 13–21. [PubMed] [Google Scholar]
- Kasai, H., Tamura, T., and Harayama, S. 2000. Intrageneric relationships among Micromonospora species deduced from gyrB-based phylogeny and DNA relatedness. Int. J. Syst. Evol. Microbiol. 50: 127–134. [DOI] [PubMed] [Google Scholar]
- Khamis, A., Colson, P., Raoult, D., and Scola, B.L. 2003. Usefulness of rpoB gene sequencing for identification of Afipia and Bosea species, including a strategy for choosing discriminative partial sequences. Appl. Environ. Microbiol. 69: 6740–6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunker, D., Haas, B., Hirtreiter, A., Figueiredo, L., Naylor, D.J., Pfeifer, G., Muller, V., Deppenmeier, U., Gottschalk, G., Hartl, F.U., et al. 2003. Coexistence of group I and group II chaperonins in the archaeon Methanosarcina mazei. J. Biol. Chem. 278: 33256–33267. [DOI] [PubMed] [Google Scholar]
- Kwok, A.Y. and Chow, A.W. 2003. Phylogenetic study of Staphylococcus and Macrococcus species based on partial hsp60 gene sequences. Int. J. Syst. Evol. Microbiol. 53: 87–92. [DOI] [PubMed] [Google Scholar]
- Kwok, A.Y., Wilson, J.T., Coulthart, M., Ng, L.K., Mutharia, L., and Chow, A.W. 2002. Phylogenetic study and identification of human pathogenic Vibrio species based on partial hsp60 gene sequences. Can. J. Microbiol. 48: 903–910. [DOI] [PubMed] [Google Scholar]
- Lew, A.E., Gale, K.R., Minchin, C.M., Shkap, V., and de Waal, D.T. 2003. Phylogenetic analysis of the erythrocytic Anaplasma species based on 16S rDNA and GroEL (HSP60) sequences of A. marginale, A. centrale, and A. ovis and the specific detection of A. centrale vaccine strain. Vet. Microbiol. 92: 145–160. [DOI] [PubMed] [Google Scholar]
- Maguire, M., Coates, A.R.M., and Henderson, B. 2002. Chaperonin 60 unfolds its secrets of cellular communication. Cell Stress & Chaperones 7: 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston, E.L., Sumner, J.W., and Regnery, R.L. 1999. Evaluation of intraspecies genetic variation within the 60 kDa heat-shock protein gene (groEL) of Bartonella species. Int. J. Syst. Bacteriol. 49: 1015–1023. [DOI] [PubMed] [Google Scholar]
- Olsen, G.J., Lane, D.J., Giovannoni, S.J., Pace, N.R., and Stahl, D.A. 1986. Microbial ecology and evolution: A ribosomal RNA approach. Annu. Rev. Microbiol. 40: 337–365. [DOI] [PubMed] [Google Scholar]
- Perna, N.T., Plunkett, G., Burland, V., Mau, B., Glasner, J.D., Rose, D.J., Mayhew, G.F., Evans, P.S., Gregor, J., Kirkpatrick, H.A., et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409: 529–533. [DOI] [PubMed] [Google Scholar]
- Saibil, H.R. and Ranson, N.A. 2002. The chaperonin folding machine. Trends Biochem. Sci. 27: 627–632. [DOI] [PubMed] [Google Scholar]
- Viale, A. 1995. GroEL (Hsp60)-based bacterial and organellar phylogenies. Mol. Microbiol. 17: 1013. [DOI] [PubMed] [Google Scholar]
- Viale, A.M. and Arakaki, A.K. 1994. The chaperone connection to the origins of the eukaryotic organelles. FEBS Lett. 341: 146–151. [DOI] [PubMed] [Google Scholar]
- Viale, A.M., Arakaki, A.K., Soncini, F.C., and Ferreyra, R.G. 1994. Evolutionary relationships among bacterial groups as inferred from GroEL (Chaperonin) sequence comparisons. Int. J. Syst. Bacteriol. 44: 527–533. [DOI] [PubMed] [Google Scholar]
- Wang, G.C. and Wang, Y. 1997. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 63: 4645–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C.R., Kandler, O., and Wheelis, M.L. 1990. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. 87: 4576–4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, S. and Harayama, S. 1995. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 61: 1104–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmasek, C.M. and Eddy, S.R. 2001. ATV: Display and manipulation of annotated phylogenetic trees. Bioinformatics. 17: 383–384. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://cpndb.cbr.nrc.ca/; cpnDB homepage.
- http://www.ncbi.nlm.nih.gov/; National Center for Biotechnology Information.
- http://www.pubcrawler.ie/; Pubcrawler homepage.
- http://cbr-rbc.nrc-cnrc.gc.ca/; Canadian Bioinformatics Resource.
- http://www.atcc.org/; American Type Culture Collection.
- http://www.bacterio.cict.fr/; List of Bacterial Names with Standing in Nomenclature. [DOI] [PubMed]
- http://www.jalview.org/; Jalview homepage.
- http://pbil.univ-lyon1.fr/bibi/; BIBI homepage.
- http://www.genetics.wustl.edu/eddy/atv/; ATV (A Tree Viewer).