Abstract
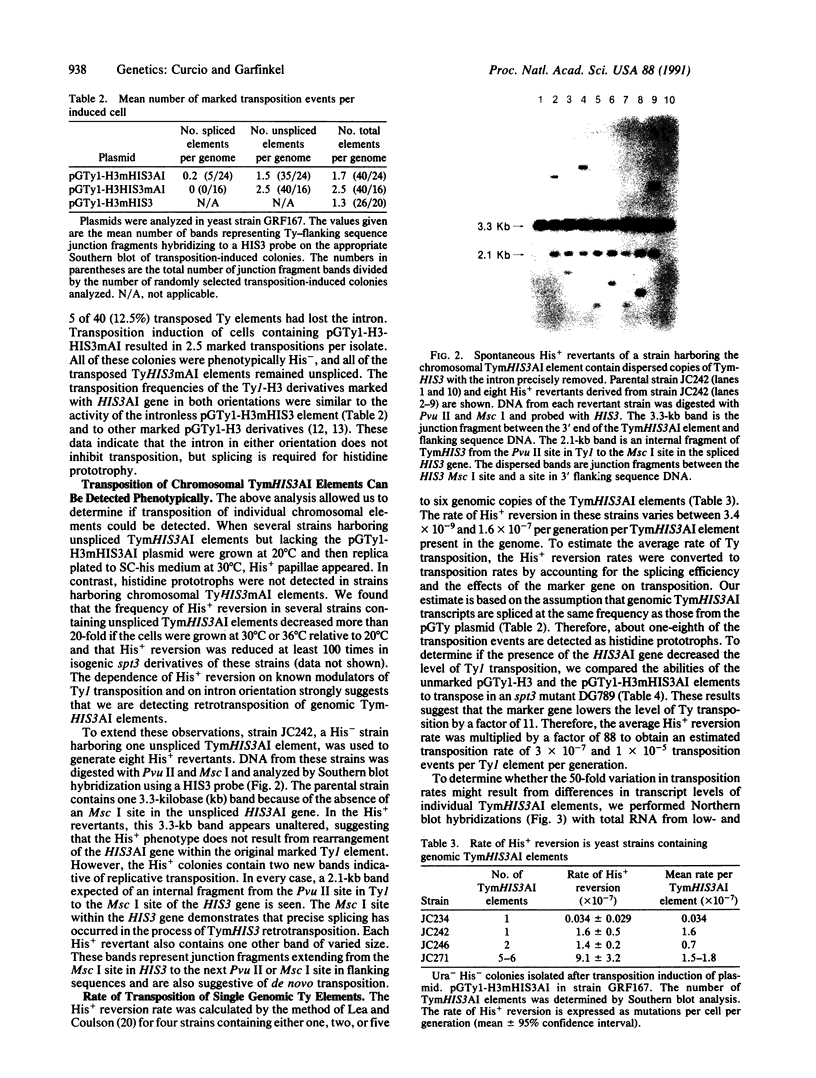
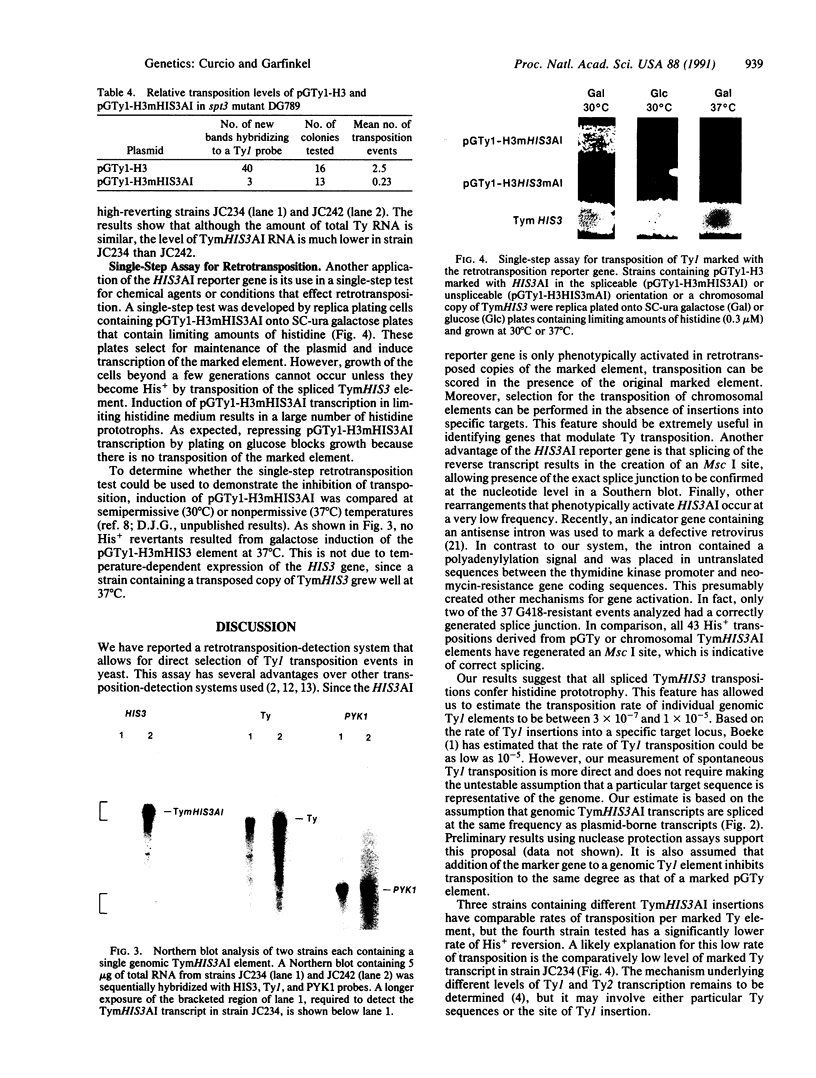
The yeast retrotransposon Ty1 has been tagged with a reporter gene that allows selection of RNA-mediated transposition events and is applicable to the study of retroelements in other organisms. The reporter gene is a yeast HIS3 gene interrupted by an artificial intron (AI) in the antisense orientation. The HIS3AI sequences were inserted into a Ty1 element such that the intron is on the sense strand of the Ty1 element; therefore, splicing and retrotransposition of marked Ty1 transcripts can give rise to His+ cells. Fusion of the Ty1-H3mHIS3AI element to the inducible GAL1 promoter resulted in a high frequency of histidine prototrophs upon galactose induction. Moreover, spontaneous His+ revertants derived from strains containing genomic TymHIS3AI elements are a result of retrotransposition. By using this assay, we estimated the Ty1 transposition rate to be between 3 x 10(-7) and 1 x 10(-5) transpositions per Ty1 element per generation. Variations in the transposition rate of individual Ty1 elements are correlated with the relative abundance of their transcripts.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Styles C. A., Fink G. R. Saccharomyces cerevisiae SPT3 gene is required for transposition and transpositional recombination of chromosomal Ty elements. Mol Cell Biol. 1986 Nov;6(11):3575–3581. doi: 10.1128/mcb.6.11.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Xu H., Fink G. R. A general method for the chromosomal amplification of genes in yeast. Science. 1988 Jan 15;239(4837):280–282. doi: 10.1126/science.2827308. [DOI] [PubMed] [Google Scholar]
- Bradshaw V. A., McEntee K. DNA damage activates transcription and transposition of yeast Ty retrotransposons. Mol Gen Genet. 1989 Sep;218(3):465–474. doi: 10.1007/BF00332411. [DOI] [PubMed] [Google Scholar]
- Cameron J. R., Loh E. Y., Davis R. W. Evidence for transposition of dispersed repetitive DNA families in yeast. Cell. 1979 Apr;16(4):739–751. doi: 10.1016/0092-8674(79)90090-4. [DOI] [PubMed] [Google Scholar]
- Curcio M. J., Hedge A. M., Boeke J. D., Garfinkel D. J. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol Gen Genet. 1990 Jan;220(2):213–221. doi: 10.1007/BF00260484. [DOI] [PubMed] [Google Scholar]
- Garfinkel D. J., Mastrangelo M. F., Sanders N. J., Shafer B. K., Strathern J. N. Transposon tagging using Ty elements in yeast. Genetics. 1988 Sep;120(1):95–108. doi: 10.1093/genetics/120.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux C. N., Mis J. R., Pierce M. K., Kohalmi S. E., Kunz B. A. DNA sequence analysis of spontaneous mutations in the SUP4-o gene of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Feb;8(2):978–981. doi: 10.1128/mcb.8.2.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann T., Heidmann O., Nicolas J. F. An indicator gene to demonstrate intracellular transposition of defective retroviruses. Proc Natl Acad Sci U S A. 1988 Apr;85(7):2219–2223. doi: 10.1073/pnas.85.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin C. E., Williamson V. M. Temperature effects on the rate of ty transposition. Science. 1984 Oct 5;226(4670):53–55. doi: 10.1126/science.226.4670.53. [DOI] [PubMed] [Google Scholar]
- Paquin C. E., Williamson V. M. Ty insertions at two loci account for most of the spontaneous antimycin A resistance mutations during growth at 15 degrees C of Saccharomyces cerevisiae strains lacking ADH1. Mol Cell Biol. 1986 Jan;6(1):70–79. doi: 10.1128/mcb.6.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picologlou S., Brown N., Liebman S. W. Mutations in RAD6, a yeast gene encoding a ubiquitin-conjugating enzyme, stimulate retrotransposition. Mol Cell Biol. 1990 Mar;10(3):1017–1022. doi: 10.1128/mcb.10.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G. S., Fink G. R. Movement of yeast transposable elements by gene conversion. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5621–5625. doi: 10.1073/pnas.79.18.5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder G. S., Smith M., Lambie E. J. Intrachromosomal movement of genetically marked Saccharomyces cerevisiae transposons by gene conversion. Mol Cell Biol. 1984 Apr;4(4):703–711. doi: 10.1128/mcb.4.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Durbin K. J., Fink G. R. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984 Dec;39(3 Pt 2):675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- Yoshimatsu T., Nagawa F. Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science. 1989 Jun 16;244(4910):1346–1348. doi: 10.1126/science.2544026. [DOI] [PubMed] [Google Scholar]











